Abstract
Objective
GPIHBP1 is an endothelial cell protein that binds lipoprotein lipase (LPL) and chylomicrons. Because GPIHBP1 deficiency causes chylomicronemia in mice, we sought to determine whether some cases of chylomicronemia in humans could be attributable to defective GPIHBP1 proteins.
Methods and Results
Patients with severe hypertriglyceridemia (n=60, with plasma triglycerides above the 95th percentile for age and gender) were screened for mutations in GPIHBP1. A homozygous GPIHBP1 mutation (c.344A>C) that changed a highly conserved glutamine at residue 115 to a proline (p.Q115P) was identified in a 33-year-old male with lifelong chylomicronemia. The patient had failure-to-thrive as a child but had no history of pancreatitis. He had no mutations in LPL, APOA5, or APOC2. The Q115P substitution did not affect the ability of GPIHBP1 to reach the cell surface. However, unlike wild-type GPIHBP1, GPIHBP1-Q115P lacked the ability to bind LPL or chylomicrons (d <1.006 g/mL lipoproteins from Gpihbp1−/− mice). Mouse GPIHBP1 with the corresponding mutation (Q114P) also could not bind LPL.
Conclusions
A homozygous missense mutation in GPIHBP1 (Q115P) was identified in a patient with chylomicronemia. The mutation eliminated the ability of GPIHBP1 to bind LPL and chylomicrons, strongly suggesting that it caused the patient’s chylomicronemia.
Keywords: lipoprotein, lipase, human, chylomicronemia, hypertriglyceridemia, GPIHBP1
Chylomicronemia can be caused by a deficiency of lipoprotein lipase (LPL) or apolipoprotein (apo) CII.1 However, many cases of chylomicronemia in humans are unexplained. In 2007, Beigneux et al2 identified a new potential cause of chylomicronemia, a deficiency of glycosylphosphatidylinositol-anchored high-density lipoprotein–binding protein 1 (GPIHBP1).
Mice lacking GPIHBP1 manifest severe chylomicronemia, even on a low-fat chow diet, with plasma triglycerides >2000 mg/dL.2 GPIHBP1 is found on the luminal surface of capillaries in heart, skeletal muscle, and adipose tissue,2 where the lipolytic processing of triglyceride-rich lipoproteins occurs.3 Transfection of a GPIHBP1 expression vector into CHO cells confers the ability to bind LPL, chylomicrons, as well as apo-AV–phospholipid disks.2 The ability of GPIHBP1-expressing cells to bind LPL and chylomicrons suggested that GPIHBP1 might function as a platform for lipolysis on endothelial cells.2
Two structural features of GPIHBP1 are important in the binding of LPL and chylomicrons. The first is an amino-terminal acidic domain, approximately 25 amino acids in length. Mutant GPIHBP1 proteins lacking all or part of the acidic domain are unable to bind LPL and chylomicrons.4 The second is a Lymphocyte antigen 6 (Ly6) domain. Ly6 motifs, which contain either 8 or 10 cysteines with a characteristic spacing pattern, are found in a number of GPI-anchored proteins, for example CD59 and the urokinase-type plasminogen activator receptor (UPAR).5 When the Ly6 domain of GPIHBP1 is replaced with the Ly6 domain from CD59, the chimeric protein reaches the cell surface but cannot bind LPL, even though the acidic domain of GPIHBP1 is intact.4
All mammalian GPIHBP1 proteins share the acidic domain and the Ly6 domain (with 10 conserved cysteines).6 The highest level of amino acid conservation lies within a portion of the Ly6 domain (residues 101 to 121 in human GPIHBP1, which contains the 6th and 7th cysteines of the Ly6 motif).6
The finding of chylomicronemia in Gpihbp1−/− mice raised the question of whether some cases of chylomicronemia in humans might be caused by GPIHBP1 mutations. Wang and Hegele7 screened GPIHBP1 coding sequences in 160 patients with chylomicronemia and identified only 1 variant, a homozygous G56R mutation in 2 siblings. Residue 56 is located in a linker segment between the acidic domain and the Ly6 domain. Recently, Gin et al8 examined the functional properties of the G56R mutant in CHO cells and could not find any defect in the ability of the mutant protein to reach the cell surface or its ability to bind LPL, chylomicrons, or apo-AV–phospholipids disks. Those findings raised doubts about whether the G56R mutation truly caused the hyperlipidemia.
In this study, we sequenced the coding regions of GPIHBP1 in 60 unrelated adults with unexplained chylomicronemia. One patient, a 33-year-old male, was homozygous for a missense mutation (Q115P) within the most highly conserved portion of the Ly6 domain.6 Cell culture studies revealed that the mutant GPIHBP1 reached the cell surface normally but could not bind LPL or chylomicrons.
Methods
Subjects
Patients with chylomicronemia (n=60; plasma triglycerides, 4464±3366 mg/dL; postheparin plasma LPL mass and activity, 79.5±48.7 ng/mL and 59.9±63.9 mU/mL, respectively) were identified within the Lipid Clinic of the Academic Medical Center Amsterdam (AMC). After excluding mutations in LPL, APOA5, and APOC2, the coding regions of GPIHBP1 (NM_178172) were amplified and sequenced. A homozygous missense mutation in GPIHBP1 (c.344A>C; p.Q115P) was identified in a 33-year-old male with severe lifelong chylomicronemia.
Three normolipidemic age-matched men and an LPL-deficient patient (a compound heterozygote for V69L and G188E LPL mutations) were used as controls. Studies were approved by the Committees on Human Research at AMC and UCLA.
Genomic DNA Analysis
Genomic DNA was prepared from blood leukocytes. The 4 exons of GPIHBP1, along with ≈50 bp of introns on either side, were amplified with primers described in supplemental Table I (available online at http://atvb.ahajournals.org). The exons of LPL, APOA5, and APOC2 were amplified with the primers listed in supplemental Table II. An M13 tail was added to each primer (forward: 5′-GTTGTAAAACGACGGCCACT-3′; reverse: 5′-CACAGGAAACAGCTATGACC-3′) to facilitate DNA sequencing.
Biochemical Measurements
Total plasma cholesterol, triglycerides, high-density lipoprotein (HDL) cholesterol, and low-density lipoprotein (LDL) cholesterol levels were determined with commercial kits (Wako). Plasma apo-B, apo-CII, and apo-CIII levels were measured with commercial assays (Randox). Plasma apo-B48 levels were determined with an ELISA (Shibayagi). Size-fractionation of plasma lipoproteins was performed by fast protein liquid chromatography (FPLC); online triglyceride measurements were obtained with a commercial assay (Biomerieux).9 Blood was obtained before and 18 minutes after an intravenous injection of heparin (50 IU/kg body weight). LPL and hepatic lipase (HL) activity levels were assessed as previously described.10 Plasma LPL mass levels were measured with an ELISA (Daiichi); HL mass levels were also measured with an ELISA.11
GPIHBP1 Constructs and Cell Transfections
Untagged and S-protein–tagged versions of mouse and human GPIHBP1 have been described previously.2 A mouse “D,E(38-48)A” GPIHBP1 mutant (in which the aspartates and glutamates between residues 38 and 48 were changed to alanine) was described previously.4 Human GPIHBP1-Q115P and mouse GPIHBP1-Q114P expression vectors were generated by site-directed mutagenesis with the QuikChange kit (Stratagene). Transient transfections of CHO pgsA-475 cells (a mutant CHO cell line with deficient sulfation of heparan sulfate proteoglycans12) were performed with Lipofectamine 2000 (Invitrogen) or by electroporation with the Nucleofector II aparatus (Lonza).
To determine whether GPIHBP1 proteins reached the cell surface, we assessed the release of GPIHBP1 into the cell culture medium after treating cells with a phosphatidylinositol-specific phospholipase C (PIPLC, 1 U/mL for 1 hour at 37°C).4 GPIHBP1 in the medium and cell extracts was assessed by Western blotting.2
Binding of Human LPL to GPIHBP1-Expressing CHO pgsA-745 Cells
In some experiments, CHO pgsA-745 cells were cotransfected with expression vectors for a V5-tagged human LPL13 (a gift from Dr Mark Doolittle, University of California, Los Angeles) and GPI-HBP1.4 After 24 hours, the cells were incubated for 15 minutes at 37°C in the absence or presence of heparin (1 U/mL). The medium was harvested and cell extracts were collected in RIPA buffer containing complete mini EDTA-free protease inhibitors (Roche). LPL in the medium and GPIHBP1 and LPL in cell extracts were assessed by Western blotting.4
In other experiments, CHO pgsA-745 cells were electroporated with GPIHBP1 (or empty vector) and then incubated for 2 hours at 4°C with 200 μL of conditioned medium from either nontransfected CHO cells or CHO cells that had been stably transfected with a V5-tagged human LPL expression vector.13 In a given experiment, each well was incubated with exactly the same amount of LPL. In some wells, heparin (500 U/mL) was added to the medium. At the end of the incubation period, cells were washed 6 times with ice-cold PBS containing 1.0 mmol/L MgCl2 and 1.0 mmol/L CaCl2. Levels of GPIHBP1 and LPL in cell extracts were assessed by Western blotting.4
The binding of avian LPL to mouse or human GPIHBP1 was performed as described.4 Levels of mouse GPIHBP1 in cell lysates were assessed with a sandwich ELISA using immunopurified rabbit antibodies against GPIHBP1; levels of human GPIHBP1 in cell lysates were assessed by western blotting.
Western Blot Analyses
Proteins were size-fractionated on 4% to 12% Bis-Tris SDS-poly-acrylamide gels, and Western blotting was performed as described.4 The antibody dilutions were 1:250 for a mouse monoclonal antibody against human apo-B; 1:2000 for an IRdye800-conjugated goat antimouse IgG (Li-Cor); 1:500 for a goat polyclonal antibody against the S-protein tag (Abcam); 1:200 for a mouse monoclonal antibody against the V5 tag (Invitrogen); 1:500 for a rabbit polyclonal antibody against β-actin (Abcam); 1:2000 for an IRdye680-conjugated donkey antirabbit IgG (Li-Cor); 1:6000 for an IRdye800-conjugated donkey antigoat IgG (Li-Cor); and 1:500 for an IRdye800-conjugated donkey antimouse IgG (Li-Cor). Antibody binding was detected with an Odyssey infrared scanner (Li-Cor).
Binding of DiI-Labeled Chylomicrons to Transfected Cells
The d<1.006 g/mL lipoproteins from Gpihbp1−/− mice (“chylomicrons”) were labeled with DiI (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate),2 and the binding of the DiI-labeled chylomicrons to human GPIHBP1-expressing CHO pgsA-745 cells was assessed by fluorescence microscopy.2,4
Results
Identification of a Missense Mutation in GPIHBP1 in a Patient With Chylomicronemia
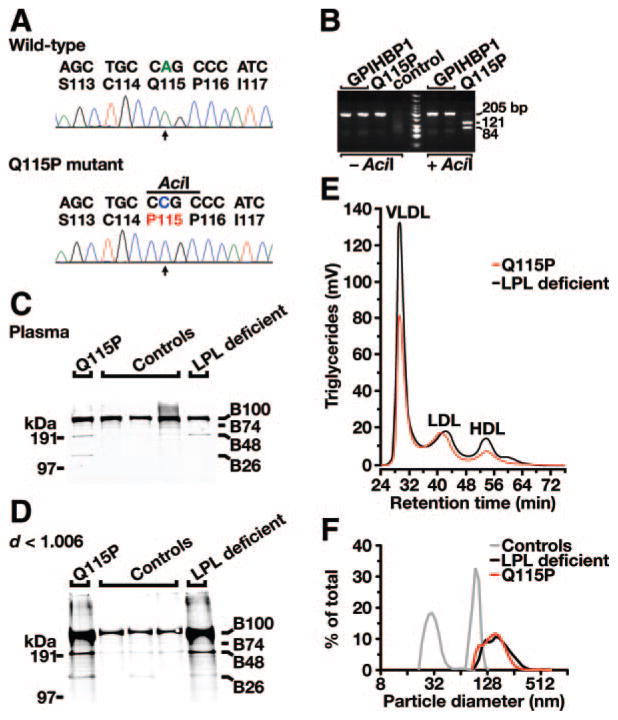
The GPIHBP1 coding sequences were examined in 60 patients with chylomicronemia; none of these patients had coding sequence variations in LPL, APOC2, or APOA5. One noteworthy GPIHBP1 mutation was identified. An A to C transversion in exon 4 of GPIHBP1 (c.344A>C, resulting in a Q115P substitution) was identified in a 33-year-old male with chylomicronemia. The mutation was documented by bidirectional DNA sequencing (Figure 1A) and confirmed by digesting DNA samples with AciI (Figure 1B). The patient was born in Columbia and adopted by a Dutch family. He exhibited hepatosplenomegaly and failure to thrive as a child and was diagnosed with type I hyperlipoproteinemia at the age of 7. The patient had a normal BMI (24.4) and normal glucose levels. Fasting chylomicronemia was documented on multiple occasions. The hyperlipidemia was partially responsive to diet; fasting plasma triglyceride levels fell from as high as 3366 mg/dL to as low as 744 mg/dL when the patient adhered to a fat-free diet (Table). The patient has had lipemia retinalis but had no history of eruptive xanthomas or pancreatitis.
Figure 1.
Identification of a homozygous mutation in GPIHBP1 (p.Q115P) in a young man with chylomicronemia. A, DNA sequence of GPIHBP1 exon 4 from a normolipidemic control subject (wild-type) and the Q115P homozygote (p.Q115P mutant). Nucleotide and amino acid sequences are shown above each chromatogram. The arrow indicates the nucleotide substitution (c.344A>C); this mutation creates a new AciI site. B, AciI digestion confirming the Q115P mutation. A 205-bp fragment of GPIHBP1 was amplified from genomic DNA with primers 5′-CCATCCTCAGCACTTGTTCCCCACTCCCC-3′ and 5′-CCTGCCCCCTTGCCTGTTGGG TCC-3′. AciI cleaves the DNA fragment from the Q115P homozygote. C and D, Increased amounts of apo-B48 in the plasma (C) and in the d<1.006 g/mL lipoproteins (D) of the Q115P proband, as judged by Western blots with an apo-B–specific monoclonal antibody (MB3).20 Samples from normolipidemic subjects and an LPL-deficient patient were included as controls. E, Distribution of triglycerides within the chylomicron-depleted lipoproteins of the proband (Q115P) and an LPL-deficient patient. Plasma lipoproteins were size-fractionated by fast protein liquid chromatography with a Superose 6 HR column. F, Distribution of lipoprotein sizes (as judged by dynamic laser light scattering) in the d<1.006 g/mL lipoproteins from the Q115P homozygote, an LPL-deficient patient, and 2 normolipidemic control subjects. 98.3% of the plasma lipoproteins in the proband had diameters of 94 to 265 nm.
Table.
Plasma Lipid and Apolipoprotein Concentrations in the Proband
| Q115P (n=1) | LPL-Deficient (n=1) | Controls (n=3) | |
|---|---|---|---|
| TG, mg/dl | 744 | 779 | 88.6±26.6 |
| TC, mg/dl | 120 | 108 | 178±34.8 |
| LDLc, mg/dl | 81.2 | 58.0 | 120±19.3 |
| HDLc, mg/dl | 8.9 | 5.5 | 42.5±7.7 |
| Apo-B, mg/dl | 98 | 103 | 117±19 |
| Apo-B48, μg/ml | 35.4 | 30.5 | 5.0±2.0 |
| Apo-CII, mg/dl | 18.5 | 14.6 | 1.5±1.3 |
| Apo-CIII, mg/dl | 17.4 | 18.5 | 7.2±2.1 |
| LPL mass, ng/ml | 36 | 273 | 421±55 |
| LPL activity, mU/ml | 40 | 19 | 275±122 |
| HL mass, ng/ml | 273 | 342 | 335±45 |
| HL activity, mU/ml | 240 | 162 | 180±50 |
Plasma concentrations of triglycerides (TG), total cholesterol (TC), low-density lipoprotein cholesterol (LDLc), high-density lipoprotein cholesterol (HDLc), and of apolipoproteins (apo) in 3 normolipidemic controls, the proband (Q115P), and a patient with a mutation in the catalytic domain of LPL (LPL-deficient). LPL and HL mass and activity were performed on plasma collected 18 minutes after an intravenous heparin injection. Data are presented as mean±SD.
The plasma of the proband contained increased amounts of apo-B48, as judged by Western blots (Figure 1C and 1D) and an ELISA (Table). Size-fractionation of the patient’s plasma revealed that most of the triglycerides were in large lipoproteins (ie, the “VLDL” peak; Figure 1E). The HDL-cholesterol levels were low (Table). The diameters of the patient’s triglyceride-rich lipoproteins were far larger than those of normolipidemic controls (Figure 1F). Postheparin LPL mass levels were 36 ng/mL (≈10% of normal), whereas HL mass levels were normal (273 ng/mL; Table). LPL activity and mass levels were comparably low, indicating that the specific activity of the LPL was normal (Table). Apo-CII levels in the plasma were elevated, compared with normolipidemic controls (18.5 mg/dL versus 7.2±2.1 mg/dL, respectively; Table). Apo-CIII levels were also elevated (Table).
GPIHBP1-Q115P Reaches the Cell Surface Normally But Cannot Bind LPL
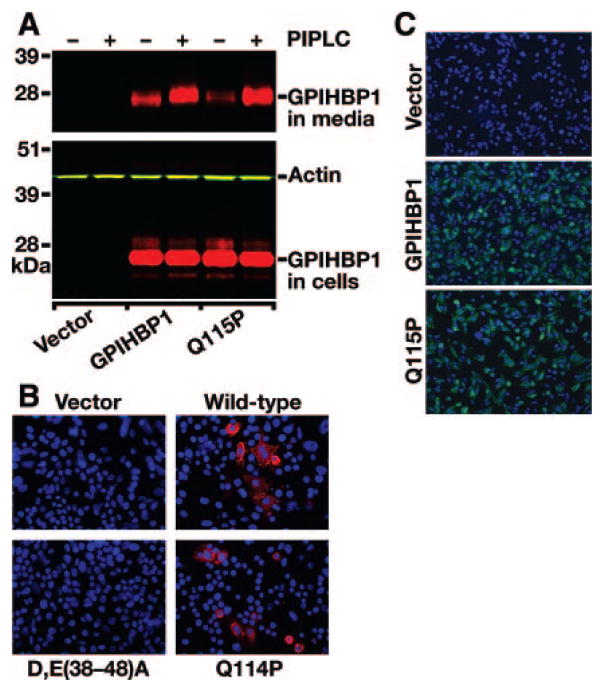
To determine whether the Q115P mutation blocks the ability of GPIHBP1 to reach the cell surface, wild-type and mutant (Q115P) GPIHBP1 constructs were transfected into CHO pgsA-745 cells. We then assessed the release of GPIHBP1 into the culture medium after treating the cells with a phosphatidylinositol-specific phospholipase C (PIPLC). Wild-type GPIHBP1 and GPIHBP1-Q115P yielded similar levels of expression in cells, and similar amounts of GPIHBP1 were released into the cell culture medium by PIPLC, showing that the Q115P mutation did not interfere with the ability of GPIHBP1 to reach the cell surface (Figure 2A). To further explore this issue, we used immunofluorescence microscopy along with an acidic domain antiserum to detect wild-type and mutant versions of GPIHBP1 on the surface of nonpermeabilized cells. For these studies, we examined the expression of wild-type mouse GPIHBP1, a mutant mouse GPIHBP1 with the analogous mutation (Q114P in the mouse sequence), and a mutant GPIHBP1 in which the acidic domain had been mutated (as a negative control).4 In side-by-side experiments, the amounts of wild-type GPIHBP1 and GPIHBP1-Q114P on the surface of cells were comparable (Figure 2B). When similar experiments were carried out with human GPIHBP1 constructs, the amounts of wild-type and mutant (Q115P) GPIHBP1 on the surface of cells were comparable (Figure 2C).
Figure 2.
Wild-type and mutant (Q115P) versions of GPIHBP1 reach the cell surface. A, Release of wild-type or mutant (Q115P) GPIHBP1 from the surface of cells after incubation of cells with a phosphatidylinositol-specific phospholipase C (PIPLC). CHO pgsA-745 cells were transiently transfected with S-protein–tagged GPIHBP1 constructs. The release of GPIHBP1 from the cell surface after PIPLC treatment (1 U/mL for 1 hour at 37°C) was assessed by Western blotting. B, Binding of antibodies against the acidic domain of mouse GPIHBP1 to GPIHBP1-transfected cells. CHO pgsA-745 cells were transfected with empty vector or vectors encoding wild-type mouse GPIHBP1, GPIHBP1-Q114P, or a mouse GPIHBP1 mutant [D,E(38-48)A] in which the aspartates and glutamates between residues 38 and 48 were changed to alanine. GPIHBP1 on the surface of nonpermeabilized cells was assessed by immunofluorescence microscopy with a rabbit antiserum against the acidic domain of mouse GPIHBP14 (red). The GPIHBP1-D,E(38-48)A mutant is not recognized by the antiserum.4 Cell nuclei were visualized with DAPI (blue). C, Binding of antibodies against the S-protein tag to cells expressing human GPIHBP1. CHO pgsA-745 cells were electroporated with empty vector or vectors encoding wild-type mutant (Q115P) GPIHBP1. GPIHBP1 on the surface of nonpermeabilized cells was assessed by immunofluorescence microscopy using a goat antiserum against the S-protein tag (green). Cell nuclei were visualized with DAPI (blue).
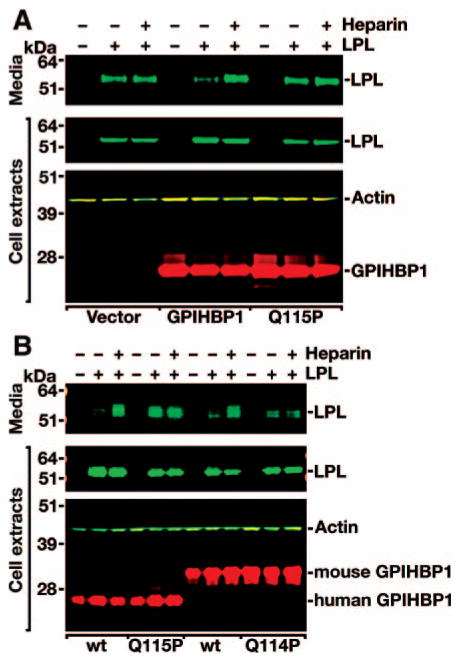
The ability of human GPIHBP1-Q115P to bind LPL was assessed with a Western blot assay. CHO pgsA-745 cells were transfected with a human LPL expression vector, alone or in combination with expression vectors for wild-type or mutant versions of human GPIHBP1. Large amounts of wild-type LPL appeared in the culture medium of cells that did not express GPIHBP1 (Figure 3A). When cells expressed wild-type GPIHBP1, little LPL was detected in the culture medium (because it was bound to GPIHBP1 on the cell surface). However, the GPIHBP1-bound LPL could be released into the medium with heparin (Figure 3A). When the same experiment was performed with the GPIHBP1-Q115P mutant, large amounts of LPL were found in the medium, and little additional LPL was released by heparin, suggesting that GPIHBP1-Q115P binds LPL poorly (Figure 3A). To further explore this issue, we tested the ability of mouse GPIHBP1-Q114P to bind LPL. When CHO pgsA-745 cells were cotransfected with human LPL and wild-type mouse GPIHBP1, little LPL was detected in the cell culture medium (because it was bound to GPIHBP1 on the cell surface), but the LPL could be released into the medium with heparin (Figure 3B). When the same experiment was performed with the GPIHBP1-Q114P mutant, significant amounts of LPL were found in the medium, and no additional LPL was released by heparin, indicating that mouse GPIHBP1-Q114P bound LPL poorly (Figure 3B). In this same experiment, human GPIHBP1-Q115P bound LPL poorly (Figure 3B).
Figure 3.
Decreased binding of LPL to human GPIHBP1-Q115P. A, Western blot representative of 7 independent experiments of cell extracts and cell culture medium from CHO pgsA-745 cells that had been transfected with a V5-tagged human LPL expression vector, alone or in combination with an empty vector, a vector for an S-protein–tagged wild-type human GPIHBP1, or a vector for a mutant human GPIHBP1 with the Q115P mutation. B, Western blot analysis of CHO pgsA-745 cells that had been transfected with a V5-tagged human LPL expression vector, alone or in combination with S-protein–tagged wild-type or mutant (Q115P) human GPIHBP1 constructs, or wild-type or mutant (Q114P) mouse GPIHBP1 constructs.
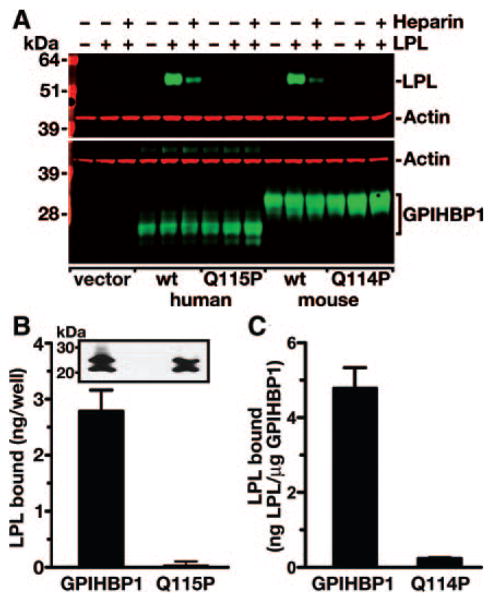
We also assessed the binding of LPL to wild-type and mutant GPIHBP1 with direct binding assays. In these studies, wild-type and mutant versions of GPIHBP1 were transiently expressed into CHO pgsA-745 cells. Cells were incubated with a V5-tagged human LPL for 2 hours at 4°C. After washing the cells, the amount of LPL bound to the cells was assessed by Western blotting. Cells expressing wild-type human or mouse GPIHBP1 bound LPL, but cells expressing mouse GPIHBP1-Q114P or human GPIHBP1-Q115P did not bind LPL (Figure 4A). In similar experiments, we incubated GPIHBP1-expressing cells with purified avian LPL, and the amount of LPL bound to cells was assessed with a monoclonal antibody–based ELISA.14 Cells expressing wild-type human GPIHBP1 bound 39.8-fold more LPL than cells expressing GPIHBP1-Q115P (Figure 4B), and cells expressing wild-type mouse GPIHBP1 bound 20.2-fold more LPL than cells expressing GPIHBP1-Q114P (Figure 4C).
Figure 4.
Decreased binding of exogenously added LPL to cells expressing wild-type or mutant versions of GPIHBP1. A, Binding of human LPL to CHO pgsA-745 cells that had been electroporated with S-protein–tagged wild-type or mutant GPIHBP1 (Q115P) expression vectors. 24 hour after the electroporation, V5-tagged human LPL was added to the cells in the presence or absence of heparin (500 U/mL). After washing the cells, the amount of LPL in cell extracts was assessed by Western blotting. B, Binding of avian LPL to CHO pgsA-745 cells that had been transiently transfected with wild-type or mutant (Q115P) human GPIHBP1 expression vectors. 24 hour after the transfection, cells were incubated for 2 hours with avian LPL (225 ng/well). After washing the cells, the amount of LPL bound to cells was quantified with an ELISA.14 Inset shows a Western blot demonstrating comparable levels of expression for wild-type GPIHBP1 and GPIHBP1-Q115P. C, Binding of avian LPL to CHO pgsA-745 cells that had been transiently transfected with wild-type or mutant (Q114P) mouse GPIHBP1 expression vectors. After incubating the cells for 2 hours with avian LPL (225 ng/well), the cells were washed, and the amount of LPL bound to cells was quantified with a sandwich ELISA. LPL binding was normalized to the amount of GPIHBP1 (as judged by an ELISA) in the same well (wild-type, 1.85±0.16 μg/well; Q114P, 1.87±0.47 μg/well). The studies shown in B and C were performed in 2 entirely independent experiments, and identical results were obtained.
GPIHBP1-Q115P Binds Chylomicrons Poorly
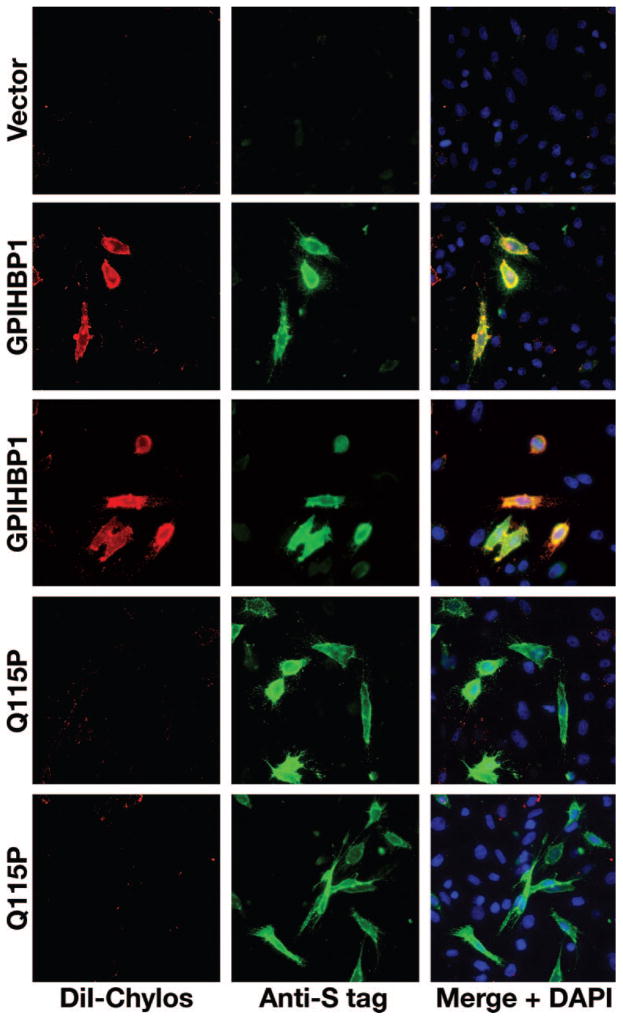
The expression of GPIHBP1 in CHO pgsA-745 cells confers on cells the ability to bind chylomicrons (d<1.006 g/mL lipoproteins from Gpihbp1−/− mice).2 To assess the impact of the Q115P mutation on chylomicron binding, we compared the binding of DiI-labeled chylomicrons to CHO pgsA-745 cells expressing wild-type GPIHBP1 or GPIHBP1-Q115P. Cells expressing wild-type GPIHBP1 bound chylomicrons avidly, whereas cells expressing GPIHBP1-Q115P did not (Figure 5).
Figure 5.
Decreased binding of DiI-labeled chylomicrons to cells expressing GPIHBP1-Q115P. CHO pgsA-745 cells were transfected with wild-type or mutant (Q115P) S-protein–tagged human GPIHBP1 expression vectors. GPIHBP1 on the surface of cells was detected by immunofluorescence microscopy with an antibody against the S-protein tag (green). Chylomicron binding was detected with DiI-fluorescence (red). Cell nuclei were visualized with DAPI (blue).
Discussion
The importance of GPIHBP1 for lipolysis in mice is undeniable; Gpihbp1−/− mice have plasma triglyceride levels of ≈3000 to 5000 mg/dL, similar to mice lacking LPL.15 To determine whether GPIHBP1 mutations might account for some cases of chylomicronemia in humans, we sequenced the coding sequences of GPIHBP1 in 60 unrelated patients with unexplained chylomicronemia. We identified one noteworthy mutation, a homozygous Q115P mutation in a 33-year-old male with Type I hyperlipoproteinemia since childhood. The amino acid substitution was located in the most highly conserved segment of the Ly6 motif of GPIHBP1.6 The Q115P mutation in human GPIHBP1 (and the Q114P mutation in mouse GPIHBP1) eliminated the ability of the molecule to bind human or avian LPL. GPIHBP1-Q115P also lacked the ability to bind chylomicrons. These results firmly establish that GPIHBP1-Q115P is dysfunctional in cell culture assays. Thus, our studies identified striking functional abnormalities in GPIHBP1 in a patient with severe lifelong chylomicronemia. It seems overwhelmingly likely that the Q115P mutation is responsible for the patient’s chylomicronemia and that we have identified the first clinically significant GPIHBP1 mutation in humans. Our findings are important because they indicate that GPIHBP1 is important for lipolysis in humans (as it is in the mouse).
Our studies identified only 1 mutation in 60 patients with chylomicronemia, reinforcing the view that mutations in GPIHBP1 are an uncommon cause of chylomicronemia in humans.7 The fact that the Q115P proband had been adopted meant that no family studies were possible. An analysis of the pedigree would have been interesting, mainly to document the impact of heterozygous GPIHBP1 mutations in humans. In mice, heterozygosity for Gpihbp1 deficiency has no effect on plasma lipid levels.2
The level of LPL in the postheparin plasma of the Q115P patient was low. Levels of LPL were also low in Gpihbp1−/− mice after an intraperitoneal injection of heparin.2 After an intravenous injection of heparin, LPL levels in Gpihbp1−/− mice were low at early time points but then increased to near-normal levels.16 Further study of LPL levels would be interesting but will require the identification of additional GPIHBP1-deficient patients.
Missense mutations in the LDL receptor often lead to retention of the protein in the ER, preventing it from reaching the cell surface.17 We considered the possibility that the Q115P mutation might interfere with the trafficking of GPIHBP1 to the cell surface, but this did not appear to be the case. Wild-type and mutant GPIHBP1 were expressed at comparable levels at the surface of cells, as judged by immunocytochemistry and by PIPLC release.
The fact that a structural alteration in the Ly6 domain would interfere with the binding of ligands is not particularly surprising. In the case of other GPI-anchored Ly6 proteins, for example UPAR or CD59, the Ly6 motif represents the ligand-binding domain,18,19 so it is quite plausible that the Ly6 domain of GPIHBP1 participates in ligand binding. Also, Gin et al4 studied a mutant GPIHBP1 containing the Ly6 domain from CD59. That chimeric protein reached the cell surface normally but was incapable of binding LPL,4 indicating that the Ly6 domain of GPIHBP1 is required for LPL binding. At this point, we do not understand how the Q115P mutation prevents LPL binding, but we suspect that the Q115P substitution alters the structure of the Ly6 motif, preventing direct interactions with LPL.
In summary, we identified a homozygous missense mutation in GPIHBP1 (p.Q115P) in a patient with lifelong chylomicronemia. This amino acid substitution blocked the ability of GPIHBP1 to bind LPL and chylomicrons, suggesting that it was responsible for the patient’s hyperlipidemia. These studies also show that GPIHBP1 is crucial for the lipolytic processing of lipoproteins in humans.
Supplementary Material
Acknowledgments
We thank the patient for participating in the study and H. Levels, A.W. Schimmel, and J.A. Sierts for help with biochemical analyses.
Sources of Funding
This work was supported by a Scientist Development Award from the American Heart Association, National Office (to A.P.B.), P01 HL090553, and R01 HL087228 (to S.G.Y.).
Footnotes
Disclosures
None.
References
- 1.Brunzell JD, Deeb SS. Familial lipoprotein lipase deficiency, apo C-II deficiency, and hepatic lipase deficiency. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill; 2001. [Google Scholar]
- 2.Beigneux AP, Davies B, Gin P, Weinstein MM, Farber E, Qiao X, Peale P, Bunting S, Walzem RL, Wong JS, Blaner WS, Ding ZM, Melford K, Wongsiriroj N, Shu X, de Sauvage F, Ryan RO, Fong LG, Bensadoun A, Young SG. Glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 2007;5:279–291. doi: 10.1016/j.cmet.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Havel RJ, Kane JP. Introduction: Structure and metabolism of plasma lipoproteins. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw-Hill; 2001. [Google Scholar]
- 4.Gin P, Yin L, Davies BSJ, Weinstein MM, Ryan RO, Bensadoun A, Fong LG, Young SG, Beigneux AP. The acidic domain of GPIHBP1 is important for the binding of lipoprotein lipase and chylomicrons. J Biol Chem. 2008;283:29554–29562. doi: 10.1074/jbc.M802579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallya M, Campbell RD, Aguado B. Characterization of the five novel Ly-6 superfamily members encoded in the MHC, and detection of cells expressing their potential ligands. Protein Sci. 2006;15:2244–2256. doi: 10.1110/ps.062242606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Young SG, Davies BS, Fong LG, Gin P, Weinstein MM, Bensadoun A, Beigneux AP. GPIHBP1: an endothelial cell molecule important for the lipolytic processing of chylomicrons. Curr Opin Lipidol. 2007;18:389–396. doi: 10.1097/MOL.0b013e3281527914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Hegele RA. Homozygous missense mutation (G56R) in glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 (GPI-HBP1) in two siblings with fasting chylomicronemia (MIM 144650) Lipids Health Dis. 2007;6:23. doi: 10.1186/1476-511X-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gin P, Beigneux AP, Davies B, Young MF, Ryan RO, Bensadoun A, Fong LG, Young SG. Normal binding of lipoprotein lipase, chylomicrons, and apo-AV to GPIHBP1 containing a G56R amino acid substitution. Biochim Biophys Acta. 2007;1771:1464–1468. doi: 10.1016/j.bbalip.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levels JH, Lemaire LC, van den Ende AE, van Deventer SJ, van Lanschot JJ. Lipid composition and lipopolysaccharide binding capacity of lipoproteins in plasma and lymph of patients with systemic inflammatory response syndrome and multiple organ failure. Crit Care Med. 2003;31:1647–1653. doi: 10.1097/01.CCM.0000063260.07222.76. [DOI] [PubMed] [Google Scholar]
- 10.Kastelein JJ, Jukema JW, Zwinderman AH, Clee S, van Boven AJ, Jansen H, Rabelink TJ, Peters RJ, Lie KI, Liu G, Bruschke AV, Hayden MR. Lipoprotein lipase activity is associated with severity of angina pectoris. REGRESS Study Group. Circulation. 2000;102:1629–1633. doi: 10.1161/01.cir.102.14.1629. [DOI] [PubMed] [Google Scholar]
- 11.Bensadoun A. Sandwich immunoassay for measurement of human hepatic lipase. Methods Enzymol. 1996;263:333–338. doi: 10.1016/s0076-6879(96)63025-0. [DOI] [PubMed] [Google Scholar]
- 12.Esko JD, Stewart TE, Taylor WH. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc Natl Acad Sci U S A. 1985;82:3197–3201. doi: 10.1073/pnas.82.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ben-Zeev O, Mao HZ, Doolittle MH. Maturation of lipoprotein lipase in the endoplasmic reticulum. concurrent formation of functional dimers and inactive aggregates. J Biol Chem. 2002;277:10727–10738. doi: 10.1074/jbc.M108128200. [DOI] [PubMed] [Google Scholar]
- 14.Cisar LA, Hoogewerf AJ, Cupp M, Rapport CA, Bensadoun A. Secretion and degradation of lipoprotein lipase in cultured adipocytes. Binding of lipoprotein lipase to membrane heparan sulfate proteoglycans is necessary for degradation. J Biol Chem. 1989;264:1767–1774. [PubMed] [Google Scholar]
- 15.Merkel M, Weinstock PH, Chajek-Shaul T, Radner H, Yin B, Breslow JL, Goldberg IJ. Lipoprotein lipase expression exclusively in liver. A mouse model for metabolism in the neonatal period and during cachexia. J Clin Invest. 1998;102:893–901. doi: 10.1172/JCI2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinstein MM, Beigneux AP, Davies BS, Gin P, Yin L, Estrada K, Melford K, Bishop JR, Dallinga-Thie GM, Esko JD, Fong LG, Bensadoun A, Young SG. Abnormal patterns of lipoprotein lipase release into the plasma in GPIHBP1-deficient mice. J Biol Chem. 2008;50:34511–34518. doi: 10.1074/jbc.M806067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobbs HH, Brown MS, Goldstein JL. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Hum Mutat. 1992;1:445–466. doi: 10.1002/humu.1380010602. [DOI] [PubMed] [Google Scholar]
- 18.Llinas P, Le Du MH, Gardsvoll H, Dano K, Ploug M, Gilquin B, Stura EA, Menez A. Crystal structure of the human urokinase plasminogen activator receptor bound to an antagonist peptide. EMBO J. 2005;24:1655–1663. doi: 10.1038/sj.emboj.7600635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bodian DL, Davis SJ, Morgan BP, Rushmere NK. Mutational analysis of the active site and antibody epitopes of the complement-inhibitory glycoprotein, CD59. J Exp Med. 1997;185:507–516. doi: 10.1084/jem.185.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curtiss LK, Edgington TS. Immunochemical heterogeneity of human plasma apolipoprotein B. I. Apolipoprotein B binding of mouse hybridoma antibodies. J Biol Chem. 1982;257:15213–15221. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.