Abstract
The use of multiple X-ray protein structures has been reported to be an efficient alternative for the representation of the binding pocket flexibility needed for accurate small molecules docking. However, the docking performance of the individual single conformations varies widely and adding certain conformations to an ensemble is even counterproductive. Here we used a very large and diverse benchmark of 1068 X-ray protein conformations of 99 therapeutically relevant proteins, first, to compare the performance of the ensemble and single conformation docking, and, secondly, to find the properties of best performing conformers that can be used to select a smaller set of conformers for ensemble docking. The conformer selection has been validated through retrospective virtual screening experiments aimed at separating known ligand binders from decoys. We found that the conformers co-crystallized with the largest ligands displayed high selectivity for binders, and when combined in ensembles they consistently provided better results than randomly chosen protein conformations. The use of ensembles encompassing between 3 to 5 experimental conformations consistently improved the docking accuracy and binders vs. decoys separation.
INTRODUCTION
Structure-based drug screening and optimization plays an essential role in the early stages of drug development.1 When the 3D structure of the protein is available, protein-ligand docking protocols are widely used to predict the bound conformation of a ligand. Presently, docking algorithms usually take into account ligand flexibility; however, their success is hampered by the difficulty of representing the conformational changes of the protein upon ligands binding.2, 3 Some of these conformational changes induced by a ligand or other components of the environment can be observed when the same protein domain is co-crystallized with different ligands or in different experimental conditions.4
Several attempts have been made to introduce protein flexibility in a docking protocol.5-8 Unfortunately, convergent and predictive molecular dynamics (MD) or any form of an exhaustive sampling of the protein-ligand conformational space is still impossible given the astronomical size of the space to be sampled and limited accuracy of the energy functions. On the other hand, it was demonstrated that docking to multiple high quality static receptor conformations and selecting the best scoring solution is a practical alternative to an exhaustive search or an MD simulation.9 It was shown that including an experimentally determined conformational ensemble into a calculation increases the docking success rate from about 50% for a single cross–docking run to 80% to 90% for an ensemble docking exemplified by the 4D grid docking or the SCARE algorithms.10, 11
To date, the most abundant source of information for structure-based drug design is X-ray crystallography.1 The X-ray structures represent 86% of the Protein data Bank (PDB), followed by nuclear magnetic resonance spectroscopy (NMR) representing 13%, and the remaining < 1% represented by electron microscopy (EM) or other techniques. Ensemble docking with multiple X-ray conformers has been reported to have higher success rates compared to the single conformation docking by several groups.12-14 However, according to these studies and our own experience, the results largely depend on the quality of the structures and the best performance is obtained when the structural ensembles include the bound form of the protein. Thus, the structures for a representative ensemble should be selected not only from the consideration of the docking speed, which increases linearly with the number of conformers, but also from the consideration of the docking success rate. Barril et al.15 showed that, although the use of ensembles could be good for the docking performance, an excess of structures increased the number of false positives. According to their cross-docking results obtained with Cyclin dependent kinase (CDK2) and Heat shock protein 90 (HSP90), the best performing single receptor structure docked 68% and 49% of ligands respectively. The best performing combinations consisting of 6 (CDK2) and 8 (HSP90) improved the results to 94% and 77%. These results suggested that a relatively small subset of conformations could efficiently represent induced fit effects. In a more realistic scenario, such as a virtual screening (VS) experiment, their results also pointed out that VS is much more sensitive to the potential artifacts introduced by ensemble docking. When only the conformations that achieved satisfactory accuracy in single receptor runs were combined, the maximum performance in terms of enrichment factors was obtained. However, when the conformations were randomly combined, the overall performance deteriorated. Some authors have reported similar results, concluding that a reduced number of selected conformations results in optimal performance.16-20 Therefore, for virtual screening experiments, it seems more appropriated to use a subset of selected conformations rather than include all the available structures. But which conformations should be selected? Can we pick the best performing set a priori without evaluating the ability of each conformation to discriminate between known binders and non binders?
In this paper, we study the specificity of the individual X-ray conformations of a protein present in the PDB according to their capability to distinguish known binders from non-binders. The goal of the study is to come up with a set of recommendations for selecting the optimal set of multiple conformations for ensemble or 4D docking. The dataset consisting of 99 therapeutically relevant proteins, containing 1068 conformations has been tested with ICM,21, 22 using single receptor conformation (SRC) and multiple receptor conformations (MRC) docking protocols. The study provides insights about selecting conformations for both single-conformation and ensemble docking.
MATERIALS AND METHODS
Benchmark: Protein−Ligand Complex Structures and Ensembles
The proposed benchmark was already used to test the performance of the Four-Dimensional Docking methodology11, 23 which contained 99 proteins with publicly available structural information.24 The sequences were initially searched against a non-redundant subset of the PDB sequences where 3D domains were annotated based on the PDB sequence boundaries and clustered to 95% sequence identity. A conformational ensemble for a protein had to represent at least three different crystal structures and include at least one co-crystallized ligand. Structures where any druggable binding site25 could not be automatically identified (indicative of the ligand binding at a crystallographic interface) were excluded as well. For the sake of uniformity with our original benchmark, members of the same ensemble had to display 100% identity in the binding site and contain the same metals and cofactors, while other positions in the sequence as well as the chain ends may vary. If more than one variant of the binding site residues could be identified, the original ensemble was split and the resulting groups assigned consecutive numbers. Consequently the number of ensembles is slightly larger than the number of proteins because some proteins need to be represented by two or three different ensembles.
A collection of ~ 3000 non trivial drug-like molecules from the PDB was built by (i) excluding ubiquitous substrates and (ii) applying relaxed Lipinski rules to filter to the entire PDB Chemical Component Dictionary. That collection was merged with the above protein domain ensemble set to obtain multiple conformation ensembles for 841 proteins structures co-crystallized with at least one relevant compound. The ligands were analyzed for correctness of their covalent geometry and checked against the electron density data from the Uppsala Electron Density Server.26 To be included in the set, ligand structures had to consist of a single fragment small organic molecule with: (i) more than 20 non-hydrogen atoms, (ii) less than 12 rotatable bonds, (iii) no rings with nine or more members, and (iv) a density fit value (defined as the fraction of heavy atoms inside one sigma of a density contour surface) > 0.8. Lastly, ligands that could not be accurately re-docked into their own cognate receptor binding site were excluded because previous studies established that the majority of those failures are indicative of crystallographic, protonation, or tautomerization errors in either ligand or receptor.27, 28 The number of ligands that passed the above filters was 291.
Preparation of Proteins
The receptors were prepared according to a minimal user intervention procedure. According to this, chains, heteroatoms, and prosthetic groups not involved in the binding site definition were deleted. The inclusion of specific crystallographic water molecules has been reported to improve the accuracy of the cross-docking predictions for some specific complexes.29 In this study, we deliberately removed all the waters molecules within the binding pocket to avoid any user-derived bias towards specific bound complexes. The assumption made here is that the role of water molecules in the binding site can be approximated after rescoring by cavities of a high distance-dependent dielectric constant. The protein atom types were assigned and hydrogen atoms and missing heavy atoms were added. The added or zero occupancy side chains and polar hydrogen atoms were optimized and assigned the lowest energy conformation. Tautomeric states of histidines and the rotations of asparagine and glutamine side chain amidic groups were optimized to improve the hydrogen bonding patterns. The cognate ligands were deleted from the complexes only after hydrogen optimization.
Preparation of Ligands
Coordinates of the 291 ligands (275 unique) were extracted from the crystallographic complexes. Bond orders, tautomeric forms, stereochemistry, hydrogen atoms, and protonation states were assigned automatically by the ICM30 chemical conversion procedure. Each ligand was assigned the MMFF31 force field atom types and charges. Ligand molecules were prepared for docking by rotational search followed by the Cartesian minimization in the absence of the receptor and the lowest-energy conformations were used as starting points for ICM docking.
ICM docking
ICM addresses the docking issue as a global optimization problem with the biased probability Monte Carlo (BPMC) global stochastic optimizer.21 Since the BPMC method was previously reported and thoroughly described, it is only briefly summarized here. During docking, the ligand torsional or roto-translational variables are randomly changed. A local refinement is carried out using the analytically differentiable energy terms by a conjugate-gradient minimization. The complete energy is calculated by adding up the contributions of the solvation energy and those of the conformational entropy, and the conformation is accepted or rejected according to the Metropolis criteria. A new random change is introduced and the whole procedure is repeated all over until the number of steps exceeds the limit pre-estimated as a function of the number of the ligand variables.
The molecular system was described using internal coordinate variables. Protein atom types and parameters were taken from a modified version of the ECEPP/3 force field.32 The binding pocket definition was based on the largest common envelope predicted in each receptor by the Pocketome Gaussian Convolution algorithm25 (tolerance value of 5.0). The binding pocket boundaries were defined by selecting all the residues with heavy atoms within 3.5 Å from the mesh. The binding pocket was described by five 0.5 Å spacing potential grid maps, representing van der Waals potentials for hydrogens and heavy atoms, electrostatics, hydrophobicity, and hydrogen bonding. Because the standard 6-12 van der Waals potential was considered too sensitive to steric clashes for the purpose of the simulations, a truncated soft van der Waals potential was introduced and the other potentials were rescaled accordingly to avoid atom overlap (e.g. for two oppositely charged atoms). The van der Waals potentials were truncated at 1.0 kcal/mol.
Virtual Screening
The 275 unique crystallographic ligands were docked in each of the 1068 protein structures. Each ligand was docked into the grid representation of the pocket using the BPMC method described above. The global optimization provided a stack of geometrically diverse poses for each ligand, and the best five were assigned a docking score by the standard ICM empirical scoring function.33, 34 Due to the stochastic nature of ICM docking procedure, the docking simulations were performed three times to ensure convergence. For each ligand, only the conformation with the lowest (best from 15) energy score was retained.
The screening performance of each protein conformation was evaluated numerically as the area under the receiver operating characteristic curve (ROC), abbreviated as AUC.35 The AUC value describes the cumulative ability of a docking procedure to recognize true positives and negatives while avoiding false positives and false negatives. A theoretically perfect performance has an AUC value of 1.0; while a random selection performance has an AUC of 0.5. In SRC docking, the ROC curves were obtained by plotting the number of top scored compounds against the number of known ligands (co-crystallized in the same protein) among them. For MRC ensemble docking, the ROC curves were built using the best ICM score for each ligand coming from the protein structures in the ensemble.
Software and Hardware
The receptor and ligand preparations, the docking simulations, and the energy evaluations were carried out with ICM 3.6-2 (Molsoft LLC, La Jolla, CA). The docking simulations ran on an Intel Core 2 Quad workstation (2.4 GHz with 3 GBytes of RAM) and a 3020 64-bit Intel XEON-EMT CPUs Linux-based cluster “Garibaldi” at The Scripps Research Institute (La Jolla, CA, USA). Each virtual screening experiment took ~ 5 hours to complete on a single CPU.
RESULTS AND DISCUSSION
Benchmark global results: Single Receptor Conformation Virtual Screening
The dataset, consisting of 1068 protein conformations from 99 independent proteins, was considered to be large and diverse enough to be representative of the therapeutically relevant targets in the PDB, naturally enriched in kinases and proteases. According to DrugBank,36 32 of the selected proteins are targeted by at least one marketed drug and 62 are targets of experimental drugs in different development stages. Among the 275 non-redundant ligands, 28 are marketed drugs and 121 experimental ones.11 The dataset has a substantial overlap with the Astex diverse set, recently reported by Hartshorn et al,29 sharing 32 proteins and 22 ligands.
The first parameter we analyzed was the global performance of the benchmark in terms of recognition specificity. For each of the 1068 individual conformations, we tested the ability of separating true binders from non-binders in virtual screening experiments, reporting the performance by means of the area under the ROC curves (AUC, see Methods). We used a stringent criterion to label binders and non-binders, by choosing as binders all the ligands that were co-crystallized with the protein target, and the rest as non-binders. On average, we had 3 active ligands per target out of 275 (~ 1% active / inactive proportion).
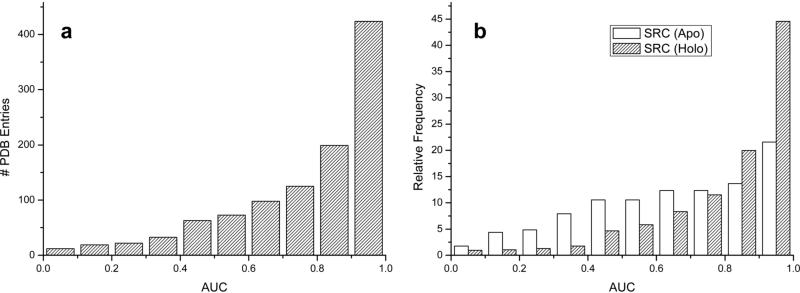
As can be seen on the histogram of the AUC values for individual 1068 conformations (Figure 1a), the majority of individual structures manifest favorable AUC values, while a small minority display a random (or worse) than random performance. In particular, 14% of the dataset had AUC values below random (0.5), 79% > 0.6, 70% > 0.7 and 58% > 0.8. If we consider values above 0.9 as a good indicator of recognition, 424 structures (40%) are above such a threshold. When we divided the benchmark into apo (227 structures) and holo (841 structures, where 291 were co-crystallized with a drug-like ligand) we observed that, in general, holo conformations had higher AUC values than apo (see Figure 1b). The distribution of AUC values for holo conformers is clearly shifted to the right part of the plot with respect to apo one, with 45% of the conformations having AUC values > 0.9, and only 10% having below random AUC values. The apo distribution of the AUCs is more spread; only 22% of the conformations have AUC values > 0.9 while 30% have values below random. It is worth mentioning that the maximum difference between the two distributions is focused in the 0.9-1.0 bin, where holo pocket conformers have a two times higher frequency than with the apo conformers.
Figure 1.
Histograms showing the performance of single receptor conformation virtual screening, a) for the 1068 protein conformations, b) for the 1068 conformations separated in apo (227) and holo (841).
According to these preliminary results, restricting the selection to only holo conformations, if available, seemed to be a reasonable choice for virtual screening. On average, apo conformers provided slightly worst ICM scores for the active ligands than holo conformers (See Supporting Figure S1). Usually, apo structures have side chains sticking inside the binding cavity thus preventing the ligand binding, whereas holo structures are more representative of a state ready for binding.
Benchmark global results: Multiple Receptor Conformations Virtual Screening
After studying the performance of each of the 1068 individual conformations, the next step was to combine all the conformers in each protein ensemble, according to ensemble docking procedure (see Methods). The aim of ensemble docking is to increase the probability of ligand binding by adding protein plasticity represented by several conformational snapshots. On average, the 106 protein ensembles contained ~10 conformations each (3 of them apo and 7 holo, see Table 1) and 3 known ligands (as described in the Methods section, we discarded all non drug-like co-crystallized ligands in the docking experiments). In 103 of the ensembles (97% of the benchmark) we had more than one holo conformation, and in 66 ensembles more than one known ligand. The average intra-ensemble heavy atom root mean square deviation (rmsd) for the binding pocket was 1.9.
Table 1.
Numbers of proteins and ligands used in this study.
| Single Receptor Conformation | Multiple Receptor Conformations | ||
|---|---|---|---|
| PDB entries | 1068 | Proteins / Ensembles | 99 / 106 |
| PDB (apo/holo) | 227 / 841 | Ensembles ≥ 1 apo | 56 |
| Drug-like ligands | 291 | Ensembles > 1 holo | 103 |
| Drug-like unique ligands | 275 | Ensembles > 1 ligand | 66 |
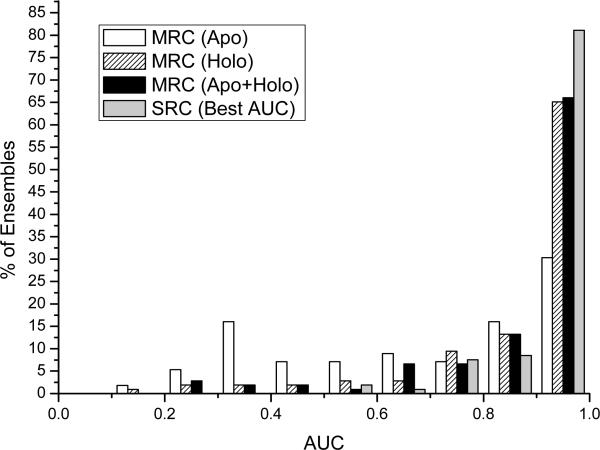
As can be observed in Figure 2, the ensemble docking and scoring has a clearly superior performance over an average single conformation. The three AUC distributions resulting from the apo, holo and apo+holo ensembles present higher AUC values in comparison with their counterparts in SRC (Figure 1, note the difference in Y axis). With SRC, the mean AUC value was 0.78 ± 0.22 and with MRC (apo+holo) increased to 0.88 ± 0.18. These results indicated that the accuracy and the specificity of discrimination between known binders from non-binders in ensemble docking systematically improved. When we used all the available conformations (including apo+holo), we found that only 7% of the ensembles had AUC values below random (0.5), and 70 of them (66%) achieved a very good separation (AUC > 0.9). Ensembles containing only the apo conformations provided less accurate results (see Figure 2), showing similar performance as apo SRC, whereas no significant difference could be noticed between holo and apo+holo ensembles.
Figure 2.
Histograms showing the distributions of values of areas under the ROC curves (AUC) values obtained with Multiple Receptor Conformation docking in 106 protein ensembles. The distributions correspond to AUCs from apo, holo or apo+holo ensembles. The best AUC values obtained from single receptor conformation are reported for comparison. The performance disparity between an average single conformation and the best a posteriori conformation will be discussed later.
In conclusion, all the results reported so far are pointing towards the benefit of using the ligand-bound protein conformations. At the benchmark level, on average, protein ensembles provided better AUC recognition values than isolated conformations. The apo conformers generated slightly worst scores for the actives than holo, and their contribution in sequential ensemble docking was limited. The low success of apo conformers suggests that the conformational substates found in bound crystals are rarely captured in apo X-ray structures.
Detailed analysis of ensemble screening performance
After reporting the average superior performance of MRC in VS, it is possible to get the optimistic impression that the use of ensembles systematically outperforms each individual conformer in terms of recognition. However, the situation is not that clear when one looks into the details of each protein, and in fact, we observed that in many cases the situation was just the opposite.
To further study the recognition properties within each ensemble, we labeled each conformation as being apo or holo, and sorted them numerically according to their AUC values (see Supporting Information Table S1). As expected,37 in 96 out of 106 ensembles the single conformation displaying the best AUC value was holo, while in only 10 out of 106 it was apo (in 4 cases we found either apo or holo). Even taking into account the bias in the dataset towards holo structures (7:3, holo:apo relation), the numbers confirm again that holo conformations have better separating power than apo ones.
After studying in detail selected cases, it was not possible to identify any strong relationship between the performance of a given conformer and some intrinsic structural descriptors. For instance, we did not find correlation between performance and binding site volume, number of atomic contacts, X-ray resolution, B-factors, or flexibility descriptors such as the ones obtained from the analysis of the eigenvectors/eigenvalues from elastic network normal mode analysis.38 Interestingly, it was possible to establish a correlation between the performance and size of the cognate ligand; in 52 out of 106 ensembles, the protein conformations co-crystallized with the largest ligands were those providing the highest individual AUC values. In particular, in 25 of the 66 ensembles having > 1 ligand (these results further improved when we included the receptor conformations with cognate ligands in the range of 25 A3 from the largest one), and, in 27 of the 40 ensembles having 1 ligand.
To assess whether or not the use of ensembles improved screening performance, we derived a simple measure based on the percentage of gain with respect to the highest AUC value obtained from a single conformation, as,
| (1) |
where type is the composition of the ensemble (i.e., apo, holo, apo+holo). A positive value means that ensemble docking increases the AUC value and a negative value means that the performance got worse. For instance, in the case that the highest AUC from a single conformation is 0.96 and the AUC from ensemble docking is 0.91, we have lost 0.05 units of AUC and the gain is −5.
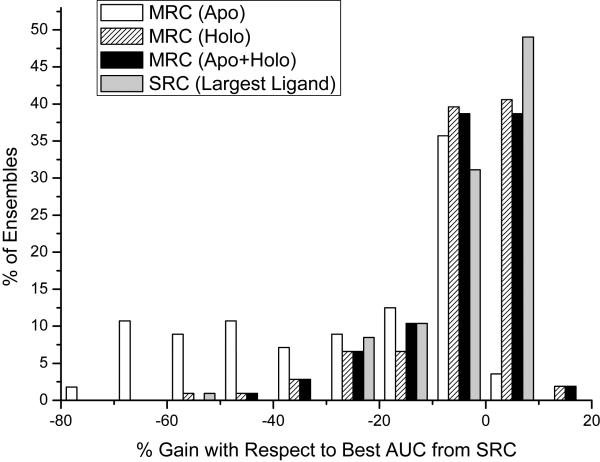
Figure 3 shows the distributions of gains when ensembles consisting of apo, holo and apo+holo were employed respectively. For comparison, the gains obtained comparing the SRC runs on the protein conformation with the largest ligand are reported. Not surprisingly, ensembles containing only apo conformations were those providing the worst results, as in 96% of the cases we obtained negative gains with respect to the best AUC. Ensembles containing apo+holo and holo again behaved similarly, improving the AUC values in 41%, and 42% of the cases respectively (47% if we consider only ensembles with > 1 ligand). On average, the use of ensembles provided gains in the range from −10 to 10, meaning that we can either gain or lose 0.1 units of AUC with respect to the best performing single conformation. Such an anticooperative behavior, where the addition of certain protein conformations can sometimes degrade the performance, has already been pointed out in the literature.15, 18, 19
Figure 3.
Histograms showing the gain obtained with ensemble docking with respect to the best AUC value obtained from a single receptor conformation.
Multiple receptor conformation advantages: Ligand profiling
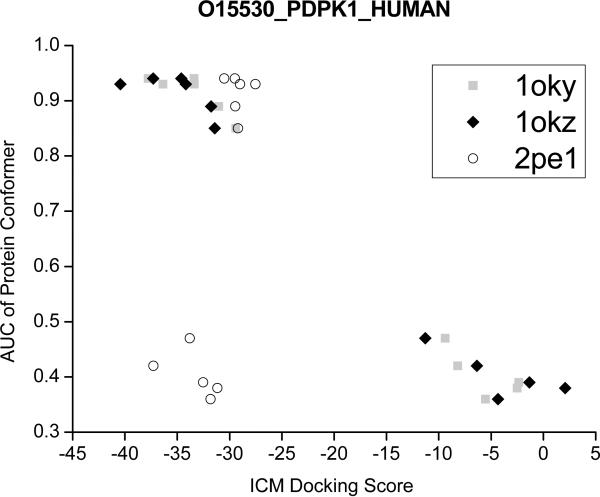
The use of ensembles had shown an improvement in the correlation between docking scores and biological activity,17, 39 suggesting that the scores coming from multiple protein conformations could help in the profiling of ligands. Often, discrimination between potential binders from non-binders is based on thresholds on the scoring energy (e.g., a score below −32 kcal/mol is regarded as good in ICM docking). Here, we considered protein 3-phosphoinositide dependent protein kinase-1, (PDPK1, Uniprot code: O14757) as a case study, and plotted the best ICM scores for the 3 active ligands (from PDB codes: 1OKY, 1OKZ and 2PE1) obtained with each protein conformer. As can be observed in the Figure 4, if we only use the single protein conformers selected according to their AUCs (i.e., AUC > 0.8), we can fail in capturing the cognate ligand of 2PE1 as a potential binder. However, when we include more holo conformers, the cognate ligand of 2PE1 obtained better cross-docking scores (e.g., −37.3 kcal/mol with conformer 2PE2). As previously suggested,15, 40 the use of several crystal structures of the same target seems to reduce the uncertainties in the docking scores results and prevented overspecialization towards single scaffolds.
Figure 4.
Example of cross-docking ICM scores (in kcal/mol units) obtained with holo conformers from 3-phosphoinositide dependent protein kinase-1 (PDPK1). The AUCs of the conformers are: 1UU8:0.94, 1OKY:0.94, 1UU9:0.93, 1OKZ:0.93, 1UU7:0.89, 1UU3:0.85, 2PE0:0.47, 2PE2:0.42, 2PE1:0.39, 1Z5M:0.38, 1H1W:0.36. The scores are shown for the cognate ligands from co-crystals 1OKY, 1OKZ and 2PE1.
Recipes for the selection of “important” protein conformations
Frequently, when multiple conformations of the same protein are available, the researcher is confronted with the decision of which protein structures to use, in a compromise between accuracy and speed. Here, a spectrum of possibilities is suggested, depending on the availability of protein conformers and ligand data. We would like to emphasize that it is not purely a matter of computational speed, because nowadays the docking calculations can be submitted in parallel to computer clusters, but rather a performance issue. Our docking experiments were performed with ICM, but we think that our principal observations are transferable to other platforms, as long as their docking accuracy had been validated.41
In a best case scenario, where activity ligand data is available for significant ligands (or at least some ligand data), a ligand guided approach seems to be the most powerful approach to validate the alternative protein conformations.9 In a ligand guided assessment, a small scale VS is performed for each of the protein conformers and the selectivity of each model is evaluated by discrimination measures, such as the enrichment factors or AUCs. Finally, the conformations are numerically sorted according to these values and only the top conformations are selected a posteriori to be used in independent dockings. Such a methodology can be applied to any kind of conformation, regardless of its origin (i.e., X-ray, NMR, homology model, or snapshots extracted from MD simulation). This approach has been successfully applied by our group in a blind test for assessing the structure of the A2a adenosine receptor, a G-protein coupled receptor.42
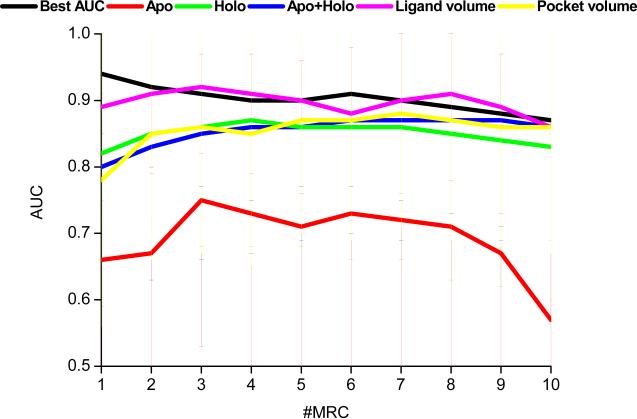
According to our results (see Figure 5 and Supporting Figure S2), the use of ensemble docking, with a few exceptions, systematically did not outperform the highest AUC obtained from a single conformation. It was not possible to outperform the best single receptor even when the conformers were handpicked according to their AUCs. The mean AUC value from the best performing single conformations was 0.94 (90% of the proteins had AUCs > 0.8 and 81% had AUCs > 0.9). Thus, there is usually a single conformation displaying an already good AUC value (see Figure 2), leaving small room for improvement. These results are consistent with those reported by Birch et al.16 and Barril et al.,15 concluding that single (“one-size-fits-all”) protein structures carefully chosen a posteriori provide optimum results in VS. Improvement made by the use of ensemble docking could become more important in cases where the initial structures have lower AUCs, as those in homology models.
Figure 5.
AUC values versus the number of conformers added to the ensemble according to different criteria. The apo, holo and apo+holo conformers were randomly added to the ensembles and the experiments were repeated three times. The results on the figure are the averages from 106 ensembles and the bars indicate the standard deviations.
In a real life scenario, where no ligand information is available to profile receptor conformations, the ensemble docking procedure seems to be an acceptable alternative. According to our results, apo+holo or holo ensembles, where the conformations were randomly chosen, improved the AUC values with respect to the original up to ~5 conformers (see Figure 5 and Supporting Figure S2). These results are in agreement with the findings presented recently by Verdonk and colleages.20 We should note that such a behavior seems quite independent on the number of X-ray conformations available for the target. Hence, in a compromise between speed and performance, the selection of only holo structures, up to 5 conformers could be a reasonable choice. Apo conformations may still be used to add diversity when they possess specific properties needed for the recognition of given scaffolds.
Recent reports suggested using non-collapsed conformations (e.g., DOLPHIN,43 active site fumigation,23 active site resin pressurization,44 SCARE,10 or Thomas et al.18 approach) in an attempt to represent a “bound state” from the structures that are currently available. On the basis of this rationale, a selection of conformers according to the volume of the pocket was tested (see Figure 5 and Supporting Figure S2). Although globally we did not observe any improvement with respect to random selection of holo or apo+holo, the selection of protein conformers according to the volume of the pocket seems to be a reasonable alternative when only apo structures are available. Often, automated predicted pockets are larger than the volume occupied by a ligand, and thus two pockets can share a similar binding site but have very different volumes.45 If the computational resources are limited, and there exist proteins with co-crystallized ligands, we recommend the use of the receptor conformation[s] having the largest[s] ligands, as, on average, we obtained results similar to those obtained when the ensembles were generated according to numerically sorted AUC values. Using the ligand as a source of information (and not the volume of the pocket) we avoid the uncertainties associated to the binding pocket volume prediction. The ligands can be sorted either by volume or number of heavy atoms indistinctly, because both parameters are correlated (see Supporting Figure S3). According to our results, a simple quality filtering of the ligands (see Methods) improves the quality of the simulations.
Future directions
To date, structure-based drug design is largely based on structures from PDB.1 In this study, we compiled an extensive benchmark consisting exclusively of X-ray structures as a source of receptor flexibility. After automatic preparation and manual filtering, the set is clean and can be safely assumed to represent the state-of-the-art recognition in PDB. Nevertheless, X-ray structures in PDB have idiosyncrasies that in some cases may interfere with their full potential in drug design applications.23, 40, 46 For instance, crystals are solved in low non-biological temperatures, where some of the ligands were added by soaking,47 and some residues are frozen in a space-averaged structure not representative of the full biological spectrum.
The success of induced fit docking with ensembles is related to our ability of represent the bound state of the proteins, which in many cases is not available in the PDB.4 For overcoming this, discrete models coming from different simulation methods such as Monte Carlo, molecular dynamics, or normal mode analysis can be used.4, 48, 49 But again the same original question arises, which models should be selected so that docking performance is not affected? These properties are currently under investigation.
CONCLUSIONS
In this study we investigated the performance of an ensemble of structures in virtual screening with respect to a single receptor conformation using a benchmark that represents the structural variability in the PDB. When ligand information is available, a ligand guided approach, where the best conformations are handpicked according to their ability to separate binders from non-binders, seems the optimal choice for a fast and efficient virtual screening experiment. When no ligand information is available, randomly chosen ensembles containing up to 3 to 5 conformers improve the recognition against a single receptor. In general, ensembles consisting of holo structures performed on par with apo+holo, suggesting that results from apo structures go unnoticed, and apo conformations might be discarded from the ensembles. The use of ensembles reduces the score-based uncertainties associated to single receptor conformation docking, generating more consistent scores for ligand profiling. If computational time is scarce, the selection of protein conformations according to the size of the co-crystallized ligand is likely to produce optimum results.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Arsen Grigoryan for helpful comments, Xavier Barril for useful discussions and suggestions on the manuscript and Karie Wright for help with manuscript preparation. MR is supported by a Spanish MEC Postdoctoral fellowship. This work was supported by NIH grant 1-R01-GM074832.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Supporting Table S1 and Figures S1 to S3 indicated in the text. This information is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Congreve M, Murray CW, Blundell TL. Keynote review: Structural biology and drug discovery. Drug Discovery Today. 2005;10:895–907. doi: 10.1016/S1359-6446(05)03484-7. [DOI] [PubMed] [Google Scholar]
- 2.Teague SJ. Implications of protein flexibility for drug discovery. Nat. Rev. Drug Discov. 2003;2:527–541. doi: 10.1038/nrd1129. [DOI] [PubMed] [Google Scholar]
- 3.Sousa SF, Fernandes PA, Ramos MJ. Protein-ligand docking: Current status and future challenges. Proteins Struct. Funct. Bioinformat. 2006;65:15–26. doi: 10.1002/prot.21082. [DOI] [PubMed] [Google Scholar]
- 4.Cozzini P, Kellogg GE, Spyrakis F, Abraham DJ, Costantino G, Emerson A, Fanelli F, Gohlke H, Kuhn LA, Morris GM, Orozco M, Pertinhez TA, Rizzi M, Sotriffer CA. Target flexibility: an emerging consideration in drug discovery and design. J. Med. Chem. 2008;51:6237–6255. doi: 10.1021/jm800562d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson HA. Protein flexibility and drug design: how to hit a moving target. Curr. Opin. Chem. Biol. 2002;6:447–452. doi: 10.1016/s1367-5931(02)00341-1. [DOI] [PubMed] [Google Scholar]
- 6.Klebe G. Virtual ligand screening: strategies, perspectives and limitations. Drug Discovery Today. 2006;11:580–594. doi: 10.1016/j.drudis.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.C BR, Subramanian J, Sharma SD. Managing protein flexibility in docking and its applications. Drug Discovery Today. 2009;14:394–400. doi: 10.1016/j.drudis.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 8.Guvench O, MacKerell AD., Jr. Computational evaluation of protein-small molecule binding. Curr. Opin. Struct. Biol. 2009;19:56–61. doi: 10.1016/j.sbi.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Totrov M, Abagyan R. Flexible ligand docking to multiple receptor conformations: a practical alternative. Curr. Opin. Struct. Biol. 2008;18:178–184. doi: 10.1016/j.sbi.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bottegoni G, Kufareva I, Totrov M, Abagyan R. A new method for ligand docking to flexible receptors by dual alanine scanning and refinement (SCARE). J. Comput.-Aided Mol. Des. 2008;22:311–325. doi: 10.1007/s10822-008-9188-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bottegoni G, Kufareva I, Totrov M, Abagyan R. Four-dimensional docking: a fast and accurate account of discrete receptor flexibility in ligand docking. J. Med. Chem. 2009;52:397–406. doi: 10.1021/jm8009958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavasotto CN, Abagyan RA. Protein flexibility in ligand docking and virtual screening to protein kinases. J. Mol. Biol. 2004;337:209–225. doi: 10.1016/j.jmb.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Ferrari AM, Wei BQ, Costantino L, Shoichet BK. Soft docking and multiple receptor conformations in virtual screening. J. Med. Chem. 2004;47:5076–5084. doi: 10.1021/jm049756p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolstad ES, Anderson AC. In pursuit of virtual lead optimization: the role of the receptor structure and ensembles in accurate docking. Proteins. 2008;73:566–580. doi: 10.1002/prot.22081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barril X, Morley S. Unveiling the full potential of flexible receptor docking using multiple crystallographic structures. J. Med. Chem. 2005;48:4432–4443. doi: 10.1021/jm048972v. [DOI] [PubMed] [Google Scholar]
- 16.Birch L, Murray CW, Hartshorn MJ, Tickle IJ, Verdonk ML. Sensitivity of molecular docking to induced fit effects in influenza virus neuraminidase. J. Comput.-Aided Mol. Des. 2002;16:855–869. doi: 10.1023/a:1023844626572. [DOI] [PubMed] [Google Scholar]
- 17.Yoon S, Welsh WJ. Identification of a minimal subset of receptor conformations for improved multiple conformation docking and two-step scoring. J. Chem. Inf. Comput. Sci. 2004;44:88–96. doi: 10.1021/ci0341619. [DOI] [PubMed] [Google Scholar]
- 18.Thomas MP, McInnes C, Fischer PM. Protein structures in virtual screening: a case study with CDK2. J. Med. Chem. 2006;49:92–104. doi: 10.1021/jm050554i. [DOI] [PubMed] [Google Scholar]
- 19.Rao S, Sanschagrin PC, Greenwood JR, Repasky MP, Sherman W, Farid R. Improving database enrichment through ensemble docking. J. Comput.-Aided Mol. Des. 2008;22:621–627. doi: 10.1007/s10822-008-9182-y. [DOI] [PubMed] [Google Scholar]
- 20.Verdonk ML, Mortenson PN, Hall RJ, Hartshorn MJ, Murray CW. Protein-ligand docking against non-native protein conformers. J. Chem. Inf. Model. 2008;48:2214–2225. doi: 10.1021/ci8002254. [DOI] [PubMed] [Google Scholar]
- 21.Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J. Mol. Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 22.Totrov M, Abagyan R. Detailed ab initio prediction of lysozyme-antibody complex with 1.6 A accuracy. Nat. Struct. Biol. 1994;1:259–263. doi: 10.1038/nsb0494-259. [DOI] [PubMed] [Google Scholar]
- 23.Abagyan R, Kufareva I. The flexible pocketome engine for structural chemogenomics. Methods. Mol. Biol. 2009;575:249–279. doi: 10.1007/978-1-60761-274-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An J, Totrov M, Abagyan R. Pocketome via comprehensive identification and classification of ligand binding envelopes. Mol. Cell. Proteomics. 2005;4:752–761. doi: 10.1074/mcp.M400159-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Kleywegt GJ, Harris MR, Zou JY, Taylor TC, Wahlby A, Jones TA. The Uppsala Electron-Density Server. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2240–2249. doi: 10.1107/S0907444904013253. [DOI] [PubMed] [Google Scholar]
- 27.Cole J, Murray CW, Willem J, Nissink M, Taylor RD, Taylor R. Comparing protein-ligand docking programs is difficult. Proteins Struct. Funct. Bioinformat. 2005;60:325–332. doi: 10.1002/prot.20497. [DOI] [PubMed] [Google Scholar]
- 28.Jain AN. Bias, reporting, and sharing: computational evaluations of docking methods. J. Comput.-Aided Mol. Des. 2008;22:201–212. doi: 10.1007/s10822-007-9151-x. [DOI] [PubMed] [Google Scholar]
- 29.Hartshorn MJ, Verdonk ML, Chessari G, Brewerton SC, Mooij WT, Mortenson PN, Murray CW. Diverse, high-quality test set for the validation of protein-ligand docking performance. J. Med. Chem. 2007;50:726–741. doi: 10.1021/jm061277y. [DOI] [PubMed] [Google Scholar]
- 30.Abagyan R, Orry A, Raush E, Budagyan L, Totrov M. ICM Manual 3.5. Molsoft LCC; La Jolla, CA: 2007. [Google Scholar]
- 31.Halgren TA. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996;17:490–519. [Google Scholar]
- 32.Nemethy G, Gibson KD, Palmer KA, Yoon CN, Paterlini G, Zagari A, Rumsey S, Scheraga HA. Energy parameters in polypeptides. 10. Improved geometrical parameters and nonbonded interactions for use in the ECEPP(SLASH)3 algorithm, with application to proline-containing peptides. J. Chem. Phys. 1992;96:6472–6484. [Google Scholar]
- 33.Totrov M, Abagyan R. Derivation of sensitive discrimination potential for virtual ligand screening. In: Istrail S, Pevzner P, Waterman M, editors. RECOMB'99: Proceedings of the third annual international conference on computational molecular biology, France, 1999; Association for Computer Machinery: France. 1999. [Google Scholar]
- 34.Totrov M, Abagyan R. Protein-ligand docking as an energy optimization problem. In: Sons JW, Raffa R, editors. Drug-receptor Thermodynamics: Introduction and experimental applications. New York: 2001. [Google Scholar]
- 35.Teramoto R, Fukunishi H. Supervised consensus scoring for docking and virtual screening. J. Chem. Inf. Model. 2007;47:526–534. doi: 10.1021/ci6004993. [DOI] [PubMed] [Google Scholar]
- 36.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGovern SL, Shoichet BK. Information decay in molecular docking screens against holo, apo, and modeled conformations of enzymes. J. Med. Chem. 2003;46:2895–2907. doi: 10.1021/jm0300330. [DOI] [PubMed] [Google Scholar]
- 38.Rueda M, Bottegoni G, Abagyan R. Consistent improvement of cross-docking results using binding site ensembles generated with elastic network normal modes. J. Chem. Inf. Model. 2009;49:716–725. doi: 10.1021/ci8003732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popov VM, Yee WA, Anderson AC. Towards in silico lead optimization: scores from ensembles of protein/ligand conformations reliably correlate with biological activity. Proteins. 2007;66:375–387. doi: 10.1002/prot.21201. [DOI] [PubMed] [Google Scholar]
- 40.Pirard B. Structure-based chemogenomics: analysis of protein family landscapes. Methods Mol. Biol. 2009;575:281–296. doi: 10.1007/978-1-60761-274-2_12. [DOI] [PubMed] [Google Scholar]
- 41.Bursulaya BD, Totrov M, Abagyan R, Brooks CL., 3rd Comparative study of several algorithms for flexible ligand docking. J. Comput.-Aided Mol. Des. 2003;17:755–763. doi: 10.1023/b:jcam.0000017496.76572.6f. [DOI] [PubMed] [Google Scholar]
- 42.Michino M, Abola E, Brooks CL, 3rd, Dixon JS, Moult J, Stevens RC. Community-wide assessment of GPCR structure modelling and ligand docking: GPCR Dock 2008. Nat. Rev. Drug Discov. 2009;8:455–463. doi: 10.1038/nrd2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kufareva I, Abagyan R. Type-II kinase inhibitor docking, screening, and profiling using modified structures of active kinase states. J. Med. Chem. 2008;51:7921–7932. doi: 10.1021/jm8010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Withers IM, Mazanetz MP, Wang H, Fischer PM, Laughton CA. Active site pressurization: a new tool for structure-guided drug design and other studies of protein flexibility. J. Chem. Inf. Model. 2008;48:1448–1454. doi: 10.1021/ci7004725. [DOI] [PubMed] [Google Scholar]
- 45.Schneider G, Baringhaus K-H. Molecular Design. Concepts and Applications. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim, Germany: 2008. Creating the design. Ligand binding sites. pp. 104–105. [Google Scholar]
- 46.Davis AM, Teague SJ, Kleywegt GJ. Application and limitations of X-ray crystallographic data in structure-based ligand and drug design. Angew. Chem. Int. Ed. Engl. 2003;42:2718–2736. doi: 10.1002/anie.200200539. [DOI] [PubMed] [Google Scholar]
- 47.Hassell AM, An G, Bledsoe RK, Bynum JM, Carter HL, 3rd, Deng SJ, Gampe RT, Grisard TE, Madauss KP, Nolte RT, Rocque WJ, Wang L, Weaver KL, Williams SP, Wisely GB, Xu R, Shewchuk LM. Crystallization of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2007;63:72–79. doi: 10.1107/S0907444906047020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cavassotto C, Kovacs J, Abagyan R. Representing receptor flexibility in ligand docking through relevant normal modes. J. Am. Chem. Soc. 2005;127:9632–9640. doi: 10.1021/ja042260c. [DOI] [PubMed] [Google Scholar]
- 49.Amaro RE, Baron R, McCammon JA. An improved relaxed complex scheme for receptor flexibility in computer-aided drug design. J. Comput.-Aided Mol. Des. 2008;22:693–705. doi: 10.1007/s10822-007-9159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.