Abstract
Many biological processes rely on protein–protein interactions. These processes include signal transduction, cell cycle regulation, gene regulation, and viral assembly and replication. Moreover, many proteins and enzymes manifest their function as oligomers. Herein is described an efficient means to sift through large combinatorial libraries and identify molecules that block the interaction of target proteins in vivo. The power of this approach is demonstrated by the identification of 9-residue peptides from a combinatorial library that inhibit the intracellular dimerization of HIV-1 protease. Fewer than 1 in 106 peptides do so. In vitro biochemical analyses of one such peptide demonstrate that it acts by dissociating HIV-1 protease into monomers, which are inactive catalysts. Inhibition is enhanced further by dimerizing the peptide. This approach enables the facile identification of new molecules that control cellular processes.
Interactions between proteins play a critical role in cell function and dysfunction. Such interactions can be identified by phage display1,2 and the yeast two-hybrid system3–6, as well as by biochemical methods. The identification of a protein–protein interaction is, however, only the first step. Modulation of the interaction is necessary to produce true insight into its biological purpose. Blocking particular protein–protein interactions with specific ligands would enable exquisite control of cellular processes and provide potential leads for novel chemotherapeutic agents7.
HIV-1 protease is critical to viral maturation and infectivity8,9, and is a primary target in AIDS chemotherapy10–12. To treat AIDS, attempts have been made to inactivate the protease using active site-directed inhibitors. This strategy has led to the appearance of numerous HIV strains with drug-resistant proteases10,13. The prevalence of these resistant strains, along with the toxicity of existing drugs, underscores the need for an alternative approach.
Disrupting the quaternary structure of HIV-1 protease is an orthoganol means of inactivating the enzyme14–25. HIV-1 protease is composed of two identical 99-residue subunits26. Dissociation of the active, dimeric form of the enzyme results in the complete loss of catalytic activity. We sought to identify new molecules that interfere with the dimerization of HIV-1 protease. In doing so, we were able to develop a genetic selection for dissociative inhibitors of virtually any designated protein–protein interaction.
Results and discussion
Strategy
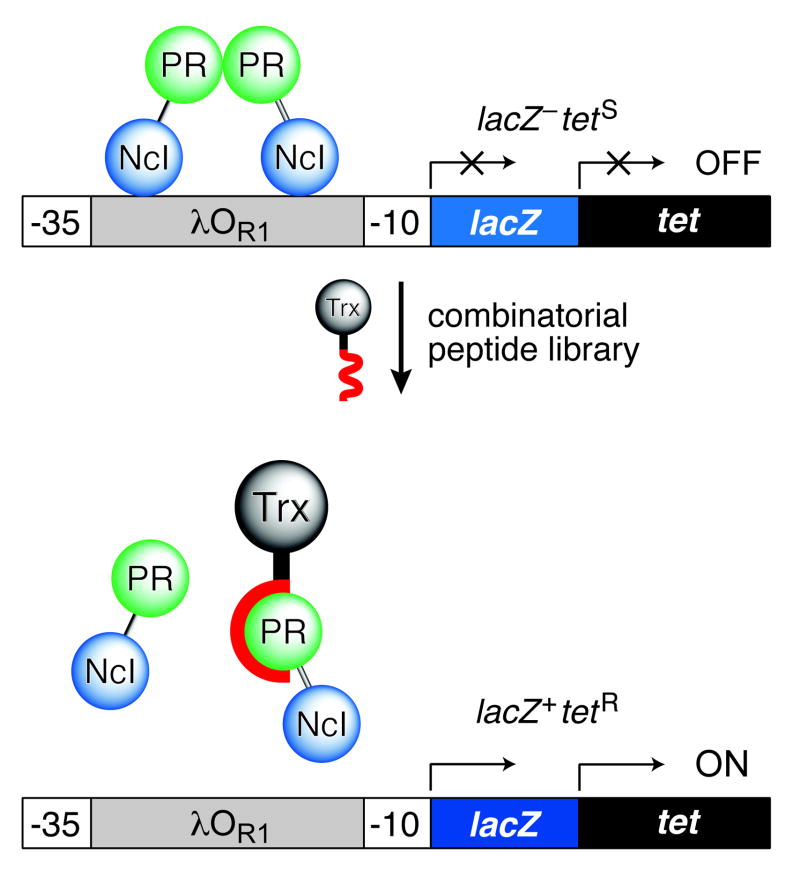
Our approach takes advantage of the properties of the repressor protein from bacteriophage λ27. The lambda repressor protein (cI) binds to its operator DNA as a homodimer. Each monomer has two distinct domains28: an N-terminal DNA-binding domain and a C-terminal dimerization domain. The role of the C-terminal domain of cI can be fulfilled by another protein29–35 or a peptide36,37.
We fused a variant protease (D25N) to the N-terminal DNA-binding domain of cI (NcI; residues 1–131) to create a hybrid repressor (NcI–PR). A dimer of the D25N variant is indistinguishable from the native protease, except for its lack of catalytic activity38. Because NcI alone is not capable of binding DNA in vivo, the function of NcI–PR relies on dimerization being mediated by its protease domain. We constructed the selection module λPR–lacZ–tet in a reporter plasmid that also directs the production of NcI–PR (Fig. 1). We found that NcI–PR and a related hybrid protein (NcI–dPR2) do efficiently repress the transcription of the lacZ and tet genes, which are under control of the λPR promoter (Table 1).
Figure 1.
Genetic selection for dissociative inhibitors of a protein–protein interaction. A. A, Strategy for selecting peptide inhibitors of HIV-1 protease (PR) dimerization. The transcription of reporter genes is turned on when a peptide fused to thioredoxin (Trx) dissociates the hybrid lambda repressor (NcI–PR). Only LacZ+TetR cells survive and form blue colonies in the presence of tetracycline and X-Gal. B, Map of the reporter plasmid (pSH26) and library plasmid used to select for peptide inhibitors of HIV-1 protease dimerization.
Table 1.
Regulatory properties of wild-type and hybrid lambda repressor proteinsa
| Plasmid | Repressor | Tetracycline Susceptibility | β-Galactosidase Activity (units) | Repression (%) |
|---|---|---|---|---|
| pSH20 | none | resistant | (1.1 ± 0.1) × 104 | 0 |
| pSH26 | NcI–PR | sensitive | (2.0 ± 0.2) × 103 | 82 |
| pSH27 | NcI–dPR2 | sensitive | (2.8 ± 0.4) × 102 | 98 |
| pSH28 | cI | sensitive | (2.0 ± 0.1) × 102 | 98 |
| pSH29 | NcI–zip | sensitive | (1.7 ± 0.1) × 103 | 84 |
NcI-PR, a fusion of cI (residues 1–131) and D25N HIV-1 protease, forms a stable dimer and functionally represses the λPR-lacZ-tet reporter cassette in vivo. The λPR promoter in the cassette contains only a single copy of λOR1 to avoid complexity arising from the cooperative binding of repressors to multiple copies of λOR. Plasmids were transformed into E. coli strain MC1061 to test for in vivo binding of hybrid repressors to λOR1. The low copy-number plasmid pSH20 contains the reporter cassette alone. pSH26–29 are derivatives of pSH20 with the expression modules of various repressor hybrids driven by the lacUV5 promoter. NcI–dPR2 is a hybrid protein of NcI and the N- and C-terminal segments of HIV-1 protease tethered by a pentapeptide linker (P1Q2I3T4L5-GGSSG-S95T96L97N98F99). NcI–zip is a hybrid of NcI and GCN4. The DNA of NcI and NcI–zip are from pJH157 and pJH370 (J. C. Hu), respectively29. Susceptibility to tetracycline (10 μg/ml) and β-galactosidase activity were measured as indicators of binding to λOR129,30. Repression is calculated as: 1 − (β-galactosidase activity with repressor/β-galactosidase activity without repressor).
We constructed a library of peptides with 9 random residues fused to the solvent-exposed C terminus of Escherichia coli thioredoxin (Trx). Trx and the nonapeptide library were separated by a SG3 tetrapeptide spacer. The genetic encoding of this library allows for its facile creation, maintenance, and reproduction39. We chose not to constrain the peptides40,5,6 so as to avoid an a priori conformational bias that could limit the number of effective inhibitors. When a cell is transformed with the reporter plasmid, NcI–PR represses the transcription of the reporter genes and the transformant shows a LacZ−TetS phenotype. Upon co-transformation with the peptide library plasmid (Fig. 1), cells that bear dissociative peptides should show a LacZ+TetR phenotype, propagating on solid medium containing tetracycline and forming blue colonies. The stringency of selection can be tuned by changing the tetracycline concentration (between 10 and 40 μg/ml). To screen the library, we transformed bacteria sequentially with the library plasmid and the reporter plasmid, and then selected for the tetR phenotype (at 20 μg/ml tetracycline).
Approximately 300 of the 3 × 108 cotransformants showed a LacZ+TetR phenotype, a selection of 1 in 106. This low frequency is surprising. Apparently, disrupting the interaction between two proteins, even small ones such as HIV-1 protease monomers, is difficult. The formidable challenge of identifying inhibitors of a protein–protein interaction highlights the need for a facile assay that can sift rapidly through large (>108-member) libraries for active molecules. Our approach meets these criteria. In contrast, genetic selections based on eukaryotic cells (such as the yeast two-hybrid system) are impaired by low transformation efficiencies and cannot reliably identify rare (1 in 106) events.
Elimination of False Positives
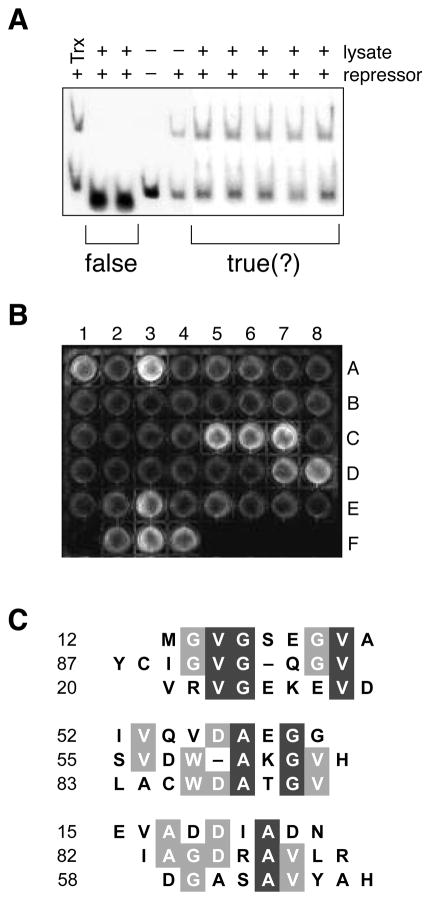
False positives are common to genetic selections and screens that rely on the transcriptional control of reporter genes. A likely false positive in our system is a Trx–peptide fusion that interferes with the interaction between λOR1 and NcI, and thus results in a LacZ+TetR phenotype regardless of the integrity of the protease dimer. We used a gel retardation assay to screen 120 positive isolates from the genetic selection for the ability to interfere with the λOR1–NcI interaction by using a gel retardation assay. Specifically, we mixed lysates from LacZ+TetR cells with wild-type cI protein and fluorescein-labeled λOR1 DNA, and subjected the mixture to electrophoresis in a polyacrylamide gel. False positives (⅕ of the cotransformants with LacZ+TetR phenotype) cause the shifted band to disappear (Fig. 2a) by interfering with the protein–DNA interaction. Additional gel retardation assays showed that the false positives bind to NcI rather than λOR1 (data not shown).
Figure 2.
In vitro screens to identify true positives from the genetic selection. A, Representative gel retardation assay. False positives that interfere with the protein–DNA interaction were identified by using a gel retardation assay to monitor the binding of cI to λOR1 in the presence of Trx–peptide fusions. B, Representative high-throughput ELISA. Trx–peptide fusions not identified as false positives by the gel retardation assay were screened for binding to immobilized monomeric protease. A1 through E8 are protease-coated surfaces to which 40 lysates were applied. Binding was detected with an anti-Trx antibody. F1 is a surface coated with free cysteine to which no lysate was applied. F2 is a protease-coated surface to which a lysate containing Trx with no fused peptide was applied. F3 is a protease-coated surface to which no lysate was applied and anti-protease antibody (S. Oroszlan) was used instead of anti-Trx antibody. F4 is a Trx-coated surface to which no lysate was applied. C, Sequence alignment of positive isolates. The 9 sequences that gave the strongest signal in the ELISA were divided into 3 subgroups according to their similarity. Residues that are conserved partially are in gray boxes; those conserved completely are in black boxes. The alignment was made with the MULTIALIGN interface of the BLOSOM62 algorithm55.
High-Throughput ELISA
A molecule that disrupts a dimer would likely have affinity for the interfacial surface of its constituent monomers. We developed a high-throughput screening method to test the ability of Trx–peptide fusions to bind to monomeric protease. Specifically, we covalently immobilized either urea-denatured or native protease on the surface of a maleimide-activated 96-well plate via its two reactive cysteine residues (Cys67 and Cys95). Free monomers were removed from immobilized monomers by extensive washing following the alkylation reaction. Positive isolates from the gel retardation assay were tested for the ability to bind to the protease monomer. Bound Trx-peptide fusions were identified by an ELISA using fluorescein diphosphate (FDP) as a substrate for the alkaline phosphatase conjugate (Fig. 2b). The affinity of a peptide for the protease should be related to the intensity of the ELISA signal. We identified the 9 isolates that produced the strongest signal, and determined the sequence of their encoded peptides. Using immobilized urea-denatured or native protease gave similar results (data not shown).
Analysis of True Positives
The sequences of the most effective peptides fall into three groups (Fig. 2c). A common feature in all three groups is the abundance of nonpolar residues, especially valine, alanine, and glycine. This overall composition is similar to that of the dimer interface of HIV-1 protease, which consists largely of an anti-parallel β-sheet composed of the four N- and C-terminal segments of the dimer: Pro1-Gln2-Ile3-Thr4 and Thr96-Leu97-Asn98-Phe99, respectively26. The lack of strong similarity among the 9 sequences is not surprising, because the clones are from a single round of selection in vivo rather than multiple rounds of selection in vitro (in which positive clones are enriched in each round1,2). Known peptide inhibitors of HIV-1 protease dimerization are based verbatim on the dimeric interface14–18,20,21,23–25. In contrast, the sequences from positive isolates (Fig. 2c) do not imitate the protease, and hence are new leads for chemotherapeutic agents.
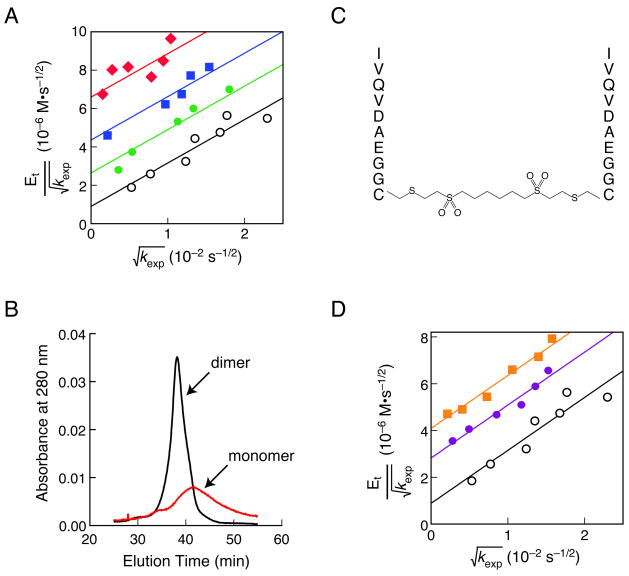
Dissociative inhibitors of multisubunit enzymes differ in their mechanism of action from that of traditional enzyme inhibitors7. An in vitro analysis of inhibition enabled us to distinguish between competitive, uncompetitive, and dissociative inhibition of HIV-1 protease. We synthesized a peptide (pep52) corresponding to a Trx-peptide fusion that gave a strong signal in the ELISA. We found that pep52 gives a series of parallel lines in a Zhang–Poorman plot, which is diagnostic of pure dissociative inhibition15. The dissociative inhibition constant (Kid) is 32 μM (Fig. 3a). We also used an independent biochemical method to test the ability of pep52 to disassemble the protease dimer. According to analytical gel filtration chromatography, pep52 causes the dimer to dissociate into monomers (Fig. 3b). The broad peak of the monomer indicates a lack of compact structure41,42, which could have the therapeutic benefit of making the enzyme vulnerable to proteolysis in vivo (vide infra).
Figure 3.
Analysis of inhibition of HIV-1 protease by pep52 and dim52. A, Zhang–Poorman plot15 for pep52. Protease activity was measured at different concentrations of pep52: 232 μM (diamonds), 116 μM (squares), 58 μM (closed circles), and 0 μM (open circles). The value of Kid for pep52 is (32 ± 1) μM; the value of Kd is (14 ± 3) nM. B, Analytical gel filtration chromatogram of wild-type HIV-1 protease in the presence or absence of pep52. C, Dim52, a dimer of pep52. D, Zhang–Poorman plot15 for dim52. Protease activity was measured at different concentrations of dim52: 2.96 μM (squares), 1.48 μM (closed circles), and 0 μM (open circles). The value of Kid for dim52 is (780 ± 20) nM.
The peptides identified by our approach are likely to bind specifically to HIV-1 protease. First, the ELISA and gel retardation assay for false positives demand that the peptides interact strongly with monomeric protease but not NcI. Second, the peptides do not impair the growth of E. coli cells (data not shown), indicating a lack of affinity for critical cellular proteins and protein complexes, such as those involved in DNA transcription.
Inhibitor Dimers
Multivalent display could be a simple way to increase the potency of a dissociative inhibitor. Creating a homodimeric inhibitor by tethering the peptide in a head-to-head or a tail-to-tail fashion would present the peptide with increased valency21,24. Accordingly, we prepared a dimeric peptide (dim52) by adding a cysteine residue to the C terminus of pep52 and crosslinking with a homobifunctional reagent (Fig. 3c). An in vitro analysis revealed that dim52 is a potent dissociative inhibitor with a Kid of 780 nM (Fig. 3d), which is 40-fold less than that of the monomer. Use of an optimized crosslinker21,23 or conformational constraint24 could decrease Kid further.
AIDS Chemotheraphy
Our approach is well-suited for developing new antiviral chemotherapeutic agents. Viruses have small genomes, conserving material by using multimeric proteins. Subunit interfaces in the many multimeric viral enzymes and the viral capsid are ideal targets for disruption. As an added benefit, dissociative inhibitors could intercept a viral polyprotein precursor before processing and assembly leads to any function20.
The high rate of viral replication and low fidelity of HIV reverse transcriptase have resulted in the emergence of HIV strains with resistance against drugs, including protease inhibitors43,44. To overcome resistance to individual drugs, combination therapy has been practiced in which multiple inhibitors with different antiviral mechanisms are used simultaneously to treat AIDS13,45,46. Recent evidence suggests, however, that the conventional triple-drug therapy can fail because of the emergence of HIV isolates with cross-resistance against different drugs47,48. Further, some of these cross-resistant HIV strains are also resistant to new compounds in clinical trials, including protease and reverse transcriptase inhibitors49. To date, all inhibitors of HIV-1 protease approved for human use are peptide mimetics that bind in the active site.
Dissociative inhibitors could be less prone to drug resistance than are active site-directed inhibitors7. Active site-directed inhibitors rely on a few high-affinity interactions with the protease. Protease variants with a change to a single active-site residue often emerge during treatment of AIDS patients with small-molecule inhibitors and can cause drug resistance13,45,46. In contrast, interactions between a dissociative inhibitor and protease monomer are likely to involve many residues encompassing a large surface area. The residues in the dimeric interface are highly conserved, even among protease variants that resist active-site–directed drugs50. The native enzyme is unlikely to tolerate changes in these residues, which are inaccessible to solvent26.
The effect of a dissociative inhibitor can be catalytic. HIV-1 protease has several sites for autoproteolysis51. The presence of these sites and the absence of compact structure (Fig. 3b)41,42 make the monomer an especially good substrate for active dimers of HIV-1 protease (as well as endogenous cellular proteases). Indeed, the asymmetry of monomer elution during gel filtration chromatography in the presence of pep52 (Fig. 3b) is consistent with its proteolysis by active dimers. It is noteworthy that proteolysis not only reduces the total number of protease molecules, but also shifts the monomer–dimer equilibrium toward the inactive monomer.
Prospectus
Many biological processes rely on noncovalent interactions between proteins. The ability to interfere with specific protein–protein interactions would expedite the dissection of complex biological processes and facilitate the development of novel chemotherapeutic agents. Our genetic selection (Fig. 1), along with our in vitro screens (Fig. 2), provides a powerful tool for the rapid identification of potent dissociative inhibitors in large combinatorial libraries. Here, we demonstrated the efficacy of our approach by identifying molecules that dissociate a homodimeric target protein (Fig. 3). Our approach is likewise applicable to the disruption of heterodimers, as effective hydrid repressor proteins can be assembled by heterodimerization31,33,37. Finally, although we panned a peptide library, our approach can also be used to sift through a library of RNA or cell-permeable small molecules for dissociative inhibitors.
Experimental protocol
Library Plasmid
A degenerate oligonucleotide, 5′-AATTTA(GGT)3(XYZ)9TAACCCGGCG-3′, was synthesized to encode a peptide of 9 random amino acid residues. The base composition of the XYZ codons, X = A(32%)/G(39%)/C(21%)/T(8%), Y = A(27%)/G(23%)/C(25%)/T(24%), and Z = G(40%)/T(60%), was as described52. The XYZ codons are flanked by codons for an SG3 spacer and ochre stop codon.
The complexity of as few as six degenerate codons (418) exceeds that of the human genome. Accordingly, producing a DNA duplex in which 9 codons are degenerate presents a special problem. (For example, much diversity would be lost by simply annealing two degenerate oligonucleotides.) To overcome this problem, we used the gapped-duplex method53, in which a degenerate oligonucleotide is annealed to two shorter oligonucleotides that are complementary to nondegenerate termini. The gap in the duplex DNA is ultimately filled in vivo. Specifically, we prepared a gapped-duplex DNA by annealing the degenerate oligonucleotide with oligonucleotides 5′-ACCACCACCTA-3′ and 5′-TCGACGCCGGGTTA-3′. The 5′ and 3′ ends of the resulting duplex are compatible with EcoRI and XhoI cleavage sites, respectively. The primers were phosphorylated at their 5′ ends by using T4 polynucleotide kinase. An NdeI site was introduced at the start codon of the glutathione S-transferase (GST) gene in the pGEX-4T3 vector (Pharmacia) by site-directed mutagenesis. The GST fragment was replaced with the trxA gene at the NdeI/EcoRI sites, resulting in plasmid pSH12. The phosphorylated gapped-duplex was ligated to pSH12 that had been digested with EcoRI and XhoI. Electroporetic transformation of MC1061 cells with the ligated DNA yielded 5 × 108 initial transformants. The transformants were grown in LB containing ampicillin (100 μg/ml) for 4 h at 37 °C before the preparation of the library plasmid (Fig. 1B). Analysis of the unamplified library indicated that 95% of clones carried inserts and that the 9 XYZ codons were indeed random.
Reporter Plasmid
DNA containing λPR, λOR1, and a start codon was prepared from plasmid pRZ4737 (W. S. Reznikoff) by the PCR using primers: 5′-CTAAGCTTGTGCGTGTTGACTATTTTACCT-3′ and 5′-AGAGAATTCCATGGACACCTCCTTAGTACATGC-3′, and inserted into the EcoRI and HindIII sites of a plasmid with the p15A origin and a chloramphenicol resistance gene. A DNA fragment of transcription terminator rrnBT2 was amplified from pMAL-p2 (New England Biolabs) by the PCR using primers 5′-CGGTCTAGAAAAACAGAATTTGCCTGG-3′ and 5′-AAAGCGGCAGAAACGCAAAAAGGCCAT-3′, and inserted into the XbaI and NotI sites, which are downstream of the λPR regulatory sequences. The lacZ and tet genes were amplified from pMC1871 (Pharmacia) and pBR322, respectively, and inserted sequentially between the start codon and rrnBT2 terminator using the NcoI and XbaI sites. PCR primers for the tet gene were designed so that the amplified gene contains the endogeneous ribosomal binding site, thereby allowing the polycistronic transcription of both lacZ and tet genes under the control of λPR and λOR1. The resulting plasmid was labeled pSH20.
A DNA fragment encoding NcI was prepared from plasmid pJH391 (J. C. Hu). A DNA fragment encoding D25N HIV-1 protease was produced by site-directed mutagenesis of plasmid pET-HIVPR (J. Tang). From these fragments, DNA directing the expression of an NcI–PR fusion protein (lacUV5–NcI–PR–stop codon) was constructed and inserted into pSH20, resulting in reporter plasmid pSH26 (Fig. 1B).
Genetic Selection
The library plasmid (1 μg) was transformed into electrocompetent MC1061 cells, yielding 3 × 108 transformants (or 0.06% of possible 9-residue sequences). Cells were diluted in LB containing ampicillin (100 μg/ml) and grown for 3.5 h at 37 °C before being prepared for electroporation. pSH26 (1 μg) was transformed into fresh library plasmid/MC1061 electrocompetent cells. The cotransformants were plated on LB agar medium containing ampicillin (100 μg/ml), chloramphenicol (50 μg/ml), X-Gal (75 μg/ml), tetracycline (20 μg/ml), and IPTG (20 μM), and incubated for 24 h at 37 °C until LacZ+TetR colonies appeared. Library plasmids were rescued from those LacZ+TetR colonies.
Cell Lysates for in vitro Assays
Each library plasmid rescued from a LacZ+TetR colony was retransformed into MC1061. Each transformant was grown at 37 °C in LB (2.0 ml) containing ampicillin (100 μg/ml) until log phase, when IPTG (to 0.5 mM) was added. IPTG-induced cells were grown for an additional 2.5 h and harvested by centrifugation. Cells were resuspended in 30 μl of 20 mM Tris-HCl buffer (pH 8.0) containing PMSF (1 mM), DTT (0.5 mM), EDTA (1 mM), and glycerol (10 % v/v). Lysozyme (2500 units; Epicentre) was added to resuspended cells with brief vortexing, and the mixture was incubated at 25 °C for 15 min for lysis to occur. After centrifugatation at 18,000 × g for 15 min at 4 °C, the cleared lysate was recovered and kept on ice or stored frozen at −80 °C.
Gel Retardation Assay
A fluorescein-labeled, double-stranded DNA corresponding to the λOR1 sequence was made by annealing oligonucleotides 5′-TTTACCTCTGGCGGTGATAG-3′ and 5′-(6-FAM)-CTATCACCGCCAGAGGTAAA-(6-FAM)-3′, where 6-FAM is 6-carboxyfluorescein. cI protein was produced from E. coli strain MC1061 transformed with plasmid pFG157 (J. C. Hu). Lysates from LacZ+TetR cells were mixed with cI protein (30 μg/ml) and incubated for 20 min at 25 °C in 10 mM Tris-HCl buffer (pH 8.0) containing MgCl2 (5 mM), CaCl2 (2 mM), BSA (50 μg/ml), sheared salmon sperm DNA (0.2 mg/ml), poly(dI-dC) (50 μg/ml), DTT (1 mM), and glycerol (5% v/v). Fluorescein-labeled λOR1 DNA was added (to 0.5 μM), and the reaction was incubated for another 15 min at 25 °C before electrophoresis in a polyacrylamide (8% w/v) gel in 1 × TBE buffer at 4 °C. After electrophoresis, the gel was scanned with a Fluorimager SI System (Molecular Dynamics).
ELISA
HIV-1 protease was produced from E. coli strain BL21(DE3)pLysS transformed with plasmid pET-HIVPR (J. Tang) as described54. Purified protease (14 μg/ml) was incubated for 3 h in 0.10 M HEPES-NaOH buffer (pH 7.0) or in 0.10 M MES-NaOH buffer (pH 6.6) containing urea (6 M) in maleimide-activated plates (Pierce). Unreacted maleimide was blocked with cysteine. Unless otherwise noted, the plate was washed (5×) by mild vortexing at 25 °C for 3 min with PBS between each of the following steps. Lysates from LacZ+TetR cells were prepared as described above, except for the use of PBS instead of Tris-HCl buffer. First, a lysate in PBS was incubated at 4 °C overnight in each well. Then, anti-Trx antibody (Sigma) in PBS containing bovine serum albumin (1 mg/ml) was incubated in each well at 25 °C for 1 h. Finally, anti-rabbit IgG-alkaline phosphatase conjugate (Sigma) in PBS was incubated at 25 °C for 1 h. The increase in the fluorescence of fluorescein diphosphate (Molecular Probes), a phosphatase substrate55, was measured by scanning the plate with a fluorimager SI System (Molecular Dynamics).
Peptide Synthesis
Peptide IVQVDAEGGC (pep52) was prepared by solid-phase peptide synthesis and purified by reverse-phase HPLC. The peptide and 1,6-hexane-bis-vinylsulfone (Pierce) were mixed in 0.10 M sodium borate buffer (pH 8.5) and incubated at 25 °C for 3 h before quenching the reaction with cysteine. The crosslinked dimer of pep52 (dim52) was purified by reverse-phase HPLC. The identity and purity of pep52 and dim52 were ascertained by electrospray ionization mass spectrometry with a Sciex API 365 System (Perkin Elmer) and amino acid analysis with an Amino Acid Analyzer 421 (Perkin Elmer), which was also used to determine solution concentrations.
HIV-1 Protease Assays
A fluorogenic substrate, R-E(EDANS)-SQNYPIVQ-K(DABCYL)-R (Molecular Probes)56, was used to assess proteolytic activity, which was measured with a QuantaMaster1 photon-counting fluorometer (Photon Technology International) as described57 with modifications. In the modifications, HIV-1 protease was preincubated with or without inhibitor for 30 min at 25 °C in 0.10 M sodium acetate buffer (pH 4.7) containing glycerol (5% v/v), PEG8000 (0.1% v/v), DTT (5 mM), EDTA (1 mM), and NaCl (0.10 M). Substrate was added to start the reaction. Data were collected for 3–5 min to obtain the initial velocity, and treated as described15 using eq 1:
| (1) |
where [Et] is the total enzyme concentration, k is the initial rate constant, [I] is the concentration of inhibitor, Kd is the equilibrium dissociation constant of the protease dimer, and Kid is the inhibition constant for dissociative inhibition. If inhibition is purely dissociative, then a plot of vs at different [I] consists of a series of parallel lines. Kinetic and thermodynamic parameters were obtained by nonlinear least-squares regression analysis of data with SIGMAPLOT 5.0 (SPSS).
Analytical Gel Filtration Chromatography
Pep52 (0 or 5 mM) was incubated for 30 min at 25°C in 50 mM sodium acetate buffer (pH 4.7) containing wild-type protease (2.8 μM). The mixture was loaded onto a column (22 cm × 20 mm2) of Superdex 75 gel filtration resin (Pharmacia), which had been pre-equilibrated with 50 mM sodium acetate buffer (pH 4.7), and eluted with the same buffer. Absorbance at 280 nm was monitored by using the program FPLC MANAGER (Pharmacia).
Acknowledgments
We thank J. C. Hu, J. Tang, and W. S. Reznikoff for providing plasmids, and L. L. Kiessling, D. H. Rich, and G. P. Roberts for advice. S. -H. P. was supported by a Korean Government Fellowship for Overseas Study. R. T. R. is an H. I. Romnes Faculty Fellow at the University of Wisconsin–Madison. This work was supported by grant GM44783 (NIH).
References
- 1.Smith GP. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 2.Matthews DJ, Wells JA. Substrate phage: Selection of protease substrates by monovalent phage display. Science. 1993;260:1113–1117. doi: 10.1126/science.8493554. [DOI] [PubMed] [Google Scholar]
- 3.Fields S, Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 4.Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: A method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geyer CR, Colman-Lerner A, Brent R. “Mutagenesis” by peptide aptamers identifies genetic network members and pathway connections. Proc Natl Acad Sci USA. 1999;96:8567–8572. doi: 10.1073/pnas.96.15.8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Norman TC, Smith DL, Sorger PK, Drees BL, O’Rourke SM, Hughes TR, Roberts CJ, Friend SH, Fields S, Murray AW. Genetic selection of peptide inhibitors of biological pathways. Science. 1999;285:591–595. doi: 10.1126/science.285.5427.591. [DOI] [PubMed] [Google Scholar]
- 7.Zutshi R, Brickner M, Chmielewski J. Inhibiting the assembly of protein–protein interfaces. Curr Opin Chem Biol. 1998;2:62–66. doi: 10.1016/s1367-5931(98)80036-7. [DOI] [PubMed] [Google Scholar]
- 8.Kohl NE, Emini EA, Schleif WA, Davis LJ, Heimbach JC, Dixon RA, Scolnick EM, Sigal IS. Active human immunodeficiency virus protease is required for viral infectivity. Proc Natl Acad Sci USA. 1988;85:4686–90. doi: 10.1073/pnas.85.13.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng C, Ho BK, Chang TW, Chang NT. Role of human immunodeficiency virus type 1-specific protease in core protein maturation and viral infectivity. J Virol. 1989;63:2550–2556. doi: 10.1128/jvi.63.6.2550-2556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flexner C. HIV-protease inhibitors. N Engl J Med. 1998;338:1281–1292. doi: 10.1056/NEJM199804303381808. [DOI] [PubMed] [Google Scholar]
- 11.Wlodawer A, Vondrasek J. Inhibitors of HIV-1 protease: A major success of structure-assisted drug design. Annu Rev Biophys Biomol Struct. 1998;27:249–284. doi: 10.1146/annurev.biophys.27.1.249. [DOI] [PubMed] [Google Scholar]
- 12.Lebon F, Ledecq M. Approaches to the design of effective HIV-1 protease inhibitors. Curr Med Chem. 2000;7:455–477. doi: 10.2174/0929867003375146. [DOI] [PubMed] [Google Scholar]
- 13.Klabe RM, Bacheler LT, Ala PJ, Erickson-Viitanen S, Meek JL. Resistance to HIV protease inhibitors: A comparison of enzyme inhibition and antiviral potency. Biochemistry. 1998;37:8735–8742. doi: 10.1021/bi972555l. [DOI] [PubMed] [Google Scholar]
- 14.Schramm HJ, Nakashima H, Schramm W, Wakayama H, Yamamoto N. HIV-1 reproduction is inhibited by peptides derived from the N- and C-termini of HIV-1 protease. Biochem Biophys Res Commun. 1991;179:847–851. doi: 10.1016/0006-291x(91)91895-j. [DOI] [PubMed] [Google Scholar]
- 15.Zhang ZY, Poorman RA, Maggiora LL, Heinrikson RL, Kezdy FJ. Dissociative inhibition of dimeric enzymes. Kinetic characterization of the inhibition of HIV-1 protease by its COOH-terminal tetrapeptide. J Biol Chem. 1991;266:15591–15594. [PubMed] [Google Scholar]
- 16.Babe LM, Rose J, Craik CS. Synthetic “interface” peptides alter dimeric assembly of the HIV 1 and 2 proteases. Protein Sci. 1992;1:1244–1253. doi: 10.1002/pro.5560011003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franciskovich J, Houseman K, Mueller R, Chmielewski J. A systematic evaluation of the inhibition of HIV-1 protease by its C-terminal and N-terminal peptides. Bioorg Med Chem Lett. 1993;3:765–768. [Google Scholar]
- 18.Schramm HJ, Billich A, Jaeger E, Rucknagel KP, Arnold G, Schramm W. The inhibition of HIV-1 protease by interface peptides. Biochem Biophys Res Commun. 1993;194:595–600. doi: 10.1006/bbrc.1993.1863. [DOI] [PubMed] [Google Scholar]
- 19.Babe LM, Rose J, Craik CS. Trans-dominant inhibitory human immunodeficiency virus type 1 protease monomers prevent protease activation and virion maturation. Proc Natl Acad Sci USA. 1995;92:10069–10073. doi: 10.1073/pnas.92.22.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schramm HJ, Boetzel J, Buttner J, Fritsche E, Gohring W, Jaeger E, Konig S, Thumfart O, Wenger T, Nagel NE, Schramm W. The inhibition of human immunodeficiency virus proteases by ‘interface peptides’. Antiviral Res. 1996;30:155–170. doi: 10.1016/0166-3542(96)00940-0. [DOI] [PubMed] [Google Scholar]
- 21.Zutshi R, Franciskovich J, Shultz M, Schweitzer B, Bishop P, Wilson M, Chmielewski J. Targeting the dimerization interface of HIV-1 protease: Inhibition with cross-linked interfacial peptides. J Am Chem Soc. 1997;119:4841–4845. [Google Scholar]
- 22.Fan X, Flentke GR, Rich DH. Inhibition of HIV-1 protease by a subunit of didemnaketal A. J Am Chem Soc. 1998;120:8893–8894. [Google Scholar]
- 23.Ulysse LG, Chmielewski J. Restricting the flexibility of crosslinked, interfacial peptide inhibitors of HIV-1 protease. Bioorg Med Chem Lett. 1998;8:3281–3286. doi: 10.1016/s0960-894x(98)00595-2. [DOI] [PubMed] [Google Scholar]
- 24.Bouras A, Boggetto N, Benatalah Z, de Rosny E, Sicsic S, Reboud-Ravaux M. Design, synthesis, and evaluation of conformationally constrained tongs, new inhibitors of HIV-1 protease dimerization. J Med Chem. 1999;42:957–962. doi: 10.1021/jm9803976. [DOI] [PubMed] [Google Scholar]
- 25.Schramm HJ, de Rosny E, Reboud-Ravaux M, Buttner J, Dick A, Schramm W. Lipopeptides as dimerization inhibitors of HIV-1 protease. Biol Chem. 1999;380:593–596. doi: 10.1515/BC.1999.076. [DOI] [PubMed] [Google Scholar]
- 26.Wlodawer A, Miller M, Jaskolski M, Sathyanarayana BK, Baldwin E, Weber IT, Selk LM, Clawson L, Schneider J, Kent SBH. Conserved folding a retroviral proteases: Crystal structure of a synthetic HIV-1 protease. Science. 1989;245:616–621. doi: 10.1126/science.2548279. [DOI] [PubMed] [Google Scholar]
- 27.Ptashne M. A Genetic Switch: Phage and Higher Organisms. Blackwell Science; Oxford, UK: 1992. [Google Scholar]
- 28.Pabo CO, Sauer RT, Sturtevant JM, Ptashne M. The lambda repressor contains two domains. Proc Natl Acad Sci USA. 1979;76:1608–1612. doi: 10.1073/pnas.76.4.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu JC, O’Shea EK, Kim PS, Sauer RT. Sequence requirements for coiled-coils: Analysis with lambda repressor–GCN4 leucine zipper fusions. Science. 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- 30.Amster-Choder O, Wright A. Modulation of the dimerization of a transcriptional antiterminator protein by phosphorylation. Science. 1992;257:1395–1398. doi: 10.1126/science.1382312. [DOI] [PubMed] [Google Scholar]
- 31.Bunker CA, Kingston RE. Identification of a cDNA for SSRP1, and HMG-box protein, by interaction with the c-Myc oncoprotein in a novel bacterial expression screen. Nucleic Acids Res. 1995;23:269–276. doi: 10.1093/nar/23.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu JC. Repressor fusions as a tool to study protein–protein interactions. Structure. 1995;15:431–433. doi: 10.1016/s0969-2126(01)00176-9. [DOI] [PubMed] [Google Scholar]
- 33.Jappelli R, Brenner S. Interaction between cAMP-dependent protein kinase catalytic subunit and peptide inhibitors analyzed with lambda repressor fusions. J Mol Biol. 1996;259:575–578. doi: 10.1006/jmbi.1996.0340. [DOI] [PubMed] [Google Scholar]
- 34.Zeng X, Herndon AM, Hu JC. Buried asparagines determine the dimerization specificities of leucine zipper mutants. Proc Natl Acad Sci USA. 1997;94:3673–3678. doi: 10.1073/pnas.94.8.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng X, Zhu H, Lashuel HA, Hu JC. Oligomerization properties of GCN4 leucine zipper e and g position mutants. Protein Sci. 1997;6:2218–2226. doi: 10.1002/pro.5560061016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, Murphy A, Hu JC, Kodadek T. Genetic selection of short peptides that support protein oligomerization in vivo. Curr Biol. 1999;9:417–420. doi: 10.1016/s0960-9822(99)80188-2. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Z, Zhu W, Kodadek T. Selection and application of peptide-binding peptides. Nature Biotechnol. 2000;18:71–74. doi: 10.1038/71951. [DOI] [PubMed] [Google Scholar]
- 38.Darke PL, Jordan SP, Hall DL, Zugay JA, Shafer JA, Kuo LC. Dissociation and association of the HIV-1 protease dimer subunits: Equilibria and rates. Biochemistry. 1994;33:98–105. doi: 10.1021/bi00167a013. [DOI] [PubMed] [Google Scholar]
- 39.Houghten RA. Peptide libraries: Criteria and trends. Trends Genet. 1993;9:235–239. doi: 10.1016/0168-9525(93)90087-x. [DOI] [PubMed] [Google Scholar]
- 40.Lu Z, Murray KS, Van Cleave V, LaVallie ER, Stahl ML, McCoy JM. Expression of thioredoxin random peptide libraries on the Escherichia coli cell surface as functional fusions to flagellin: A system designed for exploring protein–protein interactions. Biotechnology (N Y) 1995;13:366–372. doi: 10.1038/nbt0495-366. [DOI] [PubMed] [Google Scholar]
- 41.Grant SK, Deckman IC, Culp JS, Minnich MD, Brooks IS, Hensley P, Debouck C, Meek TD. Use of protein unfolding studies to determine the conformational and dimeric stabilities of HIV-1 and SIV proteases. Biochemistry. 1992;31:9491–9501. doi: 10.1021/bi00154a023. [DOI] [PubMed] [Google Scholar]
- 42.Todd MJ, Semo N, Freire E. The structural stability of the HIV-1 protease. J Mol Biol. 1998;283:475–488. doi: 10.1006/jmbi.1998.2090. [DOI] [PubMed] [Google Scholar]
- 43.Condra JH, Schleif WA, Blahy OM, Gabryelski LJ, Graham DJ, Quintero JC, Rhodes A, Robbins HL, Roth E, Shivaprakash M, Titus D, Yang T, Teppler H, Squires KE, Deutsch PJ, Emini EA. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature. 1995;374:569–571. doi: 10.1038/374569a0. [DOI] [PubMed] [Google Scholar]
- 44.Condra JH. Resistance to HIV protease inhibitors. Haemophilia. 1998;4:610–615. doi: 10.1046/j.1365-2516.1998.440610.x. [DOI] [PubMed] [Google Scholar]
- 45.Li TS, Tubiana R, Katlama C, Calvez V, Mohand HA, Autran B. Long-lasting recovery in CD4 T-cell function and viral-load reduction after highly active antiretroviral therapy in advanced HIV-1 disease. Lancet. 1998;351:1682–1686. doi: 10.1016/s0140-6736(97)10291-4. [DOI] [PubMed] [Google Scholar]
- 46.Palella FJ, Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Eng J Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 47.Piketty C, Castiel P, Belec L, Batisse D, Si Mohamed A, Gilquin J, Gonzalez-Canali G, Jayle D, Karmochkine M, Weiss L, Aboulker JP, Kazatchkine MD. Discrepant responses to triple combination antiretroviral theraphy in advanced HIV disease. AIDS. 1998;12:745–750. doi: 10.1097/00002030-199807000-00011. [DOI] [PubMed] [Google Scholar]
- 48.Rockstroh JK, Altfeld M, Kupfer B, Kaiser R, Fatkenheuer G, Salzberger B, Schneweis KE, Spengler U. Failure of double protease inhibitor therapy as a salvage therapy for HIV-infected patients resistant to conventional triple therapy. Eur J Med Res. 1999;4:271–274. [PubMed] [Google Scholar]
- 49.Palmer S, Shafer RW, Merigan TC. Highly drug-resistant HIV-1 clinical isolates are cross-resistant to many antiretroviral compounds in current clinical development. AIDS. 1999;13:661–667. doi: 10.1097/00002030-199904160-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schinazi RF, Larder BA, Mellors JW. Mutations in retroviral genes associated with drug resistance. Antivir News. 1997;5:129–142. [Google Scholar]
- 51.Rose JR, Salto R, Craik CS. Regulation of autoproteolysis of the HIV-1 and HIV-2 proteases with engineered amino acid substitutions. J Biol Chem. 1993;268:11939–11945. [PubMed] [Google Scholar]
- 52.LaBean TH, Kauffman SA. Design of synthetic gene libraries encoding random sequence proteins with desired ensemble characteristics. Protein Sci. 1993;2:1249–1254. doi: 10.1002/pro.5560020807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cwirla SE, Peters EA, Barrett RW, Dower WJ. Peptides on phage: A vast library of peptides for identifying ligands. Proc Natl Acad Sci USA. 1990;87:6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ido E, Han HP, Kezdy FJ, Tang J. Kinetic studies of human immunodeficiency virus type 1 protease and its active-site hydrogen bond mutant A28S. J Biol Chem. 1991;266:24359–24366. [PubMed] [Google Scholar]
- 55.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matayoshi ED, Wang GT, Krafft GA, Erickson J. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science. 1990;247:954–958. doi: 10.1126/science.2106161. [DOI] [PubMed] [Google Scholar]
- 57.Wondrak EM, Nashed NT, Haber MT, Jerina DM, Louis JM. A transient precursor of the HIV-1 protease. Isolation, characterization, and kinetics of maturation. J Biol Chem. 1996;271:4477–81. doi: 10.1074/jbc.271.8.4477. [DOI] [PubMed] [Google Scholar]