Abstract
CheA is a central component of the chemotaxis signal transduction pathway that allows prokaryotic cells to control their movements in response to environmental cues. This dimeric protein histidine kinase autophosphorylates via an intersubunit phosphorylation reaction in which each protomer of the dimer binds ATP, at an active site located in its P4 domain and then catalyzes transfer of the γ-phosphoryl group of ATP to the His45 side chain within the P1 domain of the trans protomer. Here we utilize the fluorescent nucleotide analogue TNP-ATP [2′(3′)-O-(2,4,6-trinitrophenyl)adenosine 5′-triphosphate] to investigate the two ATP-binding sites of the Thermotoga maritima CheA dimer (TmCheA) and the single site of the isolated TmP4 domain (a monomer). We define the affinity of CheA for TNP nucleotides and, by competition, for unmodified ATP. The two ATP-binding sites of the TmCheA dimer exhibit dramatically different affinities for TNP-ATP (Kd1TNP ∼ 0.0016 μM and Kd2TNP ∼ 22 μM at 4 °C in the presence of Mg2+) as well as for ATP (Kd1ATP ∼ 6 μM and Kd2ATP ∼ 5000 μM at 4 °C in the presence of Mg2+) and in their ability to influence the fluorescence of bound TNP-ATP. The ATP-binding site of the isolated TmP4 domain interacts with ATP and TNP-ATP in a manner similar to that of the high-affinity site of the TmCheA dimer. These results suggest that the two active sites of TmCheA homodimers exhibit large differences in their interactions with ATP. We consider possible implications of these differences for the CheA autophosphorylation mechanism and for CheA function in bacterial cells.
CheA is a protein histidine kinase (PHK)1 that functions in the chemotaxis signal transduction pathway of many bacteria and archaea (15, 17). In this pathway, CheA plays a central role in allowing membrane-spanning chemotaxis receptors to modulate the cells' swimming patterns in response to gradients of attractant and repellent chemicals encountered by the cells as they move through their environments. This modulation involves a well-characterized phosphorylation cascade that begins with CheA autophosphorylation. P-CheA then serves as a phospho-donating substrate for CheY autophosphorylation. The P-CheY generated by this reaction then interacts with the flagellar motor to effect changes in swimming patterns (55, 60). The rate of CheA autophosphorylation is regulated by the chemotaxis receptor proteins. For example, in Escherichia coli cells, binding of chemoattractants to a receptor results in a dramatic decrease in CheA autokinase activity and a subsequent decrease in the level of phosphorylated CheY (4, 5). Phosphorylation of CheB via CheA contributes to sensory adaptation by directing CheB to tune the sensitivity of the chemotaxis receptor proteins (32, 50, 52). CheA proteins from a number of bacteria (including Thermotoga maritima, E. coli, Salmonella typhimurium, and Bacillus subtilis) have been characterized extensively using a variety of biochemical approaches (1, 25, 30, 45, 58, 59). Such analysis has indicated that CheA, and other signal-transducing PHKs, function as dimers and appear to accomplish their autophosphorylation reaction via a trans mechanism in which one protomer of a dimer phosphorylates its partner protomer (15, 42, 56, 64, 65).
CheA has a modular architecture in which five distinct structural domains (P1–P5) make different contributions to the overall activity and regulation of the protein (Figure 1) (1, 3, 57). P1 functions as a phospho-transfer domain and includes the phosphorylation site (His45 for T. maritima CheA) (47, 48). P2 serves as a docking site for CheY, allowing it to bind temporarily while acquiring the phosphoryl group from the adjacent phosphorylated P1 domain (31, 33, 53, 57). P3 mediates CheA dimerization (3, 45). P4 contains the ATP-binding site and other active site components (2). P5 directs interaction of CheA with CheW and the chemoreceptors; these interactions enable receptor-mediated regulation of the CheA autophosphorylation reaction (6, 12, 44).
Figure 1.

Schematic diagram of the functional organization of CheA and the mechanism of autophosphorylation. Domains P1–P5 of CheA from T. maritima, E. coli, and S. typhimurium have been investigated by a variety of high-resolution methods, including X-ray crystallography (1, 33, 39, 47, 63) and NMR methods (34, 35, 66), as well as by lower-resolution methods such as protease sensitivity (37). The numbers written below the top protomer in the diagram indicate the domain boundaries (amino acid numbers) of T. maritima CheA. The jagged lines linking P1 to P2 and P2 to P3 are intended to depict flexible linkers. The functional role of each domain is based on this structural information as well as on biochemical and genetic experiments (17, 37, 38, 57). As depicted, CheA autophosphorylation occurs via a trans intradimer mechanism in which the ATP bound to the P4 domain of one protomer provides the phosphoryl group that is used to phosphorylate His45 on the partner protomer.
Detailed structural information is available for each of the domains of T. maritima CheA (TmCheA): there are crystal structures available for isolated individual domains (P1, P2, and P4) (2, 33, 39, 43, 47) as well as for a multidomain CheA fragment (TmCheAΔ289) that encompasses domains P3–P5 (1). How P1 and P2 are oriented relative to P3–P5 has not yet been defined in detail, but recent results (14) indicate that, in a CheA dimer, the P1–P2 segment from each protomer is constrained so that it interacts exclusively with P4 from the other protomer; this constraint could involve the flexible linker connecting P2 to P3 (37). Such an arrangement would force CheA to operate via a trans mechanism (P4 of one protomer phosphorylates P1 of the second protomer), which has been observed for CheA and several other PHKs (15, 42, 56, 64, 65).
Bilwes and co-workers have characterized the structures of TmP4 in complex with a variety of nucleotide analogues, including ADPNP, ADPCP, and TNP-ATP (2). While these structural studies have provided a wealth of information about the CheA active site and its contacts with ATP, it is important to emphasize that the TmP4 domain utilized for these studies was a monomer and that attempts to generate crystals with dimeric forms of CheA (full-length or various CheA fragments) were unsuccessful. This leaves unanswered some fundamental questions about how CheA dimers interact with ATP. Do the two active sites of the dimer function independently? Are they functionally equivalent? Do they exhibit differences that might reflect asymmetry or cooperative interactions between the two active sites within the dimer?
To begin addressing these questions, we have analyzed the binding of ATP to monomeric TmP4 and dimeric full-length TmCheA. Visualizing these binding interactions is facilitated by modifying the protein (51) or the ATP (19) so that a fluorescence signal can serve as a reporter of formation of the CheA · nucleotide complex. In previous work, we utilized TNP-ATP, a fluorescent derivative of ATP, to visualize nucleotide binding to E. coli CheA (22, 54). Our results were consistent with each dimer having two independent, noninteracting ATP-binding sites, but the level of analysis was not detailed and was hampered by (i) not having (at that time) a monomeric version CheA for comparison, (ii) not having (at that time) detailed structural information about the protein · nucleotide complex, and (iii) poor signal-to-noise ratios that limited the range of TNP-ATP concentrations over which binding could be studied. In the work reported here, we used TNP-ATP as a probe to study the active sites of TmP4 and TmCheA. By using these structurally well characterized proteins, we eliminated the first two problems listed above. In addition, we overcame sensitivity limitations by modifying our fluorescence binding assays such that we could monitor binding over a 103-fold range of ligand concentration.
Our results indicate that the two active sites of TmCheA dimers bind TNP-ATP (and unmodified ATP) with dramatically different affinities and that these binding events have different effects on the fluorescence of bound TNP-ATP. These findings suggest that the two active sites of CheA homodimers are not functionally equivalent and may not operate independently. We present our results by first describing the simple situation in which TNP-ATP binds to the isolated TmP4 domain (a monomer) and then compare these results with those generated using full-length TmCheA (a dimer).
Experimental Procedures
Chemicals and Reagents
2′(3′)-O-(2,4,6-Trinitrophenyl) adenosine 5′-triphosphate (TNP-ATP) and TNP-AMP were purchased from Invitrogen, stored at −20 °C, and used under minimal light conditions. The concentrations of the TNP-modified nucleotides were determined spectrophotometrically, using an extinction coefficient of 26.4 mM−1 cm−1 at 408 nm (20). ATP was purchased from Roche Diagnostics, and ultrapure glycerol was from MP Biomedicals. All other chemicals were reagent grade and were purchased from Sigma or Fisher Scientific Co. TEGD buffer (used for protein purification and storage) contained 50 mM Tris, 0.5 mM Na2EDTA, 10% (v/v) glycerol, and 1 mM DTT, adjusted to pH 7.5 using hydrochloric acid. TnM buffer (used for binding titrations in the presence of Mg2+) contained 25 mM Tris, 0.5 mM Na2EDTA, 10% (v/v) glycerol, 25 mM NaCl, 50 mM potassium glutamate, and 20 mM MgCl2. TnE buffer (used for binding titrations in the absence of Mg2+) contained 25 mM Tris, 10% (v/v) glycerol, 25 mM NaCl, 50 mM potassium glutamate, and 20.5 mM Na2EDTA, adjusted to pH 7.5 using hydrochloric acid. Neither titration buffer contained reducing agent because we found that DTT, β-mercaptoethanol, and TCEP all react slowly with TNP-ATP, causing a bleaching of its fluorescence. Absorbance spectra were recorded using samples in 1 cm path length cuvettes in a Cary 50 spectrophotometer.
Protein Expression and Purification
The full-length T. maritima cheA gene cloned into Novagen plasmid pET28a was kindly provided by Drs. Sang-Youn Park and Brian Crane (Cornell University, Ithaca, NY). An expression plasmid for the T. maritima P4 domain (residues 356–540) was created by cloning a corresponding PCR fragment into pET28a. The TmCheA and TmP4 proteins that we studied both carry an N-terminal His tag fusion (provided by the pET28a) to facilitate purification (23). This tag was not removed from the purified proteins prior to use in binding experiments, but previous work in this lab has demonstrated that this affinity tag does not affect the binding or kinetic properties of CheA (54). To simplify the text, the presence of the His tag is not indicated explicitly in the remainder of this paper. E. coli strain BL21 λDE3 cells (Invitrogen) carrying plasmid pET28:TmCheA or pET28:TmP4 were grown (at 32 °C) until they reached midlog phase in 4 L of Luria broth containing 50 μg/mL kanamycin, and then overproduction of the protein of interest was induced by adding IPTG (final concentration of 1 mM). Three hours after induction, the cell cultures were placed on ice, and then cell pellets were collected by centrifugation at 4 °C. Cell pellets were resuspended in TEG buffer containing 10 mM β-mercaptoethanol and then lysed (on ice) by sonication in the presence of Halt Protease Inhibitor (Pierce). The proteins were purified as described previously using a Ni-NTA column following conditions reported by Levit et al. (30) and an AffiGel Blue column using conditions summarized by Hess et al. (18). Following the final purification step, proteins were dialyzed against TEGD buffer, then concentrated to ∼400 μM using Amicon Centriprep centrifugal concentrators, and then stored in small aliquots at −80 °C. Protein concentrations were determined using the Bio-Rad protein assay and the Pierce BCA (bicinchoninic acid) assay, using bovine serum albumin as a standard.
Fluorescence-Monitored TNP-ATP Binding Titrations
These experiments were conducted at 4 °C to (i) minimize potential complications arising from dissociation of CheA dimers into monomers (45) and (ii) enable comparison of binding affinity results to binding kinetics results at low temperatures (10). Most titrations were conducted in TnM buffer (except for the no-magnesium titrations of Figure 9 which utilized TnE buffer). Buffer compositions are specified above. For each binding titration, a starting solution (2.5 mL) of protein or ligand was placed in a 1 cm × 1 cm quartz fluorescence cuvette, stirred continuously using a magnetic stir bar, and maintained at 4 °C using a circulating water bath connected to the cuvette holder. After each addition of protein or ligand, four consecutive fluorescence emission spectra were recorded and averaged using a PTI QuantaMaster instrument measuring counts per second (cps) in digital mode. Emission spectra (530–600 nm) were recorded using a λex of 520 nm (excitation and emission slits were set at 4 nm). The samples were shielded from the excitation light between additions. Each of the titrations depicted in the figures represents the average of at least two independent experiments conducted on different days using independently prepared protein and ligand solutions. Two major types of titrations were performed, “CheA-in-excess” titrations and “TNP-in-excess” titrations, and these required slightly different data manipulations, as described below.
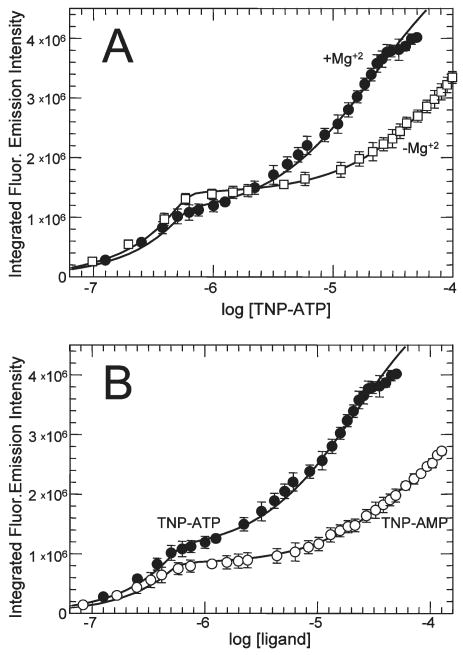
Figure 9.
TmCheA binding to TNP-AMP and to TNP-ATP in the absence of Mg2+. (A) Aliquots of TNP-ATP were added to 0.6 μM TmCheA samples (dimer concentration) in buffer either containing 20 mM MgCl2 (●) or lacking MgCl2 (and containing 20 mM Na2EDTA) (□); the resulting fluorescence signals were monitored and analyzed as described in the legend of Figure 6. The lines represent the best fit of the data using DynaFit and a two-site binding model. This analysis indicated that in the absence of Mg2+, Kd1TNP = 0.003 μM, Kd2TNP = 100 μM, IMFCT.AA = 2.4 × 106 cps/μM, and IMFCT.AA.T = 8.9 × 106 cps/μM. In the presence of Mg2+, Kd1TNP = 0.0016 μM, Kd2TNP = 22 μM, IMFCT.AA = 2 × 106 cps/μM, and IMFCT.AA.T = 9.5 × 106 cps/μM. (B) Aliquots of TNP-ATP (●) or TNP-AMP (○) were added to 0.6 μM (dimer) TmCheA samples in buffer containing 20 mM MgCl2. Binding was assessed by monitoring fluorescence. The TNP-ATP titration data are the same as those presented in panel A. The lines represent the best fit of the data using DynaFit and a two-site binding model, indicating that for TNP-AMP binding Kd1TNP = 0.003 μM, Kd2TNP = 75 μM, IMFCT.AA = 1.4 × 106 cps/μM, and IMFCT.AA.T = 6.3 × 106 cps/μM.
(i) TNP-in-Excess Titrations
A starting emission spectrum (average of fours scans) was recorded for the initial diluted protein solution. Then small aliquots (1–6 μL) of TNP-ATP solutions (5–6400 μM) were added using a calibrated micropipettor, and an emission spectrum was recorded (average of four) following each addition. To analyze the results, the starting emission spectrum was first subtracted from each emission spectrum generated in the presence of TNP-ATP. Then the area of each corrected spectrum was determined (using the integration function of QuantaMaster). These integrated fluorescence emission signals (IFES) were then corrected for the effects of dilution and for inner filter effects using the formula
| (1) |
The antilog expression in this equation is a correction for inner filter effects (28). This correction was considerably smaller using a λex of 520 nm than it was for results generated using a λex of 410 nm, as was done in previous studies (10, 21, 54). For example, for a 100 μM TNP-ATP solution, a correction factor of 1.88 was necessary using a λex of 520 nm (A520 = 0.55) compared to a correction factor of 20.9 required with a λex of 410 nm (A410 = 2.64). For “mock titrations” (in the absence of any CheA), this inner filter correction generated an IFEScor signal that was linearly dependent on TNP-ATP concentration to >100 μM. These mock titrations established the IFES signal that would be expected as background for each titration of CheA with the same concentration range of TNP-ATP. For each data point in a CheA/TNP-ATP titration, the IFES associated with the CheA · TNP-ATP complex (IFEScomplex) was calculated by subtracting the IFES value observed at the same total TNP-ATP concentration in the mock titration. IFEScomplex values generated by these manipulations are plotted in Figures 4 and 6–9 to define the relationship between integrated fluorescence emission and TNP-ATP concentration.
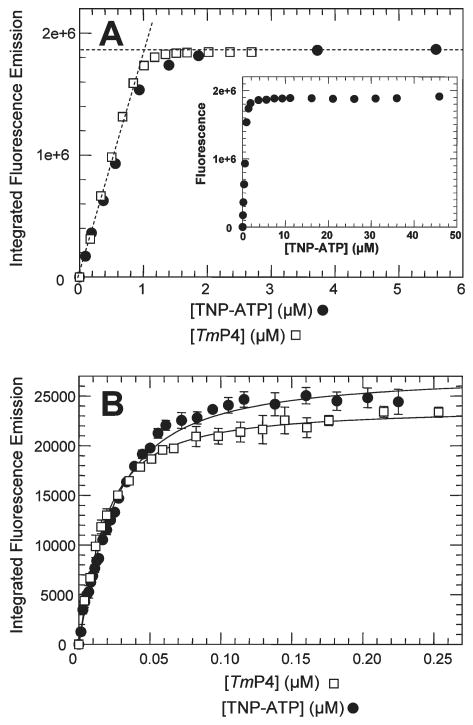
Figure 4.
Fluorescence-monitored binding of TNP-ATP to TmP4. (A) Results of titrations in which successive aliquots of TNP-ATP stock solutions were added to a 2.5 mL sample of TmP4 (1 μM) (●) or aliquots of TmP4 were added to a 2.5 mL sample of TNP-ATP (1 μM) (□). The intersection point of the dashed lines indicates a binding stoichiometry of 1:1. The inset shows the TNP-ATP titration results at higher concentrations. (B) Results of titrations at low protein and ligand concentrations. Successive aliquots of TNP-ATP stock solutions were added to a 2.5 mL sample of TmP4 (0.015 μM) (●), or aliquots of TmP4 were added to a 2.5 mL sample of TNP-ATP (0.013 μM) (□). The lines represent the best fits obtained using DynaFit to fit the data using a single-binding site model. This analysis indicated a Kd of 0.011 μM for the titration of TNP-ATP with TmP4 and a Kd of 0.016 μM for the titration of TmP4 with TNP-ATP. Error bars indicate standard deviations for three independent experiments.
Figure 6.
Fluorescence-monitored binding of TNP-ATP to TmCheA. (A) Results of titrations in which successive aliquots of TNP-ATP stock solutions were added to a 2.5 mL sample of TmCheA (0.6 μM dimer). The line represents the best fit of the data generated using DynaFit and a two-site binding model. This analysis gave the following values: Kd1TNP = 0.0029 μM, Kd2TNP = 22 μM, IMFCT.AA = 2.0 × 106 cps/μM, and IMFCT.AA.T = 9.5 × 106 cps/μM. The inset shows the same results plotted using a log scale for [TNP-ATP] to better depict the extremely biphasic nature of the binding curves. (B) Results of a reverse titration in which aliquots of a concentrated TmCheA solution were added to a 0.28 μM solution of TNP-ATP. The intersection point of the dashed lines indicates a binding stoichiometry of one TNP-ATP per CheA dimer and a value of 2.0 × 106 cps/μM for IMFCT.AA.
(ii) CheA-in-Excess Titrations
A starting emission spectrum (average of four) was recorded with the initial TNP-ATP solution. Then small aliquots of TmCheA (or TmP4) were added, and an emission spectrum was recorded (average of four) following each addition. To analyze the results, the starting emission spectrum was first subtracted from each emission spectrum generated in the presence of protein. Then the area of each corrected spectrum was determined. These IFES values were corrected for the effects of dilution, but inner filter corrections and subtraction of background signal were not necessary (control experiments demonstrated that the TmCheA and TmP4 preparations had negligible absorbance at 520 nm and exhibited negligible fluorescence when excited at 520 nm). These corrected IFES values are plotted in Figures 4, 5, and 7 for binding curves showing the relationship between integrated fluorescence emission and TmCheA or TmP4 concentration.
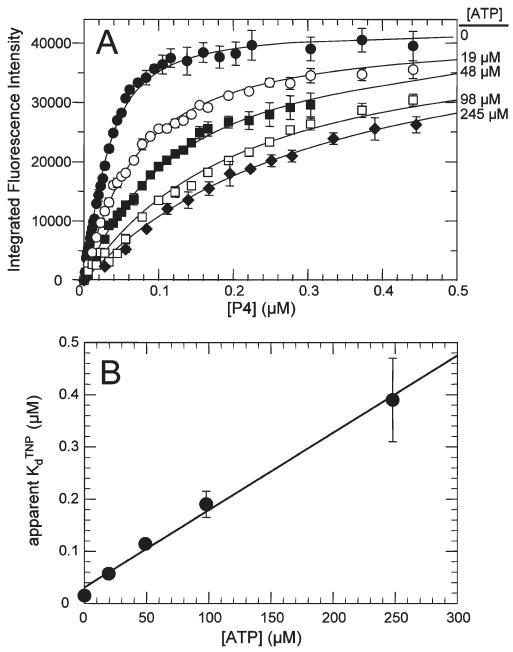
Figure 5.
Competition between ATP and TNP-ATP for the TmP4 binding site. (A) TmP4 aliquots were added to TNP-ATP samples (0.02 μM) containing 0 (●), 19 (○), 48 (■), 98 (□), and 245 μM ATP (◆). The solid lines are the best fits obtained using DynaFit to fit the data to a model of a single binding site; this analysis defined an apparent KdTNP value at each concentration of competitor (ATP). (B) Apparent dissociation constants from panel A plotted as a function of ATP concentration.
Figure 7.
Binding and ATP inhibition of binding of TNP-ATP to the high-affinity site of TmCheA. (A) Binding of TNP-ATP to TmCheA was monitored fluorometrically as successive aliquots of TNP-ATP were added to 0.003 μM TmCheA (dimer concentration) (●) and when aliquots of TmCheA were added to 0.002 μM TNP-ATP (□). The lines represent the best fits of the data generated using DynaFit and a one-site binding model. This analysis indicated a Kd1TNP of 0.0018 μM for the titration of CheA with excess TNP-ATP and a Kd1TNP of 0.0014 μM for the titration of TNP-ATP with excess TmCheA. Error bars indicate the standard deviations for three independent experiments. (B) Effect of competitor ATP on the apparent Kd1TNP of TmCheA.
To extract Kd values from titration profiles (plots of IFES vs [ligand] or [protein]), we utilized the least-squares fitting routine of DynaFit (27). This program requires input of an explicit model. For binding of TNP-ATP to TmP4, the model was a simple one-step equilibrium. For binding of TNP-ATP to TmCheA dimers, we used the following model:
| (2) |
In addition, this model assumes that the observed corrected integrated fluorescence signal adheres to the following relationship:
| (3) |
IMFCT.AA values were determined in stoichiometry titrations (Figures 4 and 6A, inset); IMFCT.AA.T values were determined using DynaFit.
Competition Binding Studies for Analyzing Interactions of CheA with ATP
Fluorescence-monitored binding experiments similar to those described above were performed to analyze binding of TNP-ATP to CheA in the presence of varying concentrations of ATP. Binding curves were analyzed using DynaFit, as described above, to generate apparent KdTNP values. These apparent Kd values are expected to conform to the following relationship:
| (4) |
Thus, a plot of apparent KdTNP versus ATP concentration should be linear with a γ-axis intercept of KdTNP and a slope of KdTNP/KdATP. Therefore, the value of KdATP can be calculated as KdTNP/ slope.
Results
TmCheA and TmP4 Alter the Absorbance and Fluorescence Excitation Spectra of TNP-ATP
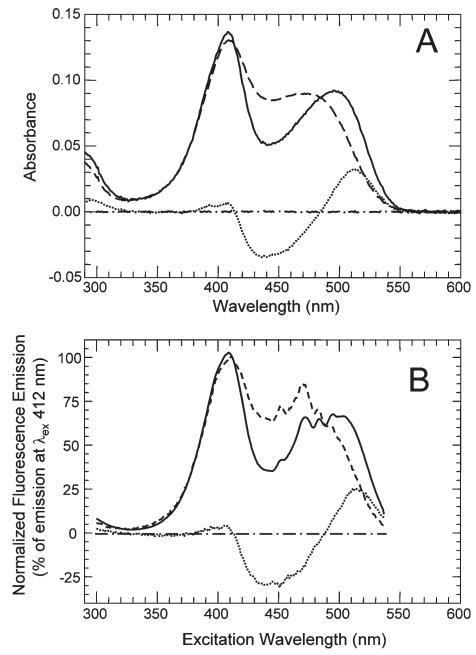
TNP-ATP binds to the active site of TmCheA and TmP4 with high affinity; this binding interaction markedly enhances its fluorescence emission intensity (2, 54), a change that likely reflects the hydrophobic environment experienced by the TNP group in the protein · nucleotide complex (19). Addition of TmP4 to TNP-ATP also altered the TNP absorbance spectrum (Figure 2A), red-shifting the longer wavelength absorbance band such that it exhibited a λmax of ∼500 nm (compared to a λmax of ∼480 nm for the uncomplexed TNP-ATP) and decreasing the absorbance in the range between the isosbestic points of 412 and 485 nm. The absorbance difference spectrum (Figure 2A, dotted line) indicates a maximal difference at ∼520 nm. Similar changes in absorbance and excitation spectra were generated when full-length TmCheA was added to TNP-ATP (results not shown). As expected, the changes in the absorbance spectrum are mirrored in the fluorescence excitation spectrum (Figure 2B).
Figure 2.
Effect of TmP4 on the absorbance and fluorescence properties of TNP-ATP. (A) The absorbance spectrum of 4.9 μM TNP-ATP (in TnM buffer) was recorded in the absence (− − −) and presence (—) of 12 μM TmP4. Also shown is the difference spectrum (⋯) calculated by subtracting the TNP-ATP spectrum from the spectrum of TNP-ATP with TmP4. Spectra were recorded vs a baseline scan of TnM buffer; a buffer vs buffer scan (− · −) is shown. (B) Fluorescence excitation spectra were recorded (λem = 540 nm) for 4.9 μM TNP-ATP in the absence (− − −) and presence (—) of 12 μM TmP4; plotted spectra were normalized relative to the emission intensity observed for a λex of 412 nm. Also shown are the calculated difference spectrum (⋯) (TNP-ATP scan subtracted from TmP4/TNP-ATP scan) and a buffer–buffer baseline (− · −).
The spectra of Figure 2B are normalized relative to the fluorescence intensity observed at 412 nm (the isosbestic point for the absorbance spectra of free and TmP4-bound TNP-ATP); this normalization is necessary because of the large increase in TNP-ATP fluorescence caused by binding to the CheA active site. The emission spectra used to generate Figure 2B are shown in Figure 3A. Analysis of these spectra indicated that the shift in the TNP-ATP absorbance/excitation spectrum caused by binding to CheA could be exploited to enhance the sensitivity of titration experiments (described below) designed to determine the affinity of the binding interaction between TNP-ATP and TmP4 or TmCheA. In such experiments, there is a mixture of bound and free TNP-ATP, each of which contributes to the overall observed fluorescence. The sensitivity of the assay is dictated by how different these contributions are, i.e., how much higher the signal (fluorescence of the CheA · TNP-ATP complex) is relative to the background (fluorescence of the free TNP-ATP). The inset of Figure 3A depicts how the excitation wavelength influences the signal-to-background ratio (S:B) for one particular combination of TmP4 and TNP-ATP. Previous work (2, 10, 54) utilized an excitation wavelength of 410 nm to generate the fluorescence signal that was analyzed to monitor binding events; however, the inset plot indicated that we could improve S:B significantly by using an excitation wavelength of >500 nm. For example, for a λex of 520 nm, S:B ∼ 30 compared to the S:B of ∼12 for a λex of 410 nm. Moreover, the decreased absorbance of the TNP-ATP at 520 nm (compared to 410 nm) reduced inner filter effects that have to be taken into account in analyzing fluorescence-monitored binding titrations (Experimental Procedures). Another change that we adopted to improve the sensitivity of detection of the CheA · TNP-ATP complex was to monitor the integrated emission signal (in the range between 530 and 600 nm) rather than relying on the emission at a single wavelength (as in our previous work). These two changes (using a λex of 520 nm and integrating the emission signals) allowed us to monitor binding over a 103-fold range of TNP-ATP concentrations, a significant improvement over our previous studies with TNP-ATP (10, 22, 54).
Figure 3.
Effect of TmP4 on the fluorescence excitation and emission spectra of TNP-ATP. (A) The main panel shows the fluorescence excitation spectrum of 4.9 μM TNP-ATP (in TnM buffer) in the absence (---) and presence (—) of 12 μM TmP4 using an emission wavelength of 540 nm. The inset plots the signal-to-background ratio as a function of the excitation wavelength for this same sample set (arrows at 410 and 520 nm). (B) Fluorescence emission spectrum of 4.9 μM TNP-ATP (in TnM buffer) recorded in the absence (---) and presence (—) of 12 μM TmP4 using an excitation wavelength of 520 nm. Also shown is the corrected buffer emission scan (− · −).
Binding of TNP-ATP to TmP4
To analyze fluorescence-monitored binding titrations of TmP4 with TNP-ATP, we needed information about the binding stoichiometry and the fluorescence coefficient of the TmP4 · TNP-ATP complex. To generate this information, we first monitored the fluorescence signal generated at relatively high protein and ligand concentrations. In these “stoichiometry titrations”, increasing concentrations of TNP-ATP were added to a fixed amount of TmP4 (1 μM in Figure 4A). We observed a linear increase in fluorescence emission intensity as the ligand concentration was increased, until a plateau was reached indicating that the binding site had been saturated. Similar results were obtained when we performed “reverse titrations” in which increasing concentrations of TmP4 were added to a fixed amount of TNP-ATP (Figure 4A). These titration results indicated a binding stoichiometry of 1:1 and defined the integrated molar fluorescence coefficient (IMFC) for the TmP4 · TNP-ATP complex (1.9 × 106 cps/μM).
The shapes of the binding “curves” in Figure 4A (linear with little or no “bowing over” prior to reaching the plateau) indicated that the Kd for the binding interaction was considerably lower than 1 μM and that these results, while useful for defining stoichiometry and IMFC, were not useful for obtaining an accurate estimate of Kd. To generate titration results capable of defining Kd, we performed fluorescence-monitored titration experiments (Figure 4B) at considerably lower protein and ligand concentrations (0.01–0.03 μM for the limiting component). Analysis of these results indicated a Kd of 0.014 ± 0.003 μM for the TmP4 · TNP-ATP complex.2 This analysis utilized the binding stoichiometry (1:1) and IMFC (1.9 × 106 cps/μM) defined as described above. Our results are consistent with previous work that indicated an upper limit of Kd of ∼0.01 μM for the TmP4–TNP-ATP binding interaction (2).
ATP Inhibition of Binding of TNP-ATP to TmP4
ATP can compete with TNP-ATP for the nucleotide-binding site of TmP4, as shown in the titration results of Figure 5A. This competition increases the apparent Kd of the TmP4–TNP-ATP binding interaction, and this relationship was analyzed (Figure 5B) to define the dissociation constant of the TmP4 ↔ ATP binding interaction (KdATP), as described in Experimental Procedures. The value indicated by this analysis (11 ± 3 μM)2 agrees well with the value (15 μM) defined by Bilwes and co-workers using a chromatographic approach (2). This agreement indicates that analysis of competition between ATP and TNP-ATP can be used reliably to define the binding affinity of the CheA active site for ATP.
Binding of TNP-ATP to Full-Length TmCheA
Using the same approaches described above to study binding of TNP-ATP to TmP4, we also investigated interaction of TNP-ATP with full-length TmCheA. One important difference between full-length CheA and P4 is that the full-length protein forms stable dimers (45), so there is the potential for complexity arising from differences between the two binding sites in each dimer. Results described below indicate that these two sites exhibit some striking differences. In discussing these results, we use the following abbreviations: AA, a full-length TmCheA dimer; T.AA, a dimer to which one molecule of TNP-ATP has bound; T.AA.T, a dimer that has bound two molecules of TNP-ATP. In addition, we refer to the high-affinity site as “site 1” and the low-affinity site as “site 2”.
In contrast to the simple, single-site binding curves observed with TmP4 (Figure 4), titration of a fixed concentration of TmCheA with excess TNP-ATP generates distinctly biphasic binding curves (Figure 6A): ∼20% of the overall fluorescence change is generated early in the titration (at ligand concentrations approximating the CheA dimer concentration), and then the remaining 80% of the fluorescence change requires considerably higher concentrations of TNP-ATP. The extremely biphasic nature of this titration is more easily visualized using a log-scale x-axis [log of TNP-ATP concentration in molar units (Figure 6A inset)]. One possible interpretation of these results is that the two TNP-ATP-binding sites of the TmCheA dimer have very different properties: one site having a tight affinity and lower fluorescence (IMFC ∼ 2× 106 cps/μM) and the second site having a weak affinity and high fluorescence (IMFC ∼ 8 × 106 cps/μM). The presence of two classes of binding sites might reflect two alternative situations: (i) negative cooperativity or (ii) preexisting asymmetry within the CheA dimers. We will consider these possibilities in greater detail in the Discussion. In the remainder of the Results, we analyze the binding curves by making use of macroscopic (Adair) binding constants without making assumptions about the underlying molecular mechanism that gives rise to a high-affinity site and a low-affinity site. From this analysis, we estimate the dissociation constant for the AA + T ↔ T.AA binding equilibrium (Kd1TNP) and for the T.AA + T ↔ T.AA.T equilibrium (Kd2TNP).
To explore binding of TNP-ATP to TmCheA in greater detail, we first examined the properties of the high-affinity binding site by titrating a limiting amount of TNP-ATP with excess TmCheA (Figure 6B). In this intital reverse titration, the TNP-ATP concentration was set at a level (0.28 μM) that we expected to be much higher than the Kd of the high-affinity site. The shape of the binding curve in Figure 6B (linear with little bowing over prior to reaching the plateau) was consistent with the assumption that Kd ≪ 0.28 μM. The results of this stoichiometry titration indicated that each TmCheA dimer can bind one molecule of TNP-ATP with high affinity and that the IMFC of the T.AA complex is ∼2× 106 cps/μM, a value that matches the IMFC observed for the presumed T.AA intermediate generated at TNP-ATP concentrations of < 1 μM in Figure 6A. To estimate the binding constant for the high-affinity binding site, we performed forward and reverse titrations at low protein and ligand concentrations (0.002–0.003 μM) (Figure 7A). Analysis of these titrations indicated a Kd1TNP value of ∼0.0016 μM and demonstrated reasonable agreement between forward and reverse titrations.
Having defined the basic characteristics of the high-affinity binding site of the TmCheA dimer, we were then able to investigate the properties of the low-affinity site by analyzing the results of titrations of TmCheA with excess TNP-ATP. Fitting the results of Figure 6A to two-site model indicates a Kd2TNP of ∼22 μM and an IMFCT.AA.T of ∼7.5 × 106 cps/μM. In Figure 6A, the highest ligand concentration (50 μM) is ∼2.3Kd2TNP which would saturate ∼70% of the low-affinity sites. In an effort to extend this range (to characterize site 2 more completely), we analyzed binding at TNP-ATP concentrations as high as 120 μM and observed Kd2 values in the range between 20 and 25 μM (results not shown) and IMFC values within 10% of 7.5 × 106 cps/μM. Above this concentration, the background correction and inner filter correction (Experimental Procedures) became prohibitively large.
In summary, results in this section indicate that the TmCheA dimer has two binding sites for TNP-ATP that differ dramatically (∼ 10000-fold) in their affinity for this ligand as well as in their ability to influence the fluorescence of the TNP group. These differences are summarized in Table 1.
Table 1.
Summary of Results from Binding Analyses
| TNP-ATP binding | ATP binding Kd2ATP (μM) | |||||
|---|---|---|---|---|---|---|
| Kd1TNP(μM) | IMFCsite 1 (cps/μM) | Kd2TNP(μM) | IMFCsite 2 (cps/μM) | Kd1ATP (μM) | ||
| TmP4 | 0.014 ± 0.003a | 1.9 × 106 | NAb | NAb | 11 ± 4a | NAb |
| TmCheA | 0.0016 ± 0.0002a | 2.0 × 106 | 22 ± 2 | 7.5 × 106 | 6 ± 2a | 5000 ± 1000a |
Uncertainties reported are standard deviations of two or three replicate measurements.
Not applicable (TmP4 has just one binding site).
ATP Inhibition of Binding of TNP-ATP to TmCheA
The large difference in affinity between the two binding sites of the CheA dimer allowed us to study ATP inhibition of TNP-ATP binding by the high-affinity site without any significant interference from the second site. Working at low concentrations of TmCheA and TNP-ATP, and using the same basic approach described above for competition experiments with TmP4, we defined the effect of competitor ATP on the observed value of Kd1TNP. This relationship (Figure 7B) indicated that the high-affinity site of the TmCheA dimer bound ATP with a Kd of ∼6 μM (Kd1ATP), a value close to that observed for TmP4 (11 μM).
ATP is also an effective competitor at the second TNP-ATP-binding site of the TmCheA dimer, as shown in Figure 8A. Analysis of the effect of ATP concentration on the observed value of Kd2TNP (Figure 8B) indicated that site 2 has a poor affinity for ATP (Kd2ATP ∼ 5000 μM). Thus, the two active sites of the TmCheA dimer exhibit dramatically different affinities for ATP, differing by ∼103-fold.
Figure 8.
Competition between ATP and TNP-ATP for TmCheA binding sites. (A) TNP-ATP aliquots were added to 0.6 μM TmCheA samples (dimer concentration) containing 0 (●), 2.34 (○), 5.9 (■), and 23.4 mM ATP (□). Binding was monitored by following fluorescence emission. Not shown (to avoid overcrowding) are titration results at 11.9 mM ATP. The solid lines are the best fits obtained using Dyna Fit to fit the data to a model in which each CheA dimer has two binding sites that make unequal contributions to the observed fluorescence change (2 × 106 cps/μM for the first site and 7.5 × 106 cps/μM for the second site, as defined in Figure 6). (B) Effect of competitor ATP on the apparent Kd2ATP (determined in panel A). Analysis of this relationship (as described in the legend of Figure 5) indicates a Kd2ATP of 5000 ± 1000 μM.
Additional Differences between the Two ATP-Binding Sites of TmCheA
To explore further possible differences between the two nucleotide-binding sites of TmCheA dimers, we examined whether they are affected by removal of Mg2+ (Figure 9A) (all of the binding studies described above were performed in the presence of 20 mM MgCl2). These results indicated that the affinity of site 2 for TNP-ATP is diminished (5-fold) in the absence of Mg2+, while site 1 is insensitive to the presence or absence of Mg2+. We also examined binding of TNP-AMP to sites 1 and 2 of TmCheA (Figure 9B). Site 1 binds TNP-AMP with an affinity close to that observed for TNP-ATP, but the affinity of site 2 for this ligand is diminished (Kd2TNP-AMP ∼ 75 μM compared \to Kd2TNP-ATP ∼ 22 μM). Overall, the results in this section suggest that the two active sites of the TmCheA dimer utilize different binding contacts in their interactions with TNP-modified nucleotides: Site 2 likely makes binding contacts involving the β- and/or γ-phosphates of the bound nucleotide and the associated divalent metal (Mg2+), but site 1 does not.
Discussion
How Do the Two Binding Sites of the TmCheA Dimer Differ?
Binding of TNP-ATP to either TmP4 or TmCheA places the TNP portion of the molecule in an environment that alters its fluorescence excitation and emission properties. We exploited this change to monitor CheA binding interactions with this nucleotide analogue and (by competition) with “regular” ATP. The results obtained with TmP4 were straightforward: Each protein molecule bound one molecule of nucleotide with tight affinity. By contrast, our results with full-length TmCheA were surprising, revealing markedly biphasic binding curves that indicated two binding sites with very different affinities and different environments for the TNP group (i.e., different IMFC values). The most straightforward way of interpreting the differences between TmP4 and full-length TmCheA is to attribute them to the different quaternary structures of the proteins: TmP4 is monomeric, while TmCheA is a dimer. Thus, the biphasic binding profiles observed with TmCheA likely reflect the different properties of the two ATP-binding sites of the dimer. Table 1 shows a compilation of the affinity and fluorescence properties for TmP4 and TmCheA. Comparisons of some of these values are informative. First, for both TNP-ATP and unmodified ATP, there is a large difference in affinity between site 1 and site 2 (Kd2TNP/Kd1TNP ∼ 104, and Kd2ATP/Kd1ATP ∼ 103), suggesting that the large affinity difference between the two binding sites is not a situation arising from peculiarities of the inhibitor TNP-ATP but rather reflects a basic feature of CheA–nucleotide interactions. Second, when considering interactions with TNP-ATP, site 1 of the dimer has properties that are similar to those of the ATP-binding site of TmP4: a similar IMFC for the protein · TNP-ATP complex and a very tight binding affinity. For binding interactions with unmodified ATP, site 1 of the dimer is also similar to the TmP4 binding site (KdATP values of 6 and 11 μM, respectively). Taken together, these results suggest that, in TmCheA dimers, one of the two ATP-binding sites is unaffected by dimer formation; this site behaves much like it would in a monomeric version of the full-length protein, exhibiting similar affinities for ATP and TNP-ATP and a similar TNP-binding environment. However, in site 2 of the dimer, these properties are altered dramatically, suggesting that dimer formation has a significant impact on the structure and conformation of this ATP-binding site.
In view of the striking differences between sites 1 and 2, one additional possibility that we considered was whether site 2 binding might reflect nonspecific association of TNP-ATP with the full-length protein (e.g., TNP interacting with segments of CheA that are distinct from the active site and that are absent from the isolated P4 domain). Although this is difficult to discount completely, we think that it is unlikely for the following reasons: (i) Unmodified ATP competed effectively with TNP-ATP binding to site 2, and (ii) site 2 binding was affected by Mg2+ and by loss of the β- and γ-phosphate groups of TNP-ATP. These observations are consistent with expectations if site 2 binding reflects TNP-ATP interacting with a bona fide ATP-binding site. If site 2 does, in fact, represent low-affinity, nonspecific association of TNP-ATP (and ATP) with TmCheA, then our overall set of results would suggest that only one of the two active sites of the TmCheA dimer is functional, i.e., that the two ATP-binding sites of TmCheA exhibit half-of-sites binding.
It is informative to relate the features of site 1, revealed in our binding studies, to the detailed three-dimensional structure of the TmP4 · TNP-ATP complex determined by Bilwes et al. (2). Several key aspects of this structure should be emphasized. First, TNP-ATP binds to the TmP4 active site in a conformation that allows the TNP portion of the molecule to occupy a hydrophobic cavity adjacent to the ATP-binding pocket. This conformation (and the unusual configuration of the ribose in TNP-ATP) places the ATP part of the molecule in an unanticipated orientation relative to the active site. Most notably, the triphosphate groups of TNP-ATP are directed outward, away from the active site, so they cannot make several of the binding contacts that are observed when ADPCP and ADPNP bind to TmP4 in what likely represents the normal active site–nucleotide orientation. Moreover, the orientation of the triphosphates of TNP-ATP relative to the CheA active site destroys its ability to serve as a substrate (phospho donor). Another notable feature of the TmP4 · TNP-ATP structure is that it lacks contacts linking active site components to the Mg2+ that is coordinated to the nucleotide phosphates. These contacts with a bridging divalent metal ion are evident in the TmP4 · ADPCP and TmP4 · ADPNP structures, but in the TmP4 · TNP-ATP complex, Mg2+ is completely absent (2). In our binding studies, we observed that site 1 of the TmCheA dimer interacts with TNP-ATP in a manner that is not affected by the presence of Mg2+ or by the removal of the β- and γ-phosphate groups of the TNP nucleotide (TNP-AMP). We also observed that the affinity and fluorescence properties of site 1 are similar to those of the isolated TmP4 domain (Table 1). Taken together, these results suggest that TNP-ATP interacts with site 1 of the TmCheA dimer in a manner similar to that observed in the TmP4 · TNP-ATP crystal structure. One important general feature of the TmP4 · TNP-ATP structure is that it indicates numerous van der Waals contacts between the TNP ring and protein side chains that line the TNP-binding cavity (2). These contacts may provide the binding energy that allows TNP-ATP to bind with an affinity that is much tighter than that observed with regular ATP despite the mispositioning of much of the ATP part of the TNP-ATP molecule in the complex. Thus, the TNP part of the TNP-ATP molecule may provide much of the thermodynamic driving force for binding to TmP4, and presumably also for binding to site 1 of TmCheA.
Site 2 of the TmCheA dimer, however, exhibits several properties that differ significantly from expectations based on the TmP4 · TNP-ATP crystal structure. For example, this low-affinity site is influenced by Mg2+ and by the absence of the β- and γ-phosphate groups of the TNP nucleotide in TNP-AMP. In this regard, site 2 behaves as if the ATP portion of the TNP-ATP molecule provides important binding determinants, a situation that differs strikingly from that described above for site 1. Thus, both the TNP and ATP components of the TNP-ATP molecule bound to site 2 may interact with the TmCheA protein in a manner that differs significantly from that observed in the TmP4 · TNP-ATP complex and from that utilized at site 1 of the dimer.
What Is Responsible for Creating Two Distinct Binding Sites in TmCheA Homodimers?
As depicted in Figure 10, there are two general scenarios to consider: (i) negative cooperativity and (ii) preexisting asymmetry (8, 11, 36). In a negative cooperativity model, the two ATP-binding sites of the CheA dimer are initially equivalent. However, when a nucleotide binds to one of the sites, this binding event is communicated to the second site, resulting in a dramatic decrease in the affinity of the second site for the nucleotide [reflecting either structural changes or changes in the entropy and dynamics of the binding site (46)]. The communication between the two active sites would also alter the TNP environment of the second active, causing site 2 to enhance the TNP fluorescence more dramatically than at site 1. In the crystal structure of TmCheAΔ289 (a dimer), the two ATP-binding sites are located on opposite sides of the dimer, separated by ∼90 Å, so any allosteric communication between them would have to involve changes propagated over this distance. Perhaps such a conformational change is responsible for Bilwes' observation that addition of nucleotides to TmCheAΔ289 crystals caused them to crack (2).
Figure 10.

Models for nucleotide binding to dimeric TmCheA. (A) Negative cooperativity model. (B) Asymmetric dimer model. Nucleotide is depicted as a black circle. CheA protomers are depicted as circles (high-affinity conformation) and squares (low-affinity conformation). Protomer colors indicate the ability of the protomer to enhance the fluorescence of bound TNP-ATP: light orange indicating a weaker ability and red indicating a stronger ability.
In a preexisting asymmetry model (Figure 10B), the two protomers of a CheA dimer would assemble into an asymmetric homodimer in a manner that creates a high-affinity (low TNP fluorescence) active site and a low-affinity (high TNP fluorescence) active site. These structural and conformational differences would exist regardless of whether ATP is present. Although the idea of an asymmetric homodimer might seem counterintuitive, there are some well-known examples of proteins that function in such a manner (7, 40). Perhaps the most well-known of these is tyrosyl-tRNA synthetase, characterized in the Fersht lab. This enzyme exhibits half-of-sites substrate binding because, in solution, each dimer has one active protomer and one inactive protomer. This functional asymmetry appears to reflect the two protomers being “frozen” into either an active or an inactive conformation in the absence of any substrate or ligand (61, 62).
What Is the Functional Relevance of CheA Having Two Active Sites with Different ATP Affinities?
Regardless of the underlying molecular mechanism, the vastly different ATP binding affinities of the two active sites of TmCheA dimers could have important implications for how this enzyme functions in the context of a cell, and such considerations lead to the following question. What benefit(s) could have driven evolution of such a situation? It is likely that the ATP concentration3 in the cytoplasm of T. maritima is greater than Kd1ATP but not greater than Kd2ATP. Given that Km = Kd for CheA (59), the different affinities of the two active sites would cause them to make unequal contributions to the overall production of P-CheY: Site 1 would operate at Vmax (because [ATP] ≫ Km1); by contrast, site 2 would operate considerably below Vmax ([ATP] < Km2). Why would an enzyme have two active sites but use primarily just one? For some oligomeric enzymes, negative cooperativity gives rise to a “flip-flop” or “alternating sites” mechanism in which equivalent active sites in the assembled protomers are forced to operate one at a time (16, 26, 29). With kinesin dimers, for example, such a mechanism enables the processive “ratcheting” movement of the motor along cytoskeletal structures (13). For other alternating sites enzymes, the advantage of the mechanism is not clear (9), and this is certainly also the case for TmCheA: It is difficult to envision a compelling reason why only one of the two active sites of CheA should catalyze P1 phosphorylation at any given time. Perhaps there are structural constraints in the large CheA/CheW/receptor signaling arrays that make such a mechanism advantageous. Another possibility is that binding of ATP to site 2 contributes to the catalytic efficiency of CheA by stimulating release of product from site 1, for example by promoting dissociation of products (ADP and/or phosphorylated P1) following an autophosphorylation event.
We hope to explore these possibilities in future work. In addition, it will be interesting to determine whether CheA proteins and other PHKs in other prokaryotic systems also exhibit negative cooperativity and asymmetry in their binding interactions with ATP and TNP-ATP in the same manner that we observe with TmCheA. It is interesting to note, for example, that the two active sites of NtrB dimers exhibit asymmetry in their interactions with ADP (24).
Acknowledgments
We thank Drs. Sang-Youn Park and Brian Crane for plasmids and Drs. Michael Gilson, Ann West, and Dorothy Beckett for helpful discussions. Ebele Okwumabua provided expert assistance purifying some of the proteins used in this work.
Footnotes
This work was supported by National Institutes of Health Grant GM052853 to R.C.S.
Abbreviations: ADPNP, 5′-adenylyl-β,γ-imidodiphosphate; ADPCP, 5′-adenyl(β,γ-methylene)diphosphonate; DTT, dithiothreitol; IPTG, isopropyl β-d-thiogalactoside; IMFC, integrated molar fluorescence coefficient (a fluorescence proportionality constant analogous to a molecular extinction coefficient); NTA, nitrilotriacetate; P-CheA, phosphorylated CheA; PHK, protein histidine kinase; TCEP, tris(2-carboxyethyl)-phosphine; TNP-ATP, 2′(3′)-O-(2,4,6-trinitrophenyl)adenosine 5′-triphosphate; λex, excitation wavelength; λmax, wavelength of maximal absorbance; AA, full-length TmCheA dimer that has no TNP-ATP bound; T.AA, CheA dimer to which one molecule of TNP-ATP has bound; T.AA.T, CheA dimer that has bound two molecules of TNP-ATP.
Uncertainties reported are standard deviations for replicate measurements.
Efforts to find information about T. maritima ATP levels in the published literature were unsuccessful. For E. coli, a value of 3 mM is often cited as the cytoplasmic ATP concentration (41); however, this number represents the total ATP pool, and the actual available concentration of ATP might be considerably lower than this, as reflected in the results of an in vivo ATP reporter assay (49).
References
- 1.Bilwes A, Alex L, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 2.Bilwes A, Quezada CM, Croal LR, Crane BR, Simon MI. Nucleotide binding by the histidine kinase CheA. Nat Struct Biol. 2001;8:353–360. doi: 10.1038/86243. [DOI] [PubMed] [Google Scholar]
- 3.Bilwes AMP, Quezada CM, Simon M, Crane BR. Histidine Kinases in Signal Transduction. Academic Press; San Diego: 2003. pp. 48–74. [Google Scholar]
- 4.Borkovich KA, Simon MI. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell. 1990;63:1339–1348. doi: 10.1016/0092-8674(90)90429-i. [DOI] [PubMed] [Google Scholar]
- 5.Bornhorst JA, Falke JJ. Attractant regulation of the aspartate receptor-kinase complex: Limited cooperative interactions between receptors and effects of the receptor modification state. Biochemistry. 2000;39:9486–9493. doi: 10.1021/bi0002737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourret RB, Davagnino J, Simon MI. The carboxy-terminal portion of the CheA kinase mediates regulation of autophosphorylation by transducer and CheW. J Bacteriol. 1993;175:2097–2101. doi: 10.1128/jb.175.7.2097-2101.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown JH. Breaking symmetry in protein dimers: Designs and functions. Protein Sci. 2006;15:1–13. doi: 10.1110/ps.051658406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Creighton TC. Proteins. Structures and Molecular Properties. 2nd. W. H. Freeman and Co.; New York: 1993. pp. 381–382. [Google Scholar]
- 9.DeQueiroz MS, Waldrop GL. Modeling and numerical simulation of biotin carboxylase kinetics: Implications for half-sites reactivity. J Theor Biol. 2007;246:167–175. doi: 10.1016/j.jtbi.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 10.Eaton AK. Ph D Thesis. University of Maryland; College Park, MD: 2008. [Google Scholar]
- 11.Fersht A. Structure and Mechanism in Protein Science. W. H. Freeman and Co.; New York: 1999. pp. 444–449. [Google Scholar]
- 12.Gegner J, Dahlquist FW. Signal transduction in bacteria: CheW forms a reversible complex with the protein kinase CheA. Proc Natl Acad Sci USA. 1991;88:750–754. doi: 10.1073/pnas.88.3.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert SP, Moyer ML, Johnson KA. Alternating site mechanism of the kinesin ATPase. Biochemistry. 1998;37:792–799. doi: 10.1021/bi971117b. [DOI] [PubMed] [Google Scholar]
- 14.Gloor SL, Falke JJ. Thermal domain motions of CheA kinase in solution: Disulfide trapping reveals the motional constraints leading to trans-autophosphorylation. Biochemistry. 2009;48:3631–3644. doi: 10.1021/bi900033r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grebe TW, Stock JB. The histidine protein kinase superfamily. Adv Microb Physiol. 1999;41:139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- 16.Gresser MJ, Myers JA, Boyer PD. Catalytic site cooperativity of beef-heart mitochondrial F, adenosine triphosphatase. Correlations of initial velocity, bound intermediate, and oxygen exchange measurements with an alternating three site model. J Biol Chem. 1982;257:12030–12038. [PubMed] [Google Scholar]
- 17.Hess JF, Bourret RB, Simon MI. Histidine phosphorylation and phosphoryl group transfer in bacterial chemotaxis. Nature. 1988;336:139–143. doi: 10.1038/336139a0. [DOI] [PubMed] [Google Scholar]
- 18.Hess JF, Bourret RB, Simon MI. Phosphorylation assays for proteins of the two-component regulatory system controlling chemotaxis in Escherichia coli. Methods Enzymol. 1991;200:188–204. doi: 10.1016/0076-6879(91)00139-n. [DOI] [PubMed] [Google Scholar]
- 19.Hiratsuka T. Fluorescent and colored trinitrophenylated analogs of ATP and GTP. Eur J Biochem. 2003;270:3479–3485. doi: 10.1046/j.1432-1033.2003.03748.x. [DOI] [PubMed] [Google Scholar]
- 20.Hiratsuka T, Uchida K. Preparation and properties of 2′ (or 3′)-O-(2,4,6-trinitrophenyl) adenosine-5′-triphosphate, an analog of adenosine triphosphate. Biochim Biophys Acta. 1973;320:635–647. doi: 10.1016/0304-4165(73)90143-8. [DOI] [PubMed] [Google Scholar]
- 21.Hirschman A. Ph D Thesis. University of Maryland; College Park, MD: 2002. [Google Scholar]
- 22.Hirschman A, Boukhvalova M, VanBruggen R, Wolfe AJ, Stewart RC. Active site mutations in CheA, the signal-transducing protein kinase of the chemotaxis system in Escherichia coli. Biochemistry. 2001;40:13876–13887. doi: 10.1021/bi0113622. [DOI] [PubMed] [Google Scholar]
- 23.Hoffman A, Roeder RG. Purification of His-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res. 1991;19:6337–6338. doi: 10.1093/nar/19.22.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang P, Peliska JS, Ninfa AJ. Asymmetry in the autophosphorylation of the two-component regulatory system transmitter protein Nitrogen Regulator II. Biochemistry. 2000;39:5057–5065. doi: 10.1021/bi992921w. [DOI] [PubMed] [Google Scholar]
- 25.Kott L, Braswell EH, Shrout AL, Weis RM. Distributed subunit interactions in CheA contribute to dimer stability: A sedimentation equilibrium study. Biochim Biophys Acta. 2004;1696:131–140. doi: 10.1016/j.bbapap.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Kovina MV, Kochetov GA. Cooperativity and flexibility of active sites in homodimeric transketolase. FEBS Lett. 1998;440:81–84. doi: 10.1016/s0014-5793(98)01423-9. [DOI] [PubMed] [Google Scholar]
- 27.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 28.Lakowicz JR. Principles of Fluorescence Spectroscopy. Plenum; New York: 1983. [Google Scholar]
- 29.Lazdunski M, Petitclerc C, Chappelet D, Lazdunski C. Flip-flop mechanisms in enzymology. Eur J Biochem. 1970;20:124–139. doi: 10.1111/j.1432-1033.1971.tb01370.x. [DOI] [PubMed] [Google Scholar]
- 30.Levit M, Liu Y, Surette M, Stock J. Active site interference and asymmetric activation in the chemotaxis protein histidine kinase CheA. J Biol Chem. 1996;271:32057–32063. doi: 10.1074/jbc.271.50.32057. [DOI] [PubMed] [Google Scholar]
- 31.Li J, Swanson RV, Simon MI, Weis RM. The response regulators CheB and CheY exhibit competitive binding to the kinase CheA. Biochemistry. 1995;34:14626–1463. doi: 10.1021/bi00045a003. [DOI] [PubMed] [Google Scholar]
- 32.Lupas A, Stock JB. Phosphorylation of an N-terminal regulatory domain activates the methylesterase in bacterial chemotaxis. J Biol Chem. 1989;264:17337–17342. [PubMed] [Google Scholar]
- 33.McEvoy MM, Hausrath AC, Randolph GB, Remington SJ, Dahlquist FW. Two binding modes reveal flexibility in kinase/response regulator interactions in the bacterial chemotaxis pathway. Proc Natl Acad Sci USA. 1998;95:7333–7338. doi: 10.1073/pnas.95.13.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McEvoy MM, Muhandiram DR, Kay LE, Dahlquist FW. Structure and dynamics of a CheY-binding domain of the chemotaxis kinase CheA determined by nuclear magnetic resonance spectroscopy. Biochemistry. 1996;35:5633–5640. doi: 10.1021/bi952707h. [DOI] [PubMed] [Google Scholar]
- 35.McEvoy MM, Zhou J, Roth AF, Lowry DF, Morrison TB, Kay LE, Dahlquist FW. Nuclear magnetic resonance assignments and global fold of a CheY-binding domain in CheA, the chemotaxis-specific kinase of Escherichia coli. Biochemistry. 1995;34:13871–13880. doi: 10.1021/bi00042a019. [DOI] [PubMed] [Google Scholar]
- 36.Miller SM, Massey V, Williams CH, Jr, Ballou DP, Walsh CT. Communication between the active sites in dimeric mercuric ion reductase: An alternating sites hypothesis for catalysis. Biochemistry. 1991;30:2600–2612. doi: 10.1021/bi00224a006. [DOI] [PubMed] [Google Scholar]
- 37.Morrison TB, Parkinson JS. Liberation of an interaction domain from the phosphotransfer region of CheA, a signaling kinase of Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5485–5489. doi: 10.1073/pnas.91.12.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morrison TB, Parkinson JS. A fragment liberated from the E. coli kinase that blocks stimulatory, but not inhibitory, chemoreceptor signaling. J Bacteriol. 1997;179:5543–5550. doi: 10.1128/jb.179.17.5543-5550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mourey L, Da Re S, Pedelacq JD, Tolstykh T, Faurie C, Guillet V, Stock JB, Samama JP. Crystal structure of the CheA histidine phosphotransfer domain that mediates response regulator phosphorylation in bacterial chemotaxis. J Biol Chem. 2001;276:31074–31082. doi: 10.1074/jbc.M101943200. [DOI] [PubMed] [Google Scholar]
- 40.Nagradova NK. Interdomain interactions in oligomeric enzymes: Creation of asymmetry in homo-oligomers and role in metabolite channeling between active centers of hetero-oligomers. FEBS Lett. 2001;487:327–332. doi: 10.1016/s0014-5793(00)02338-3. [DOI] [PubMed] [Google Scholar]
- 41.Neuhard J, Nygaard P. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. ASM Press; Washington, DC: 1987. pp. 445–473. [Google Scholar]
- 42.Ninfa EG, Atkinson MR, Kamberov ES, Ninfa AJ. Mechanism of autophosphorylation of Escherichia coli Nitrogen Regulator II (NRII or NtrB): trans-phosphorylation between subunits. J Bacteriol. 1993;175:7024–7032. doi: 10.1128/jb.175.21.7024-7032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park SY, Beel BD, Simon MI, Bilwes AM, Crane BR. In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved. Proc Natl Acad Sci USA. 2004;101:11646–11651. doi: 10.1073/pnas.0401038101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 45.Park SY, Quezada CM, Bilwes AM, Crane BR. Subunit exchange by CheA histidine kinases from mesophile Escherichia coli and the thermophile Thermotoga maritima. Biochemistry. 2004;43:2228–2240. doi: 10.1021/bi0352419. [DOI] [PubMed] [Google Scholar]
- 46.Popovych N, Sun S, Ebright RH, Kalodimos CG. Dynamically driven protein allostery. Nat Struct Mol Biol. 2006;13:831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quezada CM, Gradinaru C, Simon MI, Bilwes AM, Crane BR. Helical shifts generate two distinct conformers in the atomic resolution structure of the CheA phosphotransferase domain from Thermotoga maritima. J Mol Biol. 2004;341:1283–1294. doi: 10.1016/j.jmb.2004.06.061. [DOI] [PubMed] [Google Scholar]
- 48.Quezada CM, Hamel DJ, Gradinaru C, Bilwes AM, Dahlquist FW, Crane BR, Simon MI. Structural and chemical requirements for histidine phosphorylation by the chemotaxis kinase CheA. J Biol Chem. 2005;280:39581–30585. doi: 10.1074/jbc.M505316200. [DOI] [PubMed] [Google Scholar]
- 49.Schneider DA, Gourse RL. Relationship between growth rate and ATP concentration in Escherichia coli: A bioassay for available cellular ATP. J Biol Chem. 2004;279:8262–8268. doi: 10.1074/jbc.M311996200. [DOI] [PubMed] [Google Scholar]
- 50.Sourjik V, Berg HC. Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stewart RC. Analysis of ATP binding to CheA containing tryptophan substitutions near the active site. Biochemistry. 2005;44:4375–4385. doi: 10.1021/bi0476026. [DOI] [PubMed] [Google Scholar]
- 52.Stewart RC, Dahlquist FW. N-Terminal half of CheB is involved in methylesterase response to negative chemotactic stimuli. J Bacteriol. 1988;170:5728–5738. doi: 10.1128/jb.170.12.5728-5738.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stewart RC, Jahreis K, Parkinson JS. Rapid phosphotransfer to CheY from a CheA protein lacking the CheY-binding domain. Biochemistry. 2000;39:13157–13165. doi: 10.1021/bi001100k. [DOI] [PubMed] [Google Scholar]
- 54.Stewart RC, VanBruggen R, Ellefson DD, Wolfe AJ. TNP-ATP and TNP-ADP as probes of the nucleotide binding site of CheA, the histidine protein kinase in the chemotaxis signal transduction pathway of Escherichia coli. Biochemistry. 1998;37:12269–12279. doi: 10.1021/bi980970n. [DOI] [PubMed] [Google Scholar]
- 55.Stock J, Surette MG. In: Escherichia coli and Salmonella. Cellular and Molecular Biology. Neidhardt FC, editor. ASM Press; Washington, DC: 1996. pp. 1103–1129. [Google Scholar]
- 56.Swanson RV, Bourret RB, Simon MI. Intermolecular complementation of the kinase activity of CheA. Mol Microbiol. 1993;8:435–441. doi: 10.1111/j.1365-2958.1993.tb01588.x. [DOI] [PubMed] [Google Scholar]
- 57.Swanson RV, Schuster SC, Simon MI. Expression of CheA fragments which define domains encoding kinase, phosphotransfer, and CheY binding activities. Biochemistry. 1993;32:7623–7629. doi: 10.1021/bi00081a004. [DOI] [PubMed] [Google Scholar]
- 58.Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among Bacteria and Archaea. Microbiol Mol Biol Rev. 2004;68:301–319. doi: 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tawa P, Stewart RC. Kinetics of CheA autophosphorylation and dephosphorylation reactions. Biochemistry. 1994;33:7917–7924. doi: 10.1021/bi00191a019. [DOI] [PubMed] [Google Scholar]
- 60.Wadhams GH, Armitage JP. Making sense of it all: Bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1924–1937. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 61.Ward WHJ, Fersht AR. Asymmetry of tyrosyl-tRNA synthetase in solution. Biochemistry. 1988;27:1041–1049. doi: 10.1021/bi00403a029. [DOI] [PubMed] [Google Scholar]
- 62.Ward WHJ, Fersht AR. Tyrosyl-tRNA synthetase acts as an asymmetric dimer in charging tRNA. A rationale for half-of-sites activity Biochemistry. 1988;27:5525–5530. doi: 10.1021/bi00415a021. [DOI] [PubMed] [Google Scholar]
- 63.Welch MCN, Mourey L, Birck C, Samama JP. Structure of the CheY-binding domain of histidine kinase CheA in complex with CheY. Nat Struct Biol. 1997;5:25–29. doi: 10.1038/nsb0198-25. [DOI] [PubMed] [Google Scholar]
- 64.Wolfe AJ, Stewart RC. The short form of the CheA protein restores kinase activity an chemotactic ability to kinase-deficient mutants. Proc Natl Acad Sci USA. 1993;90:1518–1522. doi: 10.1073/pnas.90.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang Y, Inouye M. Intermolecular complementation between two defective mutant signal-transducing receptors of Escherichia coli. Proc Natl Acad Sci USA. 1991;88:11057–11061. doi: 10.1073/pnas.88.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou HJ, McEvoy MM, Lowry DF, Swanson RV, Simon MI, Dahlquist FW. Phosphotransfer and CheY-binding domains of the histidine autokinase CheA are joined by a flexible linker. Biochemistry. 1996;35:433–443. doi: 10.1021/bi951960e. [DOI] [PubMed] [Google Scholar]