Abstract
Neutrophil recruitment is pivotal to host defense against microbial infection, but also contributes to the immunopathology of disease. We investigated the mechanism of neutrophil recruitment in human infectious disease by bioinformatic pathways analysis of the gene expression profiles in the skin lesions of leprosy. In erythema nodosum leprosum (ENL), which occurs in patients with lepromatous leprosy (L-lep), and is characterized by neutrophil infiltration in lesions, the most overrepresented biologic functional group was “cell movement” including E-selectin, which was coordinately regulated with IL-1β. In vitro activation of TLR2, upregulated in ENL lesions, triggered induction of IL-1β, which together with IFN-γ, induced E-selectin expression on, and neutrophil adhesion to endothelial cells. Thalidomide, an effective treatment for ENL, inhibited this neutrophil recruitment pathway. The gene expression profile of ENL lesions comprised an integrated pathway of TLR2/FcR activation, neutrophil migration and inflammation, providing insight into mechanisms of neutrophil recruitment in human infectious disease.
Keywords: Leprosy, Mycobacterium leprae, neutrophil, immune complexes, Toll-like receptors
Introduction
The recruitment of neutrophils to the site of infection is required for host defense against many bacterial infections (1–3). The recruitment of neutrophils can also result in tissue injury. Neutrophil-mediated tissue injury occurs in immune complex diseases in which the chronic deposition of immune complexes in tissues results in inflammation. These infiltrating neutrophils are thought to release toxic oxygen intermediates and proteases leading to local tissue damage (4).
The mechanisms of neutrophil recruitment to the site of infection in humans can be investigated using leprosy as a model. The disease forms a spectrum of clinical manifestations that correlate with the immune response to the pathogen, Mycobacterium leprae. This spectrum is dynamic, with patients developing immune reactions. In particular, in patients with the disseminated form, lepromatous leprosy (L-lep), a reaction known as erythema nodosum leprosum (ENL) (5;6) is frequent, observed in up to 50% of L-lep patients receiving antimicrobial therapy (7). ENL is characterized by the eruption of erythematous painful nodules and other systemic manifestations of tissue injury (8). Histologically, neutrophils are the signature cell in ENL lesions, with the presence of granulomas similar to those in L-lep lesions (9). Immune complex deposition is thought to contribute to the pathogenesis of ENL, evidenced by granular deposits of immunoglobulin and complement in a perivascular (10) and extravascular distribution (11), detection of immune complexes in vessel walls and evidence of damaged endothelial cells (6). Given that a key clinical difference between ENL and L-lep is the characteristic infiltration of neutrophils in ENL lesions, a major goal of this study was to investigate the mechanism(s) of neutrophil recruitment at the site of disease.
Methods
Clinical Specimens
The acquisition of all specimens was approved by the committees on investigations involving human subjects of the University of California, Los Angeles. For all procedures, informed consent was obtained. Clinical classification of patients with symptomatic Mycobacterium leprae infection was performed according to the criteria of Ridley and Jopling (12). Skin biopsies were embedded in optimal cutting tissue compound (Sakura Finetek USA Inc.) and snap frozen in liquid nitrogen.
Microarray analysis
RNA was isolated from skin biopsy specimens and used for microarray analysis as previously described (13) with the following modifications. mRNA was further purified using the Rneasy MicroKit (Qiagen). To generate cRNA probes, 100 ng of total RNA was doubly amplified using the RiboAmp RNA amplification kit (Arcturus Bioscience Inc.). The probes were purified and fragmented, and hybridized to the Affymetrix U133A plus 2.0 Genechip (Affymetrix). The raw gene expression data for the leprosy patients analyzed in this study will be available online through the NCBI GEO website.
Gene expression analyses
Hierarchical clustering analysis (14) using 3,158 probe sets with significant variation across the samples (standard deviation (σ) > 5000 and a coefficient of variation (σ/mean) > 0.3) was performed. The probe sets used were also required to have Affymetrix present calls in at least half of the experiments. Cluster diagrams were generated using the Cluster and TreeView software programs http://rana.lbl.gov/. Permutation analysis was used to determine the statistical significance of our results. We systematically considered 1000 random patient groupings and determined the frequency at which a grouping yielded a result equal to or better than that of the defined ENL/L-lep grouping as described (15). For gene expression based prediction we used the weighted gene voting algorithm of Golub et al. (16) with minor modifications as previously described (15).
To identify the most informative set of differentially expressed genes between the two leprosy subclasses, each gene was ranked by the probability that the means of its expression values are statistically distinct between the ENL and L-lep patients using the Student's t-test. The fold change (FC) in mean expression of each gene was also calculated between the two groups and used as a secondary ranking. We focused our attention on genes that met our designated criteria: p ≤ 0.05 and FC ≥ 2 in the Ingenuity Pathways Analysis.
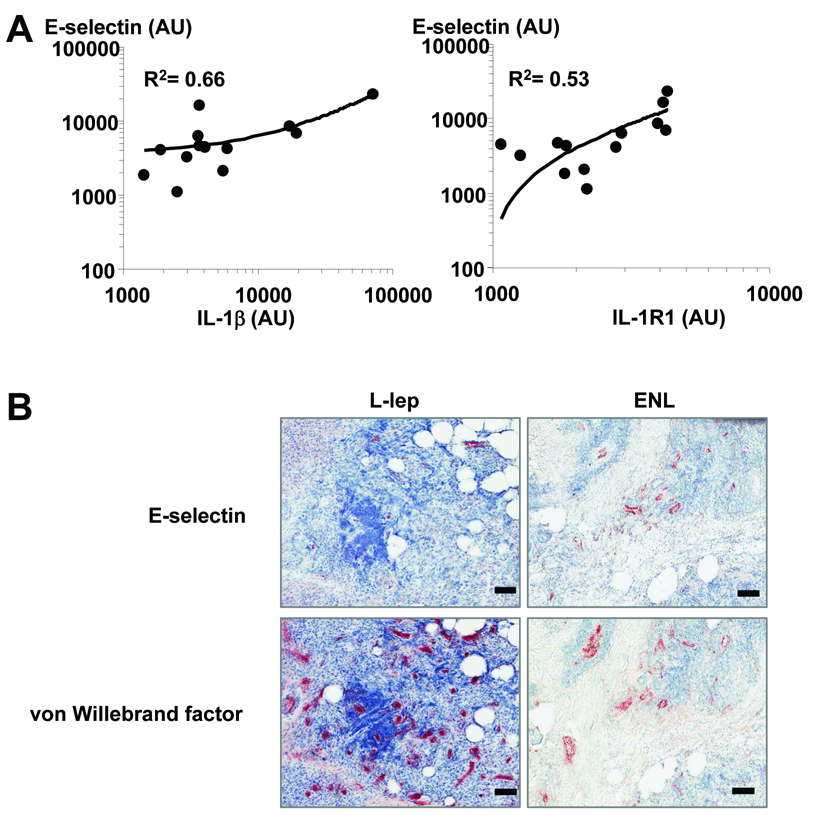
The Pearson's correlation between the expression of E-selectin and the expression of all other genes on the microarray from all samples was calculated. A threshold of R2 = 0.5 was used to obtain a list of genes that have a linear relationship in expression with E-selectin.
Functional groups and canonical pathways analyses
The functional groups and canonical pathways analyses were generated through the use of Ingenuity Pathways Analysis (Ingenuity® Systems, version 6.0, www.ingenuity.com). Probe sets comparatively increased in expression in ENL vs. L-lep that met p-value < 0.05 and FC > 2.0 were included in the analysis. Fischer’s exact test was used to calculate a p-value determining the probability that each biological function is due to chance alone.
Immunohistochemical and flow cytometry studies
Immunoperoxidase-labeling of cryostat sections was performed as described (17). Primary mAbs used were: C126C10B7 (anti-E-selectin, Biosource) and F8/86 (anti-vonWillebrand Factor, DakoCytomation). Antibodies used in flow cytometry include: 68-5H11 (anti-E-selectin, BD Pharmingen), MEM-111 (anti- ICAM-1, Caltag).
Stimulation of peripheral blood monocytes and endothelial cells
Peripheral blood mononuclear cells were purified by Ficoll-Hypaque (Pharmacia Biotech AB) gradient centrifugation followed by percoll (Amersham Biosciences) gradient separation to obtain monocytes (purity 90%). Cells were stimulated in triplicate with plate-bound human IgG (Equitech-Bio, Inc.) or the TLR2/1 agonist, a lipopeptide derived from the M. leprae 19-kDa antigen. IL-1β and IL-8 were measured by ELISA from cultured supernatant after overnight stimulation. Human umbilical vein endothelial cells (HUVECs) (Cambrex BioScience) at passage 3–6 were used for all experiments. Human umbilical vein endothelial cells were stimulated overnight with IL-1β (1 ng/ml) and IFN-γ (10 ng/ml). Cells were analyzed by flow cytometry or used in neutrophil binding assays. Thalidomide enantiomers (Sigma) in DMSO (Sigma) were added at 50 µg/ml. Statistical analysis was performed using the paired Student’s t test.
Neutrophil binding assays
Neutrophils were prepared as previously described (18). Neutrophil binding experiments were performed using the Cytoselect leukocyte-endothelium adhesion assay (Cell Biolabs, Inc.) according to manufacturer’s instructions. Relative fluorescence was measured using a Flexstation II (Molecular Devices). Statistical analysis was performed using the paired Student’s t-test.
Results
Differential gene expression profiles comparing ENL to L-lep
The gene expression profiles of the skin lesions of ENL vs. L-lep patients were investigated using Affymetrix microarrays, to identify the gene families and/or pathways involved in disease pathogenesis. Skin biopsy specimens were obtained from six ENL and seven L-lep patients. Hierarchical clustering analysis of the gene expression data separated the samples into two distinct groups that are consistent with their clinical and histopathological diagnosis (Figure 1A). Therefore, the lesions from patients with ENL vs. L-lep have distinct gene expression profiles.
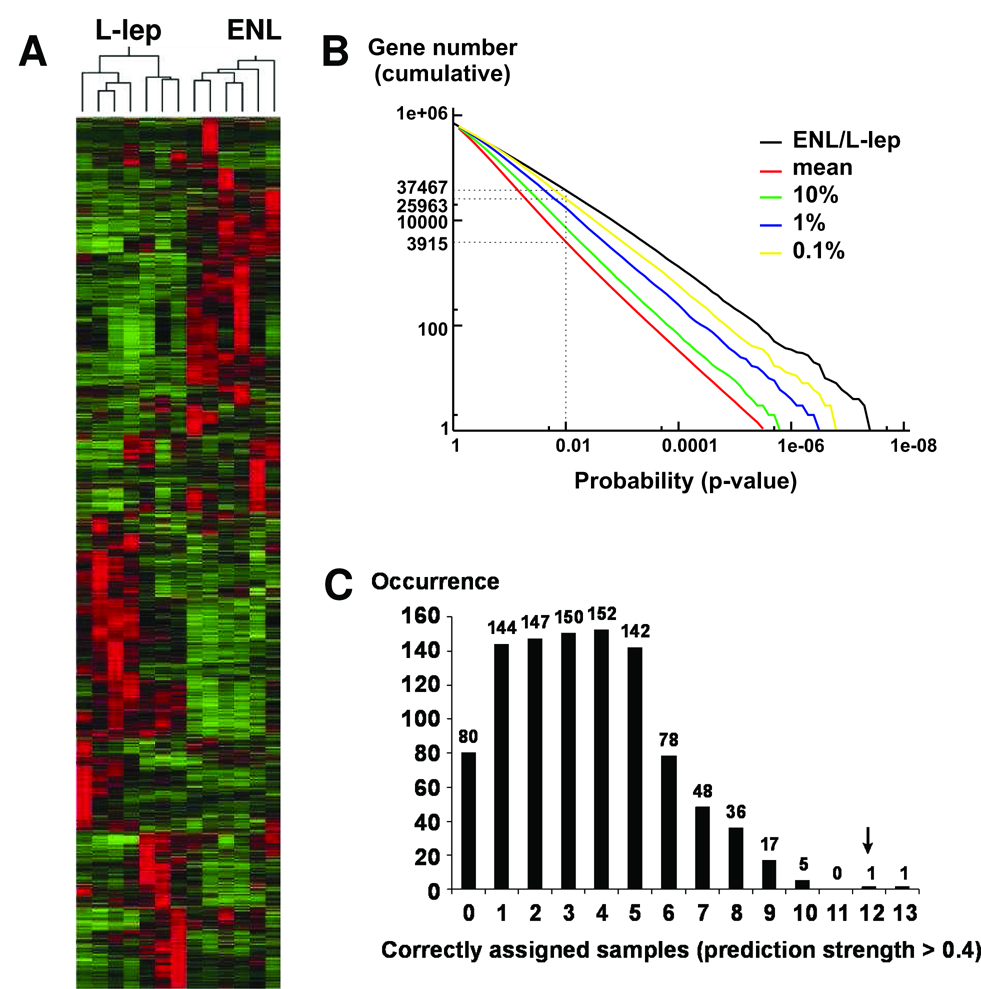
Figure 1.
(A) Unsupervised data analysis separates leprosy lesion samples into clinically relevant subclasses based on their gene expression patterns. Hierarchical clustering analysis divides 7 L-lep and 6 ENL skin lesion samples into two distinct groups that cluster on separate branches of a dendrogram. There are 3158 probe sets represented in this diagram. (B) Permutation analysis of the microarray data reveals that only less than 0.1% of the permutated groupings manifest more distinction in gene expression than the defined ENL / L-lep patient grouping. The cumulative numbers of probests (Y-axis) with Student’s t-test p-values less than various threshold levels (X-axis) were calculated for the clinically relevant ENL / L-lep grouping and plotted (black). One thousand randomly permutated groupings were also generated and tested. We plotted the mean (red), 10% (green), 1% (blue), and 0.1% (yellow) number of probesets below a given p-value among the permutated groupings and compared these to the correct ENL / L-lep grouping. Compared to the 0.1% confidence level, the ENL / L-lep grouping generally has more differentially expressed probesets with p-values below the indicated threshold, indicating that the ENL / L-lep grouping is statistically significant. (C) Prediction accuracy using leave-one-out cross-validation and weighted gene-voting. Using the ENL / L-lep grouping, our prediction algorithm correctly assigned the subclasses of 12 out of 13 samples with high confidence (prediction strength >0.4).
To determine whether the observed differences in gene expression between the ENL and L-lep patients were statistically significant, a permutation analysis was performed in which the number of probesets below each p-value threshold in the clinically relevant ENL or L-lep grouping was compared to 1000 random groupings (permutated groups). This analysis showed that fewer than 0.1% of the permutated groupings displayed more differential expression than the correct ENL–L-lep grouping (Figure 1B). The cumulative number of probesets (y-axis) with Student’s t-test p-values less than various threshold levels (x-axis) was calculated for the actual clinically diagnosed ENL– L-lep grouping and plotted. For example, at p = 0.01 the ENL– L-lep grouping had many more differentially expressed probesets (37,467) than both the mean (3,915) and the top 0.1% (25,963) of the permutated groupings. This established that, despite the relatively small number of samples examined in this study, and the cellular heterogeneity of the specimens, the differences in gene expression observed between ENL and L-lep patients are not likely to have resulted by chance.
To evaluate whether gene expression profiles were sufficiently robust to correctly assign the subclasses of unknown samples, leave-one-out cross validation analysis was performed (19). Using the clinically defined ENL vs. L-lep patient grouping, the algorithm predicted the classes of all 13 samples correctly [100% accuracy; 12 out of 13 had high confidence (prediction strength > 0.4)]. This result is statistically significant, because only 0.1% (1/1000) of the random patient groupings performed better than the clinically defined ENL–L-lep grouping at classifying the withheld sample (Figure 1C). Taken together, the results from both unsupervised and supervised prediction algorithms demonstrate that ENL and L-lep lesions have distinct gene expression profiles.
Differentially expressed functional groups in ENL vs. L-lep skin lesions
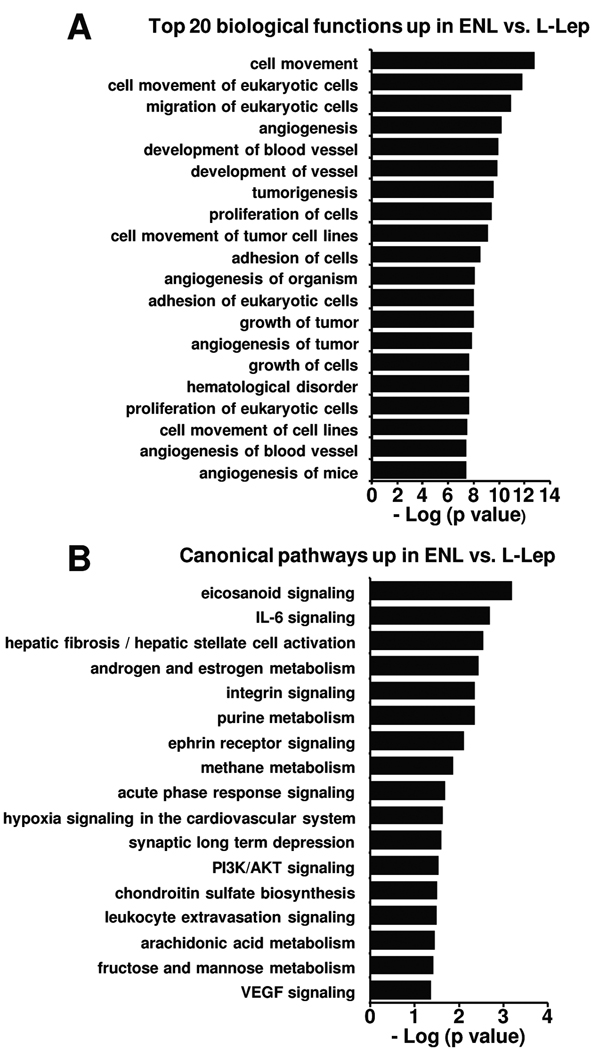
To detect which gene sets or biological pathways are overrepresented in ENL vs. L-lep that might be particularly relevant to disease pathogenesis, we compared the gene expression profile of ENL vs. L-lep skin lesions using knowledge-guided bioinformatic analysis, incorporating data on likely biologic functions, including gene ontology information and regulatory data (Ingenuity® Systems, www.ingenuity.com). This analysis identified 57 functional groups (p < 0.05) and 17 canonical pathways (p < 0.05) (Figure 2). The striking finding was the top functional pathway identified in ENL vs. L-lep lesions was “cell movement” composed of 188 genes (p = 1.98E-13, Figure 2A), including genes that are involved in recruitment of neutrophils, the “signature” cell in ENL lesions. Functional pathway analysis using DAVID (20) yielded consistent results as the top pathways all involved “cell adhesion” (data not shown). In addition, the top canonical pathway identified was “eicosanoid signaling” (Figure 2B). Using the Benjamini-Hochberg method correcting for multiple testing yields similar functional groups (not shown), however this highly stringent method leads to no statistically significant canonical pathways. Since the eicosanoid signaling pathway gene members were comparatively increased in expression, including both the enzymes for ligands and their corresponding receptors, (Figure 7) in ENL vs. L-lep lesions, we decided to use a less stringent approach that was more biologically relevant.
Figure 2.
Enriched pathways in ENL vs. L-lep. The functional and canonical pathways analyses were generated through the use of Ingenuity Pathways Analysis (www.ingenuity.com). (A) The top 20 biological functions in ENL vs. L-lep are ranked by their p-values (x-axis). (B) All statistically significant canonical pathways represented by the differentially expressed genes are shown ranked by their p-values (x-axis). Fischer’s exact test was used to calculate a p-value determining the probability that each pathway represented by the expression data is due to chance alone.
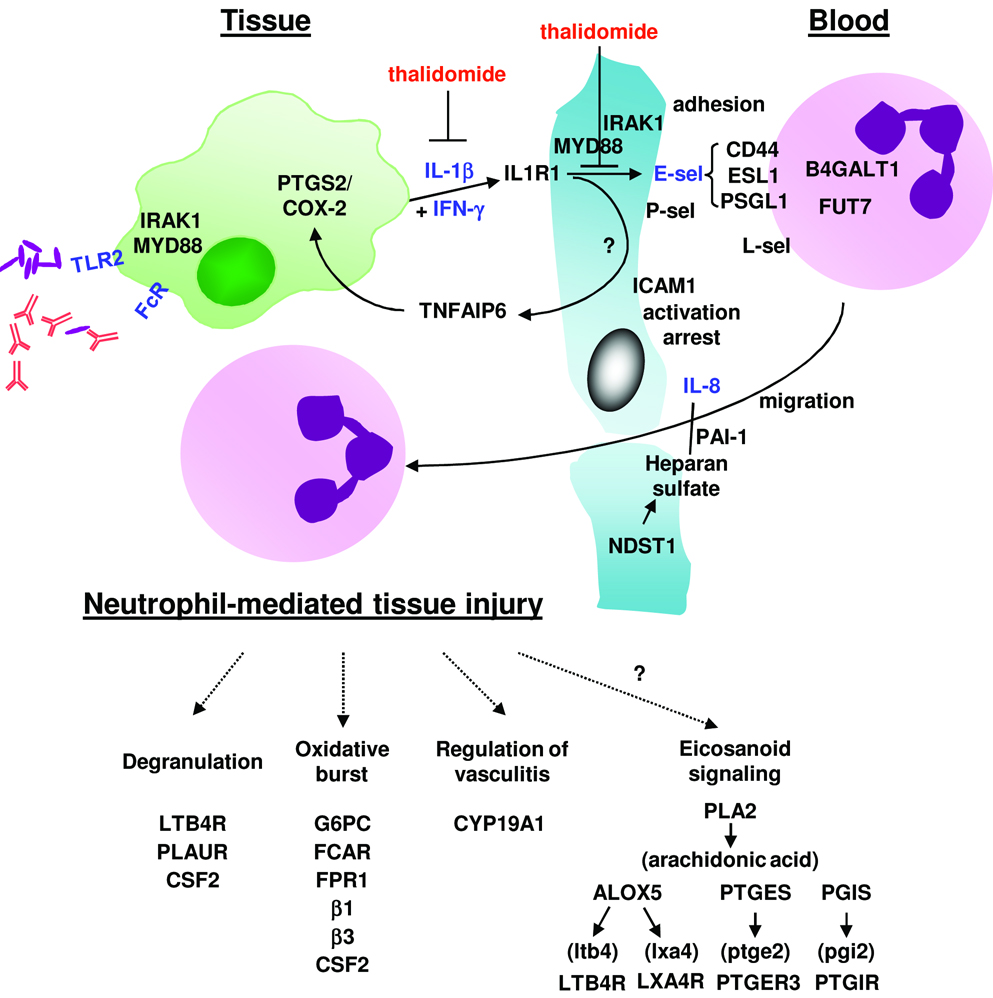
Figure 7.
A gene program of neutrophil recruitment and tissue injury based on gene expression and in vitro data. Molecules in black text represent those added to the pathway based on microarray data, while molecules/genes in blue text have supportive inmmunohistochemical and/or in vitro data. Immune complexes and/or mycobacterial components may activate FcRs and TLRs to induce pro-inflammatory cytokines such as IL-8 and IL-1β. IL-1β in combination with IFN-γ induce E-selectin expression on endothelial cells, resulting in the first step of neutrophil recruitment, adhesion and rolling along the endothelial wall. These steps are inhibited by thalidomide. Neutrophil interaction with ICAM-1 as well as other integrin-mediated activators results in arrest and activation. Glycosaminoglycans such as heparan sulfate and extracellular proteins such as plasminogen activator inhibitor 1 promote the presentation of chemokines such as IL-8 on the surface of the endothelium to mediate neutrophil activation and chemotaxis, inhibited by secretory component. As part of the innate immune response to microbial infection, the local recruitment of neutrophils may lead to tissue injury as host defense pathways are executed.
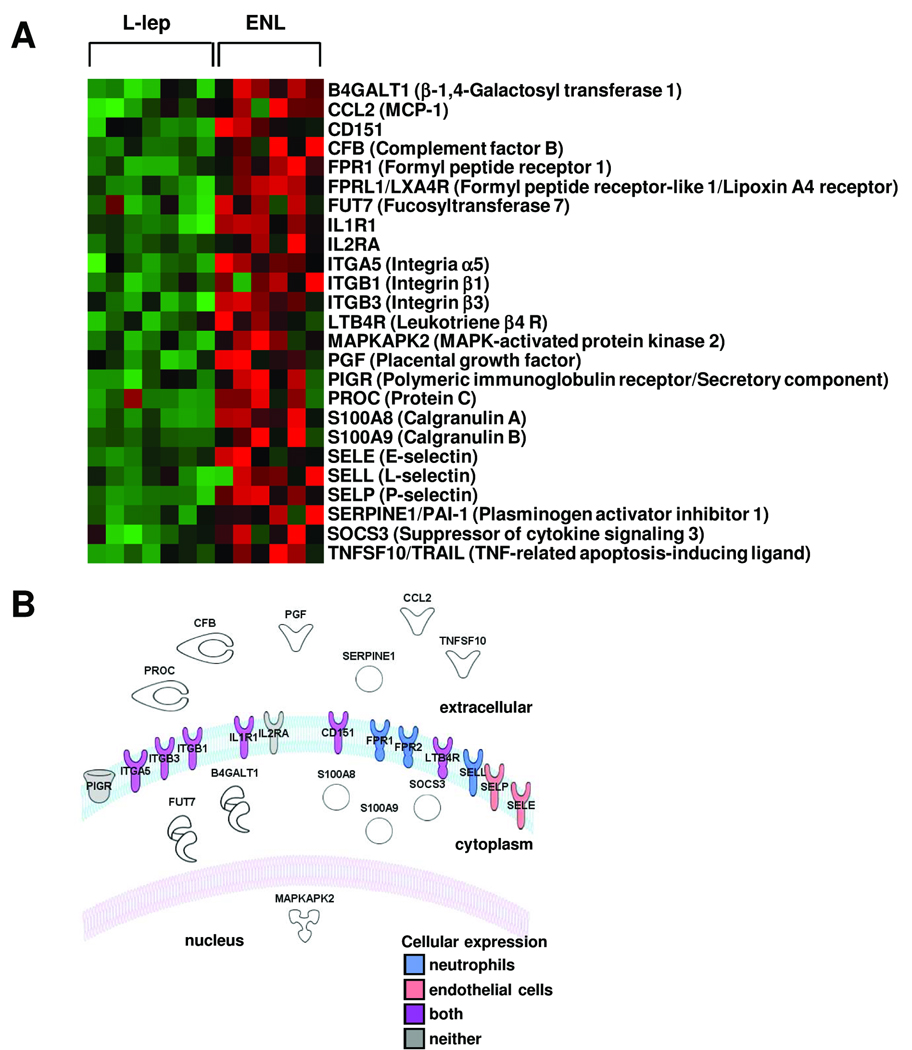
The “cell movement” list of genes that was differentially expressed in ENL lesions was further investigated by limiting the gene set to those functional groups involved in neutrophil recruitment identifying 25 genes (Figure 3A). These genes were also characterized according to cellular location based on the ingenuity pathways knowledge base (Figure 3B). Examination of the cell surface genes involved in neutrophil recruitment, revealed the differential upregulation in ENL lesions of the selectin family of adhesion molecules including E-selectin (FC 3.5, p < 0.05) and P-selectin (FC 2.8, p<0.001). E-selectin is a key molecule involved in neutrophil binding to endothelium, an initial step in neutrophil recruitment (21;22), also contributing to recruitment of other leukocytes. Analysis revealed that the E-selectin ligands, E-selectin ligand 1 (ESL1, FC 2.8, p<0.001), CD44 (FC 1.7, p<0.05) and P-selectin glycoprotein ligand 1 (PSGL-1, FC 1.7, p<0.05), were also expressed at higher levels in ENL, although for two molecules there was less than two-fold change. In summary, the bioinformatics analysis of ENL lesions according to biologic pathways revealed the upregulation of “cell movement” genes including E-selectin and its ligands, key molecules that mediate neutrophil binding to endothelial cells.
Figure 3.
(A) Genes within the top functional group represented in ENL skin lesions, cell movement, also belonging to a subset involved with neutrophil function. Each row represents the gene expression profile of a particular probe corresponding to the gene listed, individual squares represent the relative gene expression intensity of the given gene in a patient with red being relatively increased in expression and green relatively decreased in expression. (B) Diagram illustrating the cellular location of each of the genes in (A). Cell type-specific expression (neutrophil-blue, endothelial cell-pink, both-purple or neither-gray) is shown for those proteins whose cellular location is in the plasma membrane.
Regulation of E-selectin expression
To identify the inflammatory mediators that contribute to E-selectin expression in ENL lesions, we queried which genes were coordinately regulated with E-selectin expression in individual lesions. From the approximately 3000 genes identified to be coordinately regulated with E-selectin, 49 genes were identified as part of the “cell movement” biologic function related to neutrophil recruitment. Five genes were identified by pathway analysis to have a first level interaction with E-selectin, of which only IL-1β upregulates E-selectin (R2 = 0.65). In addition, the expression of IL1R1 correlated with E-selectin expression (R2 =0.54, Figure 4A). Although IL-1β was five-fold increased in ENL vs. L-lep lesions, individual variability was high and therefore the direct comparison between lesions did not achieve statistical significance. However relative increases in IL-1R1 in ENL lesions did achieve significance (two different probe sets, FC 1.6 and 2.2, p<0.001). In addition to increased expression of IL1R1 in ENL vs. L-lep lesions, other IL1R1 family members and signaling molecules were also increased such as Toll-like receptor 2 (TLR2, FC 1.9, p<0.05), myeloid differentiation response gene 88 (MYD88, FC 1.5, p<0.05), and interleukin 1 receptor-associated kinase 1 (IRAK1, FC 1.9, p<0.01), though these molecules had less than two-fold change. These data identify IL-1β and TLR2 as potential regulators of E-selectin expression on endothelial cells and subsequent neutrophil binding.
Figure 4.
(A) Correlation of E-selectin expression with IL-1β and IL-1R1. Shown are the R2 values (Pearson's correlation) between the expression of E-selectin and the expression of IL-1β and IL-1R1. (B) E-selectin and von Willebrand factor protein expression in L-lep compared to ENL skin lesions in vivo. Thin (4 µm) sections of leprosy biopsy samples were incubated with anti-E-selectin or VWF and stained secondarily with an immunoperoxidase method followed by counterstaining with hematoxylin. Each bar denotes 100 µm. The isotype controls were negative. The findings shown are representative of five patients in each group.
E-selectin expression in ENL versus L-lep skin lesions
To examine whether E-selectin protein expression reflects the differential expression of mRNA in ENL vs. L-lep, immunohistochemistry was performed on skin biopsy specimens from five ENL and five L-lep patients (Figure 4B). E-selectin was expressed in a vascular pattern and at higher levels in ENL skin lesions (20–80% of vessels) than in L-lep (0–20% of vessels). The low or absent expression of E-selectin in L-lep skin lesions was not due to a decreased number of blood vessels, as serial sections were also labeled for an endothelial cell marker, von Willebrand factor (Figure 4B).
The effect of thalidomide on IL-1β production, IL-1β–induced E-selectin expression and neutrophil binding
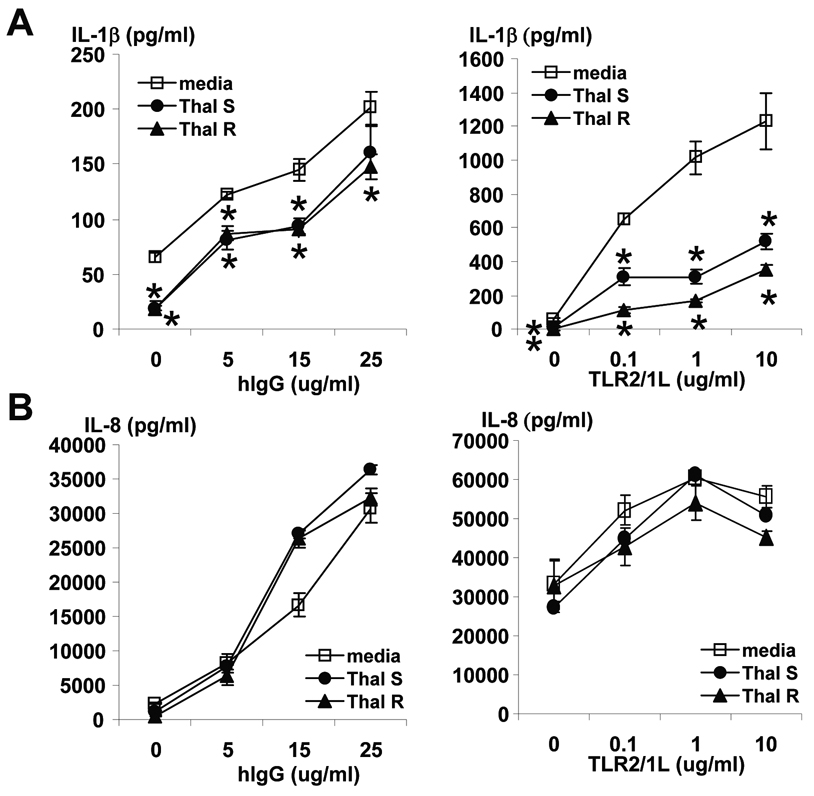
A characteristic feature of ENL is the dramatic clinical response to treatment with thalidomide, accompanied by a prompt reduction in the number of neutrophils in lesions (23;24). Given that thalidomide has been shown to inhibit induction of proinflammatory cytokines (25–27), we hypothesized that thalidomide might also inhibit the ability of immune complexes to trigger IL-1β gene expression from monocytes. In these experiments, immune complex induction of IL-1β was modeled by using immobilized or plate bound human IgG (hIgG) to activate Fc receptors, triggering monocytes to release IL-1β (Figure 5A). The racemic enantiomers of thalidomide were tested separately for their effect on IL-1β production, as it has been suggested that the (−)-(S) and (+)-(R) forms may have different effects (28). Both enantiomers diminished the ability of plate-bound hIgG to induce IL-1β by approximately 20–40% (Figure 5A).
Figure 5.
Effect of FcR or TLR2/1 activation of peripheral blood monocytes on IL-1β (A) and IL-8 (B) secretion. Percolled monocytes from healthy donors were stimulated overnight with a lipopetide known to stimulate TLR2/1 or plate-bound hIgG in media (open squares), thalidomide R (filled triangles) or thalidomide S (filled circles) enantiomers. IL-1β and IL-8 were measured from cultured supernatant by ELISA. Shown are averages of triplicate cultures +/− SEM. Asterisks denote differences between media and thalidomide-treated cells with p<0.05 calculated by paired Student’s t-test.
Given that TLR2 was also noted to be comparatively increased in expression in ENL vs. L-lep lesions, we also tested the ability of thalidomide to inhibit TLR2 induction of IL-1β (Figure 5A). A lipopeptide derived from M. leprae was used as the TLR2 ligand (29). Thalidomide diminished TLR2-induced monocyte production of IL-1β by ~50–80%, while production of IL-8 was not affected, consistent with Oliveira and colleagues (30) (Figure 5B). Thus FcR- or TLR-induced monocytes’ production of IL-1β is partially inhibited by thalidomide.
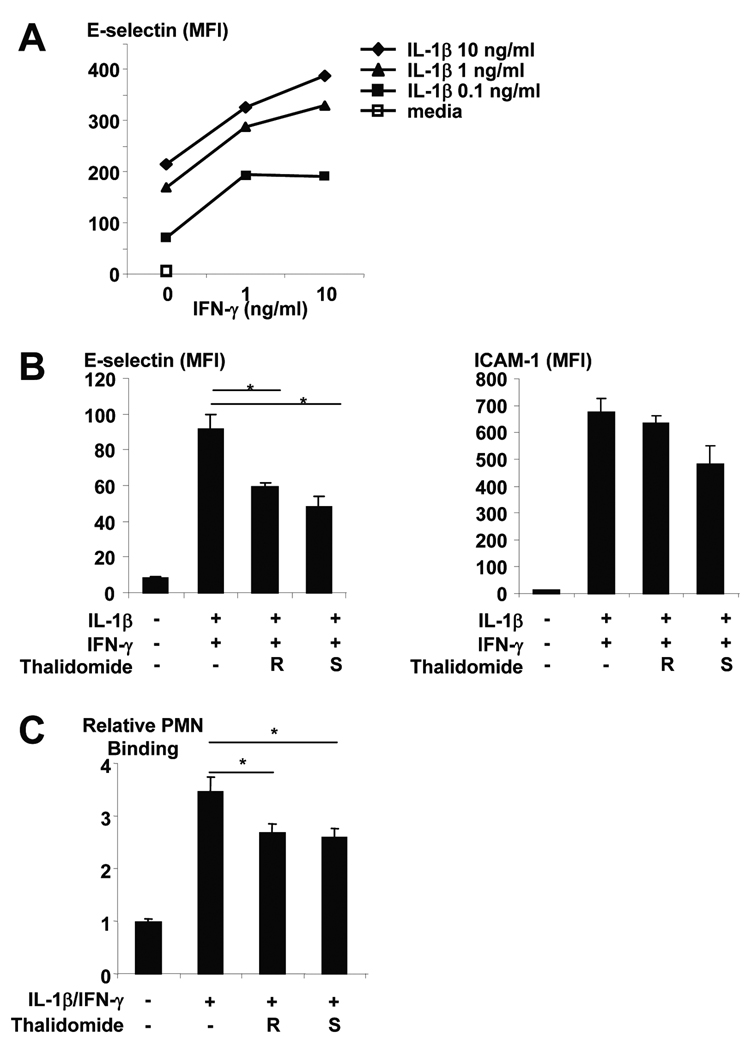
Regulation of E-selectin expression on endothelial cells is known to be mediated by various cytokines including IL-1β, IL-6, TNF-α, and IFN–γ (31). Furthermore, IL-1 acts directly on cultured HUVECs (32) rather than on leukocytes (33) to increase the adhesiveness of the endothelial cell surface for neutrophils. HUVECs cultured overnight with IL-1β showed a dose-responsive induction of E-selectin expression (Figure 6A). Since subcutaneous injections of IFN-γ in L-lep patients have been reported to induce ENL (34) and IFN–γ is known to enhance the effects of IL-1β on E-selectin expression (31) in a dose-responsive manner (Figure 6A), we tested the ability of thalidomide to inhibit the induction of E-selectin by IL-1β and IFN–γ (Figure 6B). IFN–γ alone did not induce E-selectin expression (MFI 14.3 for untreated, 15.6 with highest dose of IFN-γ). Both enantiomers of thalidomide inhibited the ability of IL-1β and IFN–γ to induce E-selectin expression by 35–47% while induction of ICAM-1 (Figure 6B) and cell viability were not affected (not shown). P-selectin expression was not induced by IL1-β and IFN–γ (P-selectin MFI on HUVECs: media-treated = 6.4, IL1-β and IFN–γ -treated = 5.4).
Figure 6.
(A) Effect of IFN-γ on IL-1β induction of E-selectin expression on HUVECs. (B) Effect of thalidomide on E-selectin and ICAM-1 expression of HUVECs induced by IL-1β and IFN-γ. (C) Neutrophil binding to HUVEC cells stimulated with IL-1β and IFN-γ in the presence or absence of thalidomide. Data shown are a compilation of 3 independent experiments. Statistical analysis was performed by paired Student’s t-test.
To test whether the effect of thalidomide on endothelial cell expression of E-selectin was functionally relevant, the effect of thalidomide on the ability of the HUVECs to bind neutrophils was examined (Figure 6C). While all cytokine-treated HUVECs bound more neutrophils than those treated with media alone, those treated with IL-1β and IFN–γ bound over three-fold more neutrophils than those cultured with media. Furthermore, the addition of thalidomide resulted in approximately 23–25% diminished neutrophil binding to HUVECs stimulated with IL-1β and IFN–γ. Together, these data suggest a novel mechanism of action for thalidomide in inhibiting IL-1β release, IL-1β induction of E-selectin and subsequent neutrophil binding to endothelial cells.
Discussion
To investigate the mechanism(s) of neutrophil recruitment and subsequent tissue injury, we determined the gene expression profile of the skin lesions of ENL, a reaction that occurs in patients with the L-lep form of leprosy. Using bioinformatics analysis of immune response pathways in combination with in situ analysis of lesions and in vitro functional experiments, an integrated pathway was identified. Major aspects of this pathway include: 1) FcR or TLR2 induction of IL-1β release, 2) endothelial activation including the upregulation of E-selectin and subsequent neutrophil binding; and, 3) the upregulation of inflammatory mediators associated with both neutrophils and monocyte/macrophages. Thalidomide, a highly effective agent used in the treatment for ENL and known to reduce neutrophil infiltration in lesions, targeted individual events of this inflammatory pathway. Together these data provide evidence that a local inflammatory mechanism linking IL-1β, E-selectin and neutrophil recruitment is part of a clinically relevant integrated pathway of tissue injury.
A key mechanism by which circulating neutrophils are recruited to disease sites involves the upregulation of E-selectin on endothelial cells, which mediates the initial adhesion and rolling required for neutrophil binding and tethering (21;35). Our data suggest macrophages may produce IL-1β that contributes to upregulated E-selectin expression in ENL lesions, given that IL-1β correlated with E-selectin expression in lesions. Importantly, we identified two distinct clinically relevant mechanisms of IL-1β induction in human disease. First, TLR2 activation of monocytes induced IL-1β release, which is relevant given that ENL develops during the initial phase of antimicrobial therapy in which bacterial breakdown is documented. Second, FcR activation of monocytes triggered IL-1β, consistent with the deposition of immune complexes containing bacterial breakdown products in lesions. The ability of IFN–γ to enhance the ability of IL-1β induction of E-selectin (31) and subsequent neutrophil binding is noteworthy, given the clinical observation that 60% of lepromatous leprosy patients who received subcutaneous injections of IFN–γ developed ENL (34). These observations suggest a role for both IL-1β and IFN–γ in the induction of E-selectin as part of the pathogenesis of ENL. While the neutrophil-recruiting leukotriene B4 synthetic pathway and its receptor were comparatively increased in ENL lesions (Figure 7), classic neutrophil chemokines (e.g. CXCL1-3, 5-8) were not.
The clinical relevance of the pathway FcR/TLR→IL-1β→E-selectin→neutrophil recruitment in the pathogenesis of tissue injury in leprosy was investigated in the context of the drug thalidomide, a particularly efficacious intervention in the management of ENL (36) and known to decrease neutrophil infiltration in disease lesions (24). Our data indicates that thalidomide can modulate neutrophil-mediated injury by diminishing FcR- and TLR2/1- induced IL-1β release, as well as moderately decreasing the ability of IL-1β and IFN-γ to induce E-selectin and subsequent neutrophil adherence to endothelial cells. Thalidomide was found to downregulate IL-1β in models of burn injury, pancreatitis, LPS-induced uveitis (26;37;38). The finding that thalidomide targets neutrophil recruitment is possibly relevant to the mechanism of action of thalidomide in the treatment of recurrent aphthous stomatitis and Behcet’s syndrome (39;40). The ability of thalidomide to target the FcR/TLR→IL-1β→E-selectin→neutrophil recruitment pathway complements studies indicating a role in inhibiting neutrophil and monocyte phagocytosis, neutrophil chemotaxis, TNF-α-induced adhesion molecules, as well as TNF-α and LPS-induced IL-1β production by monocytes (41–45). TNFα expression was not comparatively increased in ENL skin lesions (not shown). These findings warrant further serial studies of patient biopsy specimens to investigate the pathways altered by thalidomide at the site of disease.
Analyzing the gene expression profiles of disease lesions according to biologic pathways provided an unbiased method to identify mechanisms of pathogen-induced neutrophil accumulation and its inflammatory consequences. This contrasts with the ‘standard’ supervised approach of evaluating individual genes by key identifiers. Thus it was possible to integrate a large number of differentially expressed genes into biologically meaningful pathways. These genes along with those coordinately upregulated with E-selectin, were incorporated into a speculative model of neutrophil-mediated tissue injury including genes involved in neutrophil chemotaxis/migration/infiltration, degranulation, and oxidative burst (Figure 7). In addition, the results of the pathways analysis including the top functional groups and canonical pathways increased in ENL vs. L-lep indicate other inflammatory pathways that may also contribute to the pathogenesis of ENL. For example, the genes B4GALT1 and FUT7, part of the cell movement functional group, are involved in glycosylation of selectins or their ligands to mediate neutrophil recruitment (46;47). The second most represented canonical pathway, IL-6 signaling, leads to downstream genes aromatase and TNF alpha-induced protein 6, both comparatively increased in ENL vs. L-lep lesions and reported to contribute to tissue inflammation (48;49). Lastly, the top canonical pathway in ENL lesions, eicosanoid signaling, likely regulates tissue inflammation.
The model of immune complex-mediated tissue injury is relevant to the recruitment of neutrophils as part of the innate immune response to control bacterial infections suggested in animal models of infections (2;3;50). The neutrophilic abscess formed in the classical response to Staphylococcus aureus infection also requires IL-1 (2). Though neutrophil activation is important for host defense including the transfer of granules containing antimicrobial peptides to macrophages in mycobacterial infection (18), it also contributes to inflammation and tissue injury. The combination of bioinformatics pathways analysis with in situ and in vitro approaches provides new insight into disease pathogenesis and novel targets for disease intervention, in managing patients with immune complex-mediated diseases.
Acknowledgments
The authors are grateful to Dr. David Eisenberg for his insight and suggestions. The authors thank Ms. Neshat Nikoomanesh, the Ingenuity Pathways Analysis software support scientists (Drs. Julie Deschenes, Carmela Jaravata, Amy Mendelhall, Jyothi Paniyadi and Ms. Jaya Ramkumar), and the UCLA DNA Microarray and the UCLA Flow Cytometry core laboratories for technical assistance.
Financial support: This work was supported in part by NIH grants AI22553 and AI47868 (to R.L. Modlin); by NIH grant AR053104 and the Dermatology Foundation (to D.J. Lee) and the Heiser Program (to M. Tanaka).
Footnotes
Potential conflicts of interest: none.
Part of this work has been presented at the American Association of Immunologists Annual meeting in San Diego, CA in April 2008 and the Society for Investigative Dermatology Annual meeting in Montreal, Quebec, Canada in May 2009.
Reference List
- 1.Rogers HW, Unanue ER. Neutrophils are involved in acute, nonspecific resistance to Listeria monocytogenes in mice. Infect Immun. 1993 Dec;61(12):5090–5096. doi: 10.1128/iai.61.12.5090-5096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miller LS, O'Connell RM, Gutierrez MA, et al. MyD88 mediates neutrophil recruitment initiated by IL-1R but not TLR2 activation in immunity against Staphylococcus aureus. Immunity. 2006 Jan;24(1):79–91. doi: 10.1016/j.immuni.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Xu Q, Seemanapalli SV, Reif KE, Brown CR, Liang FT. Increasing the recruitment of neutrophils to the site of infection dramatically attenuates Borrelia burgdorferi infectivity. J Immunol. 2007 Apr 15;178(8):5109–5115. doi: 10.4049/jimmunol.178.8.5109. [DOI] [PubMed] [Google Scholar]
- 4.Movat HZ. Tissue injury and inflammation induced by immune complexes: the critical role of the neutrophil leukocyte. Exp Mol Pathol. 1979 Aug;31(1):201–210. doi: 10.1016/0014-4800(79)90021-2. [DOI] [PubMed] [Google Scholar]
- 5.Anthony J, Vaidya MC, Dasgupta A. Ultrastructure of skin in erythema nodosum leprosum. Cytobios. 1983;36(141):17–23. [PubMed] [Google Scholar]
- 6.Murphy GF, Sanchez NP, Flynn TC, Sanchez JL, Mihm MC, Jr, Soter NA. Erythema nodosum leprosum: nature and extent of the cutaneous microvascular alterations. J Am Acad Dermatol. 1986 Jan;14(1):59–69. doi: 10.1016/s0190-9622(86)70008-x. [DOI] [PubMed] [Google Scholar]
- 7.Waters MF, Rees RJ, Sutherland I. Chemotherapeutic trials in leprosy. 5. A study of methods used in clinical trials in lepromatous leprosy. Int J Lepr Other Mycobact Dis. 1967 Jul;35(3):311–335. [PubMed] [Google Scholar]
- 8.Vengadakrishnan K, Saraswat PK, Mathur PC. A study of rheumatological manifestations of leprosy. Indian J Dermatol Venereol Leprol. 2004 Mar;70(2):76–78. [PubMed] [Google Scholar]
- 9.Rea TH, Levan NE. Erythema nodosum leprosum in a general hospital. Arch Dermatol. 1975;111:1575–1580. [PubMed] [Google Scholar]
- 10.Wemambu SNC, Turk JL, Waters MFR, Rees RJW. Erythema nodosum leprosum: a clinical manifestation of the Arthus phenomenon. Lancet. 1969;2:933–935. doi: 10.1016/s0140-6736(69)90592-3. [DOI] [PubMed] [Google Scholar]
- 11.Ridley MJ, Ridley DS. The immunopathology of erythema nodosum leprosum: the role of extravascular complexes. Lepr Rev. 1983 Jun;54(2):95–107. doi: 10.5935/0305-7518.19830015. [DOI] [PubMed] [Google Scholar]
- 12.Ridley DS, Jopling WH. Classification of leprosy according to immunity. A five-group system. Int J Lepr. 1966;34:255–273. [PubMed] [Google Scholar]
- 13.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 14.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998 Dec 8;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bleharski JR, Li H, Meinken C, et al. Use of genetic profiling in leprosy to discriminate clinical forms of the disease. Science. 2003 Sep 12;301(5639):1527–1530. doi: 10.1126/science.1087785. [DOI] [PubMed] [Google Scholar]
- 16.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999 Oct 15;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 17.Modlin RL, Hofman FM, Horwitz DA, et al. In situ identification of cells in human leprosy granulomas with monoclonal antibodies to interleukin 2 and its receptor. J Immunol. 1984 Jun;132(6):3085–3090. [PubMed] [Google Scholar]
- 18.Tan BH, Meinken C, Bastian M, et al. Macrophages acquire neutrophil granules for antimicrobial activity against intracellular pathogens. J Immunol. 2006 Aug 1;177(3):1864–1871. doi: 10.4049/jimmunol.177.3.1864. [DOI] [PubMed] [Google Scholar]
- 19.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999 Oct 15;286(5439):531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 20.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):3. [PubMed] [Google Scholar]
- 21.Mulligan MS, Varani J, Dame MK, et al. Role of endothelial-leukocyte adhesion molecule 1 (ELAM-1) in neutrophil-mediated lung injury in rats. J Clin Invest. 1991 Oct;88(4):1396–1406. doi: 10.1172/JCI115446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrence MB, Springer TA. Neutrophils roll on E-selectin. J Immunol. 1993 Dec 1;151(11):6338–6346. [PubMed] [Google Scholar]
- 23.Sheskin J. Thalidomide in the treatment of lepra reactions. Clin Pharmacol Ther. 1965 May;6:303–306. doi: 10.1002/cpt196563303. [DOI] [PubMed] [Google Scholar]
- 24.Sampaio EP, Kaplan G, Miranda A, et al. The influence of thalidomide on the clinical and immunologic manifestation of erythema nodosum leprosum. J Infect Dis. 1993 Aug;168(2):408–414. doi: 10.1093/infdis/168.2.408. [DOI] [PubMed] [Google Scholar]
- 25.Moreira AL, Sampaio EP, Zmuidzinas A, Frindt P, Smith KA, Kaplan G. Thalidomide exerts its inhibitory action on tumor necrosis factor alpha by enhancing mRNA degradation. J Exp Med. 1993 Jun 1;177(6):1675–1680. doi: 10.1084/jem.177.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodrigues GB, Passos GF, Di GG, et al. Preventive and therapeutic anti-inflammatory effects of systemic and topical thalidomide on endotoxin-induced uveitis in rats. Exp Eye Res. 2007 Mar;84(3):553–560. doi: 10.1016/j.exer.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 27.van CR, Vonk AG, Netea MG, Kullberg BJ, van der Meer JW. Modulation of LPS- , PHA- and M. tuberculosis-mediated cytokine production by pentoxifylline and thalidomide. Eur Cytokine Netw. 2000 Dec;11(4):574–579. [PubMed] [Google Scholar]
- 28.Wnendt S, Finkam M, Winter W, Ossig J, Raabe G, Zwingenberger K. Enantioselective inhibition of TNF-alpha release by thalidomide and thalidomide-analogues. Chirality. 1996;8(5):390–396. doi: 10.1002/(SICI)1520-636X(1996)8:5<390::AID-CHIR6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 29.Krutzik SR, Ochoa MT, Sieling PA, et al. Activation and regulation of Toll-like receptors 2 and 1 in human leprosy. Nat Med. 2003 Apr 14;9(5):525–532. doi: 10.1038/nm864. [DOI] [PubMed] [Google Scholar]
- 30.Oliveira RB, Moraes MO, Oliveira EB, Sarno EN, Nery JA, Sampaio EP. Neutrophils isolated from leprosy patients release TNF-alpha and exhibit accelerated apoptosis in vitro. J Leukoc Biol. 1999 Mar;65(3):364–371. doi: 10.1002/jlb.65.3.364. [DOI] [PubMed] [Google Scholar]
- 31.Raab M, Daxecker H, Markovic S, Karimi A, Griesmacher A, Mueller MM. Variation of adhesion molecule expression on human umbilical vein endothelial cells upon multiple cytokine application. Clin Chim Acta. 2002 Jul;321(1–2):11–16. doi: 10.1016/s0009-8981(02)00048-7. [DOI] [PubMed] [Google Scholar]
- 32.Bevilacqua MP, Pober JS, Wheeler ME, Cotran RS, Gimbrone MA., Jr Interleukin-1 activation of vascular endothelium. Effects on procoagulant activity and leukocyte adhesion. Am J Pathol. 1985 Dec;121(3):394–403. [PMC free article] [PubMed] [Google Scholar]
- 33.Bevilacqua MP, Pober JS, Wheeler ME, Cotran RS, Gimbrone MA., Jr Interleukin 1 acts on cultured human vascular endothelium to increase the adhesion of polymorphonuclear leukocytes, monocytes, and related leukocyte cell lines. J Clin Invest. 1985 Nov;76(5):2003–2011. doi: 10.1172/JCI112200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sampaio EP, Moreira AL, Sarno EN, Malta AM, Kaplan G. Prolonged treatment with recombinant interferon gamma induces erythema nodosum leprosum in lepromatous leprosy patients. J Exp Med. 1992 Jun 1;175:1729–1737. doi: 10.1084/jem.175.6.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulligan MS, Watson SR, Fennie C, Ward PA. Protective effects of selectin chimeras in neutrophil-mediated lung injury. J Immunol. 1993 Dec 1;151(11):6410–6417. [PubMed] [Google Scholar]
- 36.Sheskin J, Convit J. Results of a double blind study of the influence of thalidomide on the lepra reaction. Int J Lepr. 1969;37:135–146. [PubMed] [Google Scholar]
- 37.Eski M, Sahin I, Sengezer M, Serdar M, Ifran A. Thalidomide decreases the plasma levels of IL-1 and TNF following burn injury: is it a new drug for modulation of systemic inflammatory response. Burns. 2008 Feb;34(1):104–108. doi: 10.1016/j.burns.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Malleo G, Mazzon E, Genovese T, et al. Effects of thalidomide in a mouse model of cerulein-induced acute pancreatitis. Shock. 2008 Jan;29(1):89–97. doi: 10.1097/shk.0b013e318067df68. [DOI] [PubMed] [Google Scholar]
- 39.Letsinger JA, McCarty MA, Jorizzo JL. Complex aphthosis: a large case series with evaluation algorithm and therapeutic ladder from topicals to thalidomide. J Am Acad Dermatol. 2005 Mar;52(3 Pt 1):500–508. doi: 10.1016/j.jaad.2004.10.863. [DOI] [PubMed] [Google Scholar]
- 40.Hamuryudan V, Mat C, Saip S, et al. Thalidomide in the treatment of the mucocutaneous lesions of the Behcet syndrome. A randomized, double-blind, placebo-controlled trial. Ann Intern Med. 1998 Mar 15;128(6):443–450. doi: 10.7326/0003-4819-128-6-199803150-00004. [DOI] [PubMed] [Google Scholar]
- 41.Barnhill RL, Doll NJ, Millikan LE, Hastings RC. Studies on the anti-inflammatory properties of thalidomide: effects on polymorphonuclear leukocytes and monocytes. J Am Acad Dermatol. 1984 Nov;11(5 Pt 1):814–819. doi: 10.1016/s0190-9622(84)80458-2. [DOI] [PubMed] [Google Scholar]
- 42.Faure M, Thivolet J, Gaucherand M. Inhibition of PMN leukocytes chemotaxis by thalidomide. Arch Dermatol Res. 1980;269(3):275–280. doi: 10.1007/BF00406421. [DOI] [PubMed] [Google Scholar]
- 43.Geitz H, Handt S, Zwingenberger K. Thalidomide selectively modulates the density of cell surface molecules involved in the adhesion cascade. Immunopharmacology. 1996 Mar;31(2–3):213–221. doi: 10.1016/0162-3109(95)00050-x. [DOI] [PubMed] [Google Scholar]
- 44.Sampaio EP, Sarno EN, Galilly R, Cohn ZA, Kaplan G. Thalidomide selectively inhibits tumor necrosis factor alpha production by stimulated human monocytes. J Exp Med. 1991;173:699–703. doi: 10.1084/jem.173.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shannon E, Noveck R, Sandoval F, Kamath B. Thalidomide suppressed IL-1beta while enhancing TNF-alpha and IL-10, when cells in whole blood were stimulated with lipopolysaccharide. Immunopharmacol Immunotoxicol. 2008;30(3):447–457. doi: 10.1080/08923970802135161. [DOI] [PubMed] [Google Scholar]
- 46.Burne MJ, Rabb H. Pathophysiological contributions of fucosyltransferases in renal ischemia reperfusion injury. J Immunol. 2002 Sep 1;169(5):2648–2652. doi: 10.4049/jimmunol.169.5.2648. [DOI] [PubMed] [Google Scholar]
- 47.Asano M, Nakae S, Kotani N, et al. Impaired selectin-ligand biosynthesis and reduced inflammatory responses in beta-1,4-galactosyltransferase-I-deficient mice. Blood. 2003 Sep 1;102(5):1678–1685. doi: 10.1182/blood-2003-03-0836. [DOI] [PubMed] [Google Scholar]
- 48.Shoda H, Inokuma S, Yajima N, Tanaka Y, Setoguchi K. Cutaneous vasculitis developed in a patient with breast cancer undergoing aromatase inhibitor treatment. Ann Rheum Dis. 2005 Apr;64(4):651–652. doi: 10.1136/ard.2004.023150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mindrescu C, Le J, Wisniewski HG, Vilcek J. Up-regulation of cyclooxygenase-2 expression by TSG-6 protein in macrophage cell line. Biochem Biophys Res Commun. 2005 May 13;330(3):737–745. doi: 10.1016/j.bbrc.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 50.Rogers HW, Tripp CS, Schreiber RD, Unanue ER. Endogenous IL-1 is required for neutrophil recruitment and macrophage activation during murine listeriosis. J Immunol. 1994 Sep 1;153(5):2093–2101. [PubMed] [Google Scholar]