Abstract
To examine the influence of insulin-like growth factor (IGF) pathway gene polymorphisms on muscle mass and strength responses to strength training (ST), we studied 128 White and Black men and women before and after a 10-wk single-leg knee extension ST program. One-repetition maximum strength, muscle volume (MV) via computed tomography, and muscle quality (MQ) were assessed at baseline and after 10 wk of ST. There was a significant combined IGF1 cytosine adenine (CA) repeat gene effect, which included both the IGF1 CA repeat main effect and IGF1 CA repeat × PPP3R1 insertion-deletion (I/D) gene × gene interaction effect, on the changes in strength (P < 0.01) and MQ (P < 0.05) with ST. There was a trend for a significant gene × gene interaction between IGF1 CA repeat and PPP3R1 I/D for changes in strength (P = 0.07) and MQ (P = 0.06) with ST. The influence of the PPP3R1 A-202C gene polymorphism on change in MV with ST approached significance (P = 0.06). The IGF1 CA repeat polymorphism had a significant influence on the change in strength and MV combined with ST (P < 0.05), whereas the influence of the PPP3R1 I/D polymorphism approached significance (P = 0.08). There were no associations between the IGFBP3 A-202C gene polymorphism and the muscle phenotypic responses to ST. These data suggest that two of the three IGF pathway gene polymorphisms identified in this study influence muscle phenotypic responses to ST in both Black and White older men and women.
Keywords: genetics, muscle strength, muscle volume, muscle quality
Losses of muscle mass and strength with aging, referred to as sarcopenia, have been well documented (25, 27, 28) and have been associated with many adverse health consequences, including increased mortality (29–31). Strength training (ST) has been shown to be an effective intervention for the prevention and treatment of the adverse consequences of sarcopenia with few side effects (17, 18, 42). Nevertheless, increases in muscle mass and strength are highly variable among individuals (20, 26), suggesting a genetic influence. Further support for a genetic influence on muscle phenotypes comes from twin studies, which have shown that up to 90% of the variance in baseline muscle mass and ∼60% of the variance in baseline muscle strength are heritable (19). Although it can account for a smaller percentage of the variance, response of these muscle phenotypes to ST also appears to be heritable (51). However, there have been few candidate genes identified that influence muscle responses to ST (2, 10, 20, 24, 39), and there are no reports, that we are aware of, on the influence of candidate genes that are linked in a common physiological pathway.
Insulin-like growth factor-I (IGF-I) is a potent mitogen and anabolic agent important in the growth of various body tissues, including skeletal muscle (9, 48). The decline of circulating IGF-I levels with advancing age is related to the losses of muscle mass and strength that occur with age (52). ST increases skeletal muscle IGF-I protein and IGF-I mRNA levels, even in the elderly (8, 12), and IGF-I produced in skeletal muscle can stimulate muscle hypertrophy through activation of satellite cells and increased protein synthesis rates (15, 16). Nevertheless, significant variability in skeletal muscle IGF-I levels have been reported (8, 13).
Some of this variability in response to ST may be accounted for by a cytosine adenine (CA) dinucleotide repeat polymorphism in the promoter region of the IGF1 gene, which encodes for the IGF-I protein (24). It is plausible that other polymorphisms within the IGF pathway of genes could be involved in muscle hypertrophy and strength response to ST, but no reports are available, to our knowledge, on the influences of these genes on muscle phenotypic responses to ST. Two examples of such genes within the IGF pathway are insulin-like growth factor binding protein 3 (IGFBP3) and calcineurin B (PPP3R1).
Most circulating IGF-I is bound by IGFBP-3 (1), which can potentiate or inhibit the action of IGF-I on skeletal muscle (48). Although it is unclear whether IGFBP-3 is the primary carrier of IGF-I in skeletal muscle, there is evidence that it does exist in skeletal muscle (11, 43, 47) and that secretion of IGFBP-3 in primary adult human skeletal muscle cell models is stimulated by IGF-I (11). There have been several reports that the A-202C polymorphism in the promoter region of the IGFBP3 gene can influence the levels of this protein in circulation (3, 22, 46), which may in turn modulate the activity of IGF-I. Deal et al. (3) showed that this polymorphism was directly related to promoter activity of the IGFBP3 gene, suggesting a functional association that potentially affects IGFBP-3 protein levels.
IGF-I-induced muscle hypertrophy in skeletal muscle cells in culture is at least partially mediated by a Ca2+-dependent calcineurin signaling pathway (35, 44). Calcineurin is a Ca2+/calmodulin-dependent protein phosphatase, which consists of a catalytic subunit, designated as calcineurin A, and a regulatory Ca2+-binding subunit, designated as calcineurin B (55). Calcineurin plays a role in both cardiac (34, 40) and skeletal muscle hypertrophy (6, 7, 35, 44, 49). Tang et al. (50) reported that the 5-bp insertion-deletion (I/D) polymorphism in the promoter region of the PPP3R1 gene was associated with the incidence of inappropriately high left ventricular mass in severely hypertensive individuals. Although conflicting results have been reported on the role of calcineurin on skeletal muscle hypertrophy (38, 45), it is conceivable that the PPP3R1 I/D polymorphism may influence hypertrophic responses of skeletal muscle, based on findings in cardiac muscle, given that these two tissues share common hypertrophic pathways (36).
The purpose of the present study was to test the hypothesis that polymorphisms in the promoter regions of the IGF1, IGFBP3, and PPP3R1 genes, which may be linked in a common biological pathway, will significantly influence the changes in muscle volume (MV), strength, and muscle quality (MQ) with ST in older White and Black men and women. To test this association, the CA dinucleotide repeat polymorphism in the promoter region of the IGF1 gene, the −202 locus polymorphism in the promoter region of the IGFBP3 gene, and the 5-bp I/D polymorphism in the promoter region of the PPP3R1 gene were studied.
Methods
Subjects
One hundred twenty-eight previously physically inactive relatively healthy men (n = 58) and women (n = 70) between the ages of 50 and 85 yr volunteered to participate in this study. A small portion of the subjects (n = 10) were from a previous study cohort in our laboratory who underwent the identical ST intervention program (20). Before participation, all subjects underwent a phone-screening interview, received medical clearance from their primary care physicians, and completed a detailed medical history. They were nonsmokers and free of significant cardiovascular disease and metabolic or musculoskeletal disorders that would affect their ability to safely perform heavy-resistance exercise. Subjects who were already taking medications for at least 3 wk before the start of the study were permitted into the study as long as they did not change their medications or dosages at any time throughout the study. After all methods and procedures were explained, subjects read and signed a written consent form that had been approved by the Institutional Review Board of the University of Maryland (College Park, MD). All subjects were continually reminded throughout the study not to alter physical activity levels or dietary habits for the duration of the study. Body weight was monitored weekly throughout the study to ensure compliance in maintaining a stable diet.
Body composition assessment
Body composition was measured by dual-energy x-ray absorptiometry using fan-beam technology (model QDR 4500A, version 8.21 software; Hologic, Waltham, MA). A total body scan was performed at baseline and again after the ST program to assess total body and thigh fat-free mass (FFM), fat mass, and percent body fat as described in a previous study from our laboratory (20). The coefficients of variation (CV) for all dual-energy x-ray absorptiometry measures of body composition were calculated from repeated scans of 10 subjects who were scanned three consecutive times with repositioning. The CV was 0.6% for FFM and 1.0% for percent body fat.
Strength testing
One-repetition maximum (1 RM) strength tests were performed before and after the ST program using an air-powered knee-extension resistance machine (Keiser A-300 leg extension machine) as described previously (20). At least two familiarization training sessions were performed before the 1 RM strength testing and the ST program, in which the exercises were performed with little or no resistance. These low-resistance training sessions were conducted to familiarize the subjects with the equipment, to help control for the large 1-RM strength gains that commonly result from skill (motor learning) acquisition during the initial stages of training, and to help prevent injuries and soreness after the ST program. The criterion for a successful attempt during the strength test was extending the lower leg to a knee joint extension of ∼165°, which was assessed on every trial by a system that turned on a red light each time the leg was extended to this knee joint angle.
ST program
The ST program consisted of unilateral (one-legged) training of the knee extensors of the dominant leg, three times per week, for ∼10 wk on a Keiser A-300 air-powered leg extension machine as described previously (20). The training protocol was specifically designed to elicit near-maximal effort in an individualized manner on all repetitions after warm-up, while maintaining high volume, as we have described previously in detail (5, 24). The untrained control leg was kept in a relaxed position throughout the training program. One-on-one supervision by qualified exercise leaders was provided for all exercise sessions.
MV and MQ
To quantify quadriceps MV, computed tomography imaging of the trained and untrained thighs was performed (GE Lightspeed Qxi; General Electric, Milwaukee, WI) at baseline and during the last week of the 10-wk unilateral ST program with Medical Image Processing, Analysis, and Visualization software (National Institutes of Health, Bethesda, MD) used to analyze all images, as previously described by our group (20). The final MV was calculated with the truncated cone formula, as described previously by our group (53). The changes in MV in the untrained leg (measured pre- and posttraining) were subtracted from the changes in the trained leg (measured before and after) to correct for those factors that were not being investigated in this study (i.e., seasonal variation). In addition, data were used in this analysis from 10 subjects from a previous cohort. All methods for testing and training these subjects were the same as for the cohort in the present study, except that MV was measured by MRI. However, Mitsiopoulos et al. (32) have shown a correlation of 0.99 between computed tomography and MRI for the measurement of skeletal MV. Investigators were blinded to subject identification, date of scan, and training status, for both baseline and after-training analysis. Repeated-measure CV was calculated for each investigator based on axial assessments on 2 separate days. Intrainvestigator CV averaged 1.7 and 2.3%, whereas, interinvestigator CV was <4.3%. MQ for the quadriceps was defined as strength per unit of muscle mass (MV).
Genotyping
Genomic DNA was prepared from EDTA-anticoagulated whole blood samples by standard salting-out procedures (Gentra Systems). The CA microsatellite of IGF1 (rs no. 10665874) was amplified by PCR of genomic DNA using fluorescence-tagged primers (24, 41). The ABI 3100 DNA sequencer (PE Applied Biosystems) and ABI Genescan/Genotyper 2.5 software program (PE Applied Biosystems) were used to determine the genotype of the CA repeat microsatellite in the promoter region of the IGF1 gene. Genotype assignment was based on the method described by Rosen et al. (41) (e.g., 19 CA repeats = 192 bp), in which these authors found the 192 allele to be the most common and thus compared it with other alleles for this microsatellite. Genotyping of the IGFBP3 A-202C polymorphism (rs no. 2854744) was performed by PCR and −202 restriction digest of the PCR product with Alw21I as described by Deal et al. (3). The 5-bp I/D polymorphism located at the −1059 to −1063 loci relative to the transcription site of the calcineurin B gene (PPP1R3) (rs no. 3039851) was genotyped by standard PCR and enzyme digest with AseI as described by Tang et al. (50). Direct sequencing was used to confirm the accuracy of all genotyping methods.
Statistical analyses
All statistical analyses were performed with SAS software (SAS version 9.1, SAS Institute, Cary, NC). Changes in body weight, percent body fat, and FFM with ST within each sex, race, and genotype group were tested by paired t-tests. Fixed-effect linear models were used to test differences in baseline muscle phenotypes (1 RM strength, MV, and MQ) among the categorical variables: sex and hormone replacement therapy status (male, female on hormone replacement therapy, or female not on hormone replacement therapy), race, and genotype groups and to test for differences in the change in muscle phenotypes with ST among sex and hormone replacement therapy status, race, and genotype groups. Initial linear models for each muscle phenotype (dependent variable) included the main effect of the following independent variables: the CA dinucleotide repeat polymorphism of IGF1, the −202 locus polymorphism in the promoter region of IGFBP3, and the 5-bp I/D polymorphism of calcineurin B (PPP3R1). The initial models also included their two-way interactions with each other, as well as with race (White and Black). There were insufficient data (n < 5) for certain genotype by sex and hormone replacement therapy status groups, so these gene by sex and hormone replacement therapy status interactions were not tested in the models.
Because sample sizes for each genotype were a function of different allelic frequencies, the experiment was unbalanced. Therefore, nonindependent sources of variation were removed using a backward elimination process similar to that previously described (14). The final statistical model for the change in strength with ST included baseline strength, age, height, body-mass index, race, a class variable that combined sex difference and hormone replacement therapy status, IGF1 CA repeat polymorphism × PPP3R1 I/D polymorphism interaction, IGFBP3 A-202C polymorphism × race interaction, IGF1 CA repeat polymorphism, IGFBP3 A-202C polymorphism, and PPP3R1 I/D polymorphism. The final statistical model for the change in MV with ST included baseline MV, age, height, body-mass index, race, a class variable that combined sex and hormone replacement therapy status, IGF1 CA repeat polymorphism, IGFBP3 A-202C polymorphism, and PPP3R1 I/D polymorphism. The final statistical model for change in MQ with ST included baseline MQ, age, height, body-mass index, race, a class variable that combined sex and hormone replacement therapy status, IGF1 CA repeat polymorphism × PPP3R1 I/D polymorphism interaction, IGF1 CA repeat polymorphism, IGFBP3 A-202C polymorphism, and PPP3R1 I/D polymorphism. For those final models in which interaction terms were present, the significance of the contribution for the combined gene effect, which included both main effect and interaction effects, was tested by comparing the error term sums of squares for the full model (all gene effects and error term) with the error term sums of squares for the model in which the gene effects of interest were removed from the model. The combined gene effect for a particular gene included the main effect for that gene and its interactions with other genes that were tested. For example, the combined gene effect for IGF1 CA repeat would include the IGF1 CA repeat main effect and either the IGF1 CA repeat × IGFBP3 A-202C interaction or the IGF1 CA repeat × PPP3R1 I/D interaction. Levels of significance for combined gene effects were tested in the cases in which the gene × gene interaction was significant (P < 0.05) or approached significance (P < 0.10). Results are presented as means (SD) for age, height, body weight, percent body fat, and FFM and as least-squares means ± SE for muscle phenotypes.
For all analyses, the initial threshold of significance was set to P < 0.05. Mean comparisons were made by t-tests, with P values adjusted using a Bonferroni correction to reduce the chance of a type I error.
Hardy-Weinberg equilibrium
Genotype distributions were evaluated for conformity to Hardy-Weinberg equilibrium using χ2 tests for the IGFBP3 A-202C and PPP3R1 5-bp I/D loci and using exact tests for the IGF1 CA repeat microsatellite.
Race by genotype
To determine whether data for change in muscle phenotypes with ST for Black and White subjects could be pooled, gene × race interactions were tested in each linear model for the IGF1, IGFBP3, and PPP3R1 gene polymorphisms. For the IGF1 gene polymorphism, there was insufficient data for Black subjects homozygous for the IGF1 192 allele. Therefore, initially for each of the linear models, subjects homozygous for the IGF1 192 allele and subjects heterozygous for the 192 allele were combined so that sufficient data existed to test the IGF1 × race interaction. However, once this interaction term was no longer significant in the model and removed from the model, the IGF1 gene effects were tested with all IGF1 genotype groups (subjects homozygous for the 192 allele, subjects heterozygous for the 192 allele, and noncarriers of the 192 allele). In addition, to control for the potential influence of race on muscle phenotype responses to ST, race was used as a covariate in all linear models.
Percent variability explained by genotype
To estimate the percent variability for the change in strength, MV, and MQ with ST attributable to IGF1 CA repeat, IGFBP3 A-202C, and PPP3R1 I/D genotypes and any relevant gene × gene interactions, the sums of squares of the gene or gene × gene interaction of interest were divided by the sums of squares of all gene effects present in the model and the error sums of squares. With this procedure, it was not possible to determine the contribution of each gene to the gene × gene interaction. Therefore, it was assumed that each gene involved contributes an equal portion to the percent variability.
A multivariate analysis was used to test for the influence of IGF1 CA repeat, IGFBP3 A-202C, and PPP3R1 I/D gene polymorphisms on the combined change of strength and MV with ST.
Results
Allele and genotype frequencies
Table 1 shows the allele and genotype frequencies for the IGF1 CA repeat, IGFBP3 A-202C, and PPP3R1 I/D promoter polymorphisms. The allele and genotype frequencies for the IGF1 CA repeat microsatellite, IGFBP3 A-202C, and PPP3R1 5 bp I/D loci did not differ significantly from Hardy-Weinberg expectations, and these frequencies were similar to those reported previously (3, 50). There was a significant difference between races in the allele frequencies for the IGF1 CA repeat (P < 0.05) and PPP3R1 I/D variant (P < 0.001). There was no significant difference between race in allele frequency for the IGFBP3 A-202C variant. Data for the PPP3R1 DD genotype group were combined with the PPP3R1 I/D genotype group, as D allele carriers, and were compared with data from subjects homozygous for PPP3R1 II because only one subject was a PPP3R1 DD homozygote.
Table 1. IGF1 CA promoter, IGFBP3 A-202C promoter, and PPP3R1 5-base pair I/D promoter allele and genotype frequencies for all subjects.
| IGF1 CA Promoter Gene Polymorphism | |||
|---|---|---|---|
| Total No. (%) | Total No. for White Subjects (%) | Total No. for Black Subjects (%) | |
| Allele | |||
| 192 | 141 (55) | 115 (61) | 26 (39) |
| – | 115 (45) | 75 (39) | 40 (61) |
| Genotype | |||
| 192/192 | 39 (30) | 34 (36) | 5 (15) |
| 192/− | 63 (49) | 47 (49) | 16 (48) |
| −/− | 26 (20) | 14 (15) | 12 (36) |
| IGFBP3 A-202C Promoter Gene Polymorphism | |||
| Total No. (%) | Total No. for White Subjects (%) | Total No. for Black Subjects (%) | |
| Allele | |||
| A | 128 (50) | 90 (47) | 38 (58) |
| C | 128 (50) | 100 (53) | 28 (42) |
| Genotype | |||
| A/A | 33 (26) | 22 (23) | 11 (33) |
| A/C | 62 (48) | 46 (48) | 16 (48) |
| C/C | 33 (26) | 27 (28) | 6 (18) |
| PPP3R1 5-bp I/D Promoter Gene Polymorphism | |||
| Total No. (%) | Total No. for White Subjects (%) | Total No. for Black Subjects (%) | |
| Allele | |||
| I | 227 (89) | 177 (93) | 50 (76) |
| D | 29 (11) | 13 (7) | 16 (24) |
| Genotype | |||
| I/I | 100 (78) | 82 (86) | 18 (55) |
| I/D | 27 (21) | 13 (14) | 14 (42) |
| D/D | 1 (1) | 0 (0) | 1 (3) |
IGF1, insulin-like growth factor 1 gene; IGFBP3, insulin-like growth factor binding protein 3 gene; PPP3R1, calcineurin B gene; CA, cytosine adenine; I/D, insertion-deletion; 192 allele is equivalent to 19 CA repeats; − represents non-192 allele.
Physical characteristics
Tables 2 and 3 show that there were no significant changes in body weight, percent body fat, or FFM with ST within sex or race groups. Similarly, there were no significant within genotype group differences for change in body weight, percent body fat, or FFM with ST, except those with MV data who were IGF1 ca carriers of the 192 allele, who had a significant decrease in percent body fat with ST (P < 0.001). However, these data are not reported in Tables 2 and 3. Men had greater mean values than women for baseline 1 RM strength (P < 0.001), MV (P < 0.001), and MQ (P < 0.01), and Black subjects had greater MV results than White subjects at baseline (P < 0.001).
Table 2. Physical characteristics for all men (n = 58) and women (n = 70) at baseline and after ST.
| Men (n = 53–58) | Women (n = 61–70) | |||
|---|---|---|---|---|
| Baseline | After ST | Baseline | After ST | |
| Age, yr | 65 (8) | 63 (9) | ||
| Height, cm | 174 (7) | 162 (7) | ||
| Weight, kg | 85.8 (13.5) | 86.0 (13.4) | 72.1 (12.6) | 72.3 (13.2) |
| Body fat, % | 28.0 (4.9) | 27.6 (4.6) | 38.6 (5.7) | 38.1 (5.7) |
| Fat-free mass, kg | 61.2 (8.1) | 61.7 (7.9) | 43.8 (5.7) | 44.2 (5.9) |
| 1 RM, kg | 33±1.0† | 41±1.2‡ | 22±1.0 | 27±1.2 |
| MV, cm3 | 1,770±34† | 1,930±38 | 1,330±38 | 1,430±43 |
| MQ, kg/cm3 × 10−3 | 18.8±0.56* | 21.6±0.53 | 16.2±0.63 | 19.2±0.59 |
| Blacks/Whites | 12/46 | 21/49 | ||
Values are means (SD). Values for knee extension one-repetition maximum (1 RM), muscle volume (MV), and muscle quality (MQ) are least square means ± SE. Sample size variability in number of subjects was because of missing data for muscle phenotypes. MQ is strength (1 RM) per unit of muscle mass (MV). ST, strength training.
Significantly greater than for women (P < 0.01);
significantly greater than for women (P < 0.001);
significantly greater increase than the increase in women (P < 0.05).
Table 3. Physical characteristics for all White subjects (n = 95) and Black subjects (n = 33) at baseline and after ST.
| White (n = 85–95) | Black (n = 29–33) | |||
|---|---|---|---|---|
| Baseline | After ST | Baseline | After ST | |
| Age, yr | 65 (8) | 62 (8) | ||
| Height, cm | 168 (9) | 166 (7) | ||
| Weight, kg | 78.2 (15.3) | 78.4 (15.6) | 78.5 (12.7) | 78.9 (12.8) |
| Body fat, % | 34.1 (7.6) | 33.7 (7.5) | 33.0 (7.3) | 32.5 (7.2) |
| Fat-free mass, kg | 51.4 (11.3) | 51.8 (11.3) | 52.6 (10.5) | 53.3 (10.6) |
| 1 RM, kg | 24±0.9 | 29±1.0 | 27±1.2 | 35±1.4 |
| MV, cm3 | 1,380±31 | 1,490±35 | 1,580±39† | 1,700±48 |
| MQ, kg/cm3 × 10−3 | 16.9±0.49 | 19.4±0.46 | 17.1 ±0.66 | 20.7±0.62‡ |
| Men/women | 46/49 | 12/21 | ||
Values are means (SD). Values for 1 RM, MV, and MQ are least square means ± SE. Sample size variability in number of subjects was because of missing data for muscle phenotypes.
Significantly greater than for White subjects (P < 0.001);
significantly greater increase than the increase in White subjects (P < 0.05).
Genotype associations with muscle phenotypes at baseline
There were no significant differences in baseline muscle phenotypes among IGF1 CA repeat, IGFBP3 A-202C, and PPP3R1 I/D genotype groups. The IGFBP3 A-202C × race interaction for baseline 1 RM strength approached significance (P = 0.06), and there was also a significant IGFBP3 A-202C × race interaction for baseline MQ (P < 0.05).
Muscle phenotype responses to ST for sex and race groups
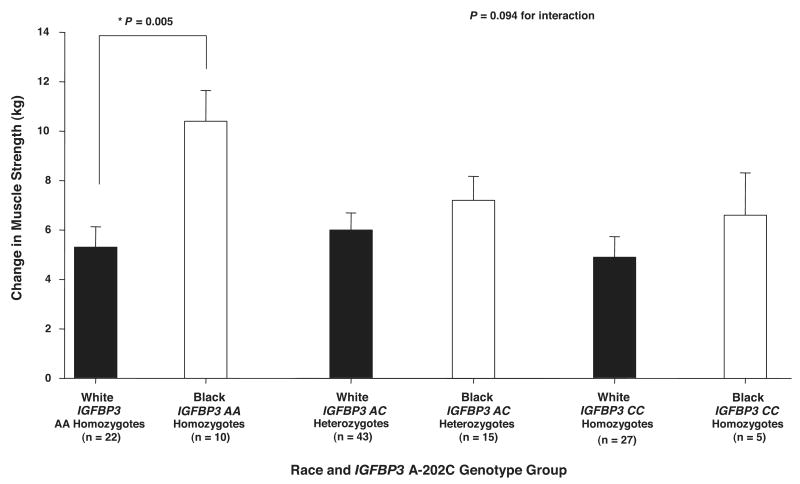
Men had significantly greater absolute (9.2 ± 0.8 vs. 5.1 ± 0.7 kg; P < 0.001) and relative (%) increases (37.0 ± 3.4 vs. 27.0 ± 3.1%; P < 0.05) in knee extension 1 RM strength with ST than women. It was not possible to determine whether there was a significant difference among races for absolute and relative change in strength because of a trend for a significant IGFBP3 × race interaction (see Fig. 2). There was no significant difference between the absolute or relative changes in MV or MQ with ST between men and women. In addition, there were no significant differences between Black and White individuals for absolute or relative changes in MV with ST. There was a significant difference in the absolute change in MQ in Black compared with White individuals (3.5 ± 0.4 vs. 2.6 ± 0.3 kg/cm3 × 10−3; P ≤ 0.04). The relative difference in MQ between Black and White individuals could not be determined because of an IGFBP3 A-202C × race interaction for this phenotype.
Fig. 2.
Influence of insulin-like growth factor binding protein 3 (IGFBP3) A-202C genotype × race groups on change in 1 RM strength with ST. There was a trend for a significant IGFBP3 A-202C gene × race interaction (P = 0.094). Black subjects homozygous for IGFBP3 AA had significantly greater increases in 1 RM strength with ST than White subjects homozygous for IGFBP3 AA (*P = 0.005). Values are covaried for age, hormone replacement therapy status and sex, race, height, body mass index, and baseline 1 RM strength. Values are means ± SE.
Race × sex interactions for change in muscle phenotypes with ST
There was a significant race × sex interaction for change in muscle strength with ST (P < 0.05). However, there were no significant race × sex interactions for changes in MV (P = 0.29) or MQ (P = 0.46).
Race × gene interaction for change in muscle phenotypes with ST
There were no significant gene × race interactions for the changes in 1 RM strength, MV, or MQ with ST, except for a trend for a significant race × IGFBP3 A-202C interaction for change in strength. For this reason, data from Black and White subjects were combined for all other genotype analyses to improve statistical power.
Genotype influence on 1 RM strength responses to ST
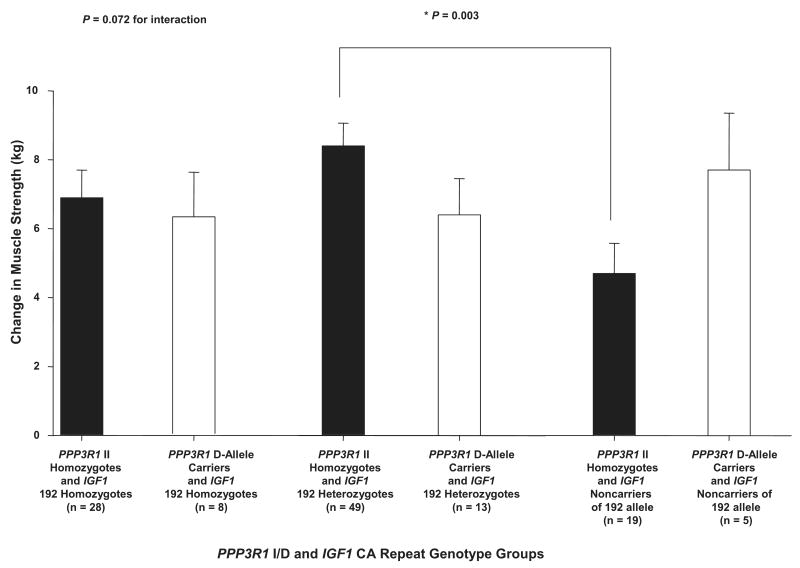
There was a significant combined gene effect, including both IGF1 CA repeat main effect and IGF1 CA repeat × PPP3R1 I/D gene × gene interaction effect, on change in strength with ST (P < 0.01). Table 4 shows that this total gene effect accounted for 3.5% of the variability in change in muscle strength with ST. Figure 1 shows that the gene × gene interaction between IGF1 CA repeat and PPP3R1 I/D alone approached significance for change in strength with ST (P = 0.07). In this case, individuals homozygous and heterozygous for IGF1 192 responded similarly, whereas IGF1 noncarriers of the 192 allele responded differently with respect to PPP3R1 genotype groups. In fact, individuals homozygous for PPP3R1 II who were also heterozygous for 192 allele for IGF1 had significantly greater increases in strength with ST than those homozygous for PPP3R1 II, who were also noncarriers of the 192 allele for IGF1 (8.4 ± 0.7 vs. 4.7 ± 0.9 kg; P < 0.01). However, for PPP3R1 D allele carriers, there was no significant difference in change in strength with ST among IGF1 genotype groups. There was no significant combined gene effect for IGFBP3 A-202C, which included both the IGFBP3 A-202C main effect and the IGFBP3 A-202C × race interaction effect, although the IGFBP3 A-202C × race interaction approached significance (P = 0.09). However, Black individuals who were in the AA genotype group for the IGFBP3 A-202C polymorphism had a significantly greater increase in strength with ST than White individuals who were also homozygous for IGFBP3 AA (10.1 ± 1.2 vs. 5.3 ± 0.8 kg; P < 0.01), whereas there were no significant differences between races for change in strength for the IGFBP3 AC and CC genotype groups (Fig. 2). There was no significant combined gene effect for PPP3R1 I/D, including both PPP3R1 I/D main effect and PPP3R1 I/D × IGF1 CA repeat gene × gene interaction effect, on the change in strength with ST. The IGF1 CA repeat × IGFBP3 A-202C, IGFBP3 A-202C ×PPP3R1 I/D, IGF1 CA repeat × race, and PPP3R1 I/D × race interactions for change in strength with ST were not significant. Also, there was no significant influence of the IGF1 CA repeat, IGFBP3 A-202C, and PPP3R1 I/D main effects on change in strength with ST.
Table 4. Percent variability for muscle phenotypes attributable to IGF1 CA repeat, IGFBP3 A-202C, and PPP3R1 I/D gene polymorphisms.
| Genotype | Individual Source | Combined Gene Effect | P Value |
|---|---|---|---|
| Percent variability for genotype for change in muscle strength with ST | |||
| IGF1 | 1.22 | 3.46 = 1.22 + ½ (4.48) | <0.01 |
| IGFBP3 | 2.83 | 4.84 = 2.83 + ½ (4.02) | >0.05 |
| PPP3R1 | 0.03 | 2.27 = 0.03 + ½ (4.48) | >0.05 |
| IGF1 × PPP3R1 | 4.48 | 0.07 | |
| IGFBP3 × race | 4.02 | 0.09 | |
| Percent variability for genotype for change in MV with ST | |||
| IGF1 | 2.04 | N/A | 0.32 |
| IGFBP3 | 0.16 | N/A | 0.91 |
| PPP3R1 | 3.23 | N/A | 0.06 |
| Percent variability for genotype for change in MQ with ST | |||
| IGF1 | 0.95 | 3.71 = 0.95 + ½ (5.54) | <0.05 |
| IGFBP3 | 1.28 | N/A | 0.51 |
| PPP3R1 | 0.22 | 2.99 = 0.22 + ½ (5.54) | >0.05 |
| IGF1 × PPP3R1 | 5.54 | 0.06 | |
“Combined gene effect” was computed as the main effect plus one-half of any gene × gene interaction or gene × race interaction. For example for IGF1, combined gene effect is the IGF1 main effect (1.22) plus one-half of the IGF1 × PPP3R1 gene × gene interaction [1/2(4.48)]. The other half of the gene × gene interaction is credited to PPP3R1. N/A, combined gene effect could not be determined because there was not at least a trend for a significant gene × gene or gene × race interaction.
Fig. 1.
Influence of calcineurin B (PPP3R1) insertion-deletion (I/D) × insulin-like growth factor 1 (IGF1) cytosine adenine (CA) repeat genotype groups on change in 1-repetition maximum (1 RM) strength with strength training (ST). There was a trend for a significant gene × gene interaction between IGF1 CA repeat and PPP3R1 I/D (P = 0.072). Individuals homozygous for PPP3R1 II who were also heterozygous for IGF1 192 allele had significantly greater increases in 1 RM strength with ST than individuals homozygous for PPP3R1 II who were also IGF1 noncarriers of the 192 allele (*P = 0.003). Values are covaried for age, hormone replacement therapy status and sex, race, height, body mass index, and baseline 1 RM strength. Values are means ± SE.
Genotype influence on MV responses to ST
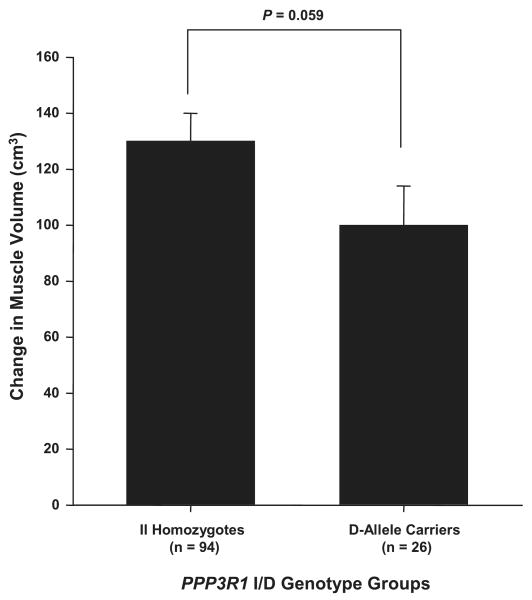
Because of no significant gene × gene or gene × race interactions for change in MV with ST, combined gene effects were not determined. However, Fig. 3 shows that individuals homozygous for II of the I/D polymorphism in the promoter region of the PPP3R1 gene approached significance for having a greater increase in MV with ST than D allele carriers (130 ± 10 vs. 100 ± 14 cm3; P = 0.06). There was no significant influence of the IGF1 CA repeat and IGFBP3 A-202C main effects on change in MV with ST.
Fig. 3.
Influence of calcineurin B (PPP3R1) I/D genotype groups on change in muscle volume (MV) with ST. There was a trend for individuals homozygous for PPP3R1 II to have greater increases in MV with ST than the PPP3R1 D allele carriers (P = 0.059). Values are covaried for age, hormone replacement therapy status and sex, race, height, body mass index, and baseline MV. Values are means ± SE.
Genotype influence on MQ responses to ST
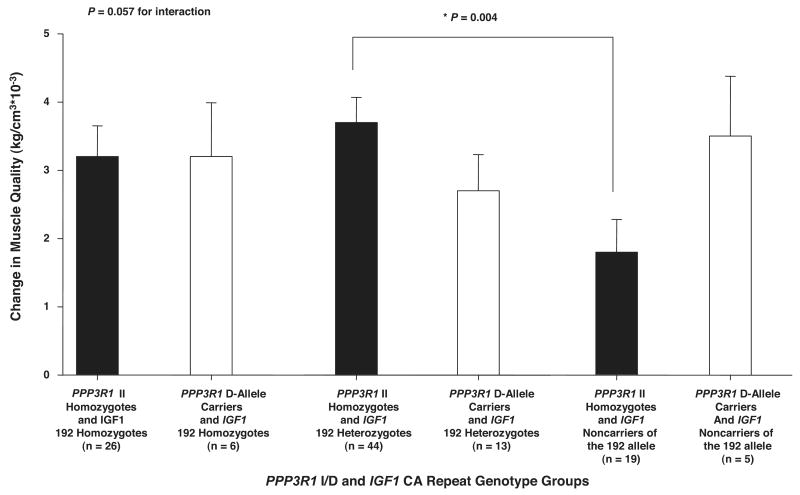
There was a significant combined gene effect for IGF1 CA repeat, including both IGF1 CA repeat main effect and IGF1 CA repeat × PPP3R1 I/D gene × gene interaction effect, for change in MQ with ST (P < 0.05). Figure 4 shows that the IGF1 CA repeat × PPP3R1 I/D gene × gene interaction alone approached significance for change in MQ with ST (P = 0.06). Because of this interaction, there was no consistent difference between PPP3R1 I/D genotype groups in MQ response to ST among IGF1 CA repeat genotype groups. Those who were both homozygous for PPP3R1 II and heterozygous for IGF1 192 allele had a significantly greater increase in MQ with ST than those homozygous for PPP3R1 II who were IGF1 noncarriers of the 192 allele (3.7 ± 0.4 vs. 1.8 ± 0.5 kg/cm3 × 10−3; P < 0.01). However, for PPP3R1 D allele carriers, there was no significant difference in the change in MQ with ST among IGF1 CA repeat genotype groups. There was no significant combined gene effect for PPP3R1 I/D, including both PPP3R1 I/D main effect and PPP3R1 I/D × IGF1 CA repeat gene × gene interaction effect, for change in MQ with ST. Also, the IGF1 CA repeat × IGFBP3 A-202C, IGFBP3 A-202C × PPP3R1 I/D, IGF1 CA repeat × race, IGFBP3 A-202C × race, and PPP3R1 A-202C × race interactions for change in MQ with ST were not significant. Finally, there was no significant influence of the IGF1 CA repeat, IGFBP3 A-202C, and PPP3R1 I/D main effects on change in MQ with ST.
Fig. 4.
Influence of calcineurin B (PPP3R1) I/D × IGF1 CA repeat genotype groups on change in muscle quality (MQ) with ST. The gene × gene interaction between IGF1 CA repeat and PPP3R1 I/D approached significance (P =0.057). Individuals homozygous for PPP3R1 II who were also heterozygous for IGF1 192 allele had significantly greater increases in 1 RM strength with ST than PPP3R1 II homozygote individuals who were also IGF1 noncarriers of the 192 allele (*P = 0.004). Values are covaried for age, hormone replacement therapy status and sex, race, height, body mass index, and baseline MQ. Values are means ± SE.
Gene polymorphism contribution to each muscle phenotype
Table 4 shows the estimated percent variability attributable to IGF1 CA repeat, IGFBP3 A-202C, and PPP3R1 I/D genotypes and to each relevant gene × gene interaction for changes in strength, MV, and MQ with ST. The contributions to the percent variability for change in strength and MQ with ST for the IGF1 CA repeat and PPP3R1 I/D gene × gene interactions was ∼4.5 and 5.5%, respectively. The single gene contributions of IGF1 CA repeat, IGFBP3 A-202C, and PPP3R1 I/D to percent variability for change in strength and MQ were ∼2–5% and 1–4%, respectively. For change in MV with ST, the single gene contributions were 2–3% for IGF1 CA repeat and PPP3R1 I/D, whereas the contribution of IGFBP3 A-202C was <1%.
A correlation matrix analysis resulted in a correlation between change in strength and MV with ST of r = 0.25. Results from a multivariate analysis showed that there was a significant influence of IGF1 CA repeat (P < 0.05) and a nonsignificant trend for an influence of PPP3R1 I/D (P = 0.08) on the combined change in strength and MV with ST.
Discussion
To our knowledge, this is the first report that has investigated the influence of genes linked in a common biological pathway on muscle phenotypic responses to ST. The results offer partial support to the hypothesis that IGF pathway gene polymorphisms, as operationally defined for this study, influence changes in muscle phenotypes with ST and suggest that the IGF1 and PPP3R1 genes may be linked to influence muscle strength and MQ responses to ST.
The major finding of this study was that change in strength with ST was influenced by the combination of gene polymorphisms for IGF1 CA repeat main effects and IGF1 CA repeat × PPP3R1 I/D interaction effects. This combination also influenced change in MQ. This finding was expected, given that the change in MQ is the change in strength per change in MV and that there was only a marginal main effect for PPP3R1 I/D on change in MV. The findings of this study complement those of a previous study from our laboratory (24), which used some of the same subjects who were enrolled in this investigation and found a significant IGF1 CA repeat main effect on the strength response to ST. However, the present study extends these findings by showing that a combined gene effect for IGF1 CA repeat, which includes both IGF1 CA repeat main effect and IGF1 CA repeat × PPP3R1 I/D interaction effect, influenced the strength and MQ responses to ST. In addition, our data support the need for further study to investigate interactions of genes in common biological pathways that may influence skeletal muscle phenotypic responses to ST.
Two previous studies, which investigated individual genes of this biological pathway, have shown an individual influence of the IGF1 dinucleotide repeat polymorphism (24) and the PPP3R1 5-bp I/D polymorphism (50) on muscle phenotypes. In addition, other studies have shown an influence of IGF-I on calcineurin to promote skeletal muscle cell hypertrophy in culture (35, 44). However, these studies did not test the influence of IGF pathway genes on responses to ST. The muscle phenotypes that we investigated in the present study are complex phenotypes and would likely be influenced by several genes in several different pathways. However, we found that the contribution to percent variability attributable to the IGF1 CA repeat × PPP3R1 I/D interaction was ∼4.5% and 5.5%, respectively, for the change in strength and MQ with ST. These contributions are larger than the contributions of single genes (∼2%) reported to influence other muscle phenotypic responses to ST (2).
The fact that we observed no significant combined gene effects for PPP3R1 I/D, including both PPP3R1 I/D main effects and PPP3R1 I/D × IGF1 CA repeat interaction effects, on changes in strength and MQ with ST was unexpected. However, the influence of calcineurin on the muscle phenotypic response to ST may be less than that of IGF-I because of the presence of other IGF-I-linked signaling pathways influencing muscle phenotypic responses to ST. In addition, the role of calcineurin in the muscle phenotypic response to ST remains unclear. Although there have been studies that have suggested calcineurin plays a role in hypertrophy, other findings suggest calcineurin plays an important role in fiber-type switching (33, 38). Nonetheless, in the present study, there was a significant difference between PPP3R1 I/D genotype groups that approached significance for change in MV with ST. Tang et al. (50) reported that the 5-bp I/D polymorphism of the PPP3R1 gene influenced left ventricular muscle mass in White and Black individuals who were severely hypertensive (50). These authors reported that those individuals possessing at least one D allele had a greater risk of developing inappropriately high left ventricular mass than those with two copies of the I allele. The functional significance of the variant (D) allele of the PPP3R1 I/D polymorphism is unknown. However, these authors suggested that this variant eliminates a transcription factor binding site, and they hypothesized that this is an important binding site for a repressor or inhibitor of PPP3R1 transcription. Our results differed from those of Tang et al. (50) in that II homozygotes tended to increase their skeletal muscle mass with overload more than D allele carriers. These discrepancies could be due to 1) differences in the function of calcineurin B, especially for the variant allele, in different tissues, 2) the nature of the load (ST vs. hemodynamic overload) inducing hypertrophy, 3) differences in population being studied, or 4) a combination of two or more of these factors. We are unaware of any other studies that have compared variations at this locus to the response of interventions designed to change muscle mass or strength.
We hypothesized a significant influence of IGFBP3 A-202C because previous studies have shown that levels of IGFBP-3, a major carrier of IGF-I in circulation (1), can be influenced by IGF-I (54), and several studies have shown IGFBP-3 to be present in skeletal muscle. In addition, Foulstone et al. (11) have shown that increased secretion of IGFBP-3 in primary adult human skeletal muscle cells is stimulated by IGF-I (11). However, our results showed no significant influence of the IGFBP3 A-202C gene polymorphism on muscle phenotypic responses to ST, although we did observe a trend for a significant race × IGFBP3 A-202C interaction to influence change in strength. Previous studies have shown that the −202 polymorphism in the promoter region of the IGFBP3 gene influences levels of the IGFBP-3 protein (3, 22, 46), although these studies have shown that several factors can interact with this polymorphism to influence protein levels, including female hormone levels, height, and body-mass index (3, 22, 46). In an in vitro study, Deal et al. (3) showed that the −202 polymorphism influenced the promoter activity of the gene, suggesting the possibility that this polymorphism could influence the levels of the protein in skeletal muscle. However, it is also possible that the isoforms of IGF-I in skeletal muscle may be carried by a different binding protein than IGFBP-3. Therefore, although the −202 locus of the IGFBP3 gene may influence the levels of IGFBP-3 in skeletal muscle, this protein may not be the major potentiator of IGF-I action in skeletal muscle.
In light of the trend for a significant IGFBP3 A-202C × race interaction for influencing change in strength with ST, the influence of IGF pathway gene polymorphisms on responses to ST should be studied more extensively in Black individuals. On the basis of the different frequencies for the IGF1 CA repeat, IGFBP3 A-202C, and PPP3R1 I/D genes between Black and White individuals, it is possible that race effects may have played a greater role than genotype effects for the findings that we observed. For example, Black individuals had a higher frequency of the non-192 allele for the IGF1 CA repeat gene polymorphism than White individuals. In contrast, White individuals had a higher frequency of the variant (C) allele for the IGFBP3 A-202C gene polymorphism than Black individuals. Finally, Black individuals had a higher frequency of the deletion allele for the PPP3R1 I/D gene polymorphism than White individuals. Similar differences between Black and White individuals in allele frequencies for the IGF1 CA repeat and PPP3R1 I/D gene polymorphisms have been observed in previous studies (4, 21, 23, 50). There are no reports that we are aware of on the frequency difference between races for the IGFBP3 A-202C polymorphism.
There are several limitations to the present study. One limitation, due to the sophisticated genetic analysis that is being performed, is low statistical power from small sample size for some genotype groups, particularly for MV and MQ assessments. The lower statistical power for detecting differences among genotype groups for changes in MV and MQ with ST was, in part, due to smaller effect sizes projected for these phenotypes compared with changes in muscle strength. Because of small sample sizes in the groups compared in this study, the possibility that positive false findings could have occurred cannot be ruled out. Additionally, the use of an untrained control leg in the design of the present study may have reduced the effect size needed for MV. However, the use of a control leg allowed for a better assurance that the results represent the independent effects of ST by controlling for variation due to methodological, biological, or seasonal factors. Thus future studies should consider changes in MV and MQ with larger sample sizes to test for gene × gene interactions, as well as to investigate other genes in this biological pathway. Also, the present study is limited in that there is insufficient data to test for sex × gene interactions. Thus the possibility exists that the polymorphisms tested may have a different impact on the muscle phenotypic responses in men and women.
Future studies should be performed with larger sample sizes to better determine the influence of IGF1 CA repeat, IGFBP3 A-202C, and PPP3R1 I/D, especially for gene × gene interactions, on muscle phenotypic responses and to investigate whether other polymorphisms in the IGF pathway play a larger role in influencing muscle phenotypes. For example, it is possible that other polymorphisms in the PPP3R1 gene or a polymorphism in the catalytic subunit of calcineurin may be more responsible for influencing muscle phenotypic responses to ST. Second, there is a need to investigate other IGF-I-dependent mechanical signaling pathways that influence muscle phenotypic responses to ST. For example, it is conceivable that the IGF1-PI3K/Akt/mTOR pathway (37) may compensate for some of the effects of a potentially detrimental allele for the PPP3R1 I/D gene polymorphism. Finally, measurements should be made on transcription and protein levels of the IGF pathway gene polymorphisms investigated in the present study to better understand how they may influence muscle phenotypic responses to ST.
In conclusion, this is the first study to examine the effects of IGF pathway gene polymorphisms on muscle phenotypic responses to ST in older adults. The results suggest that combined IGF1 CA repeat effects, i.e., the main effect for IGF1 CA repeat plus the interaction effect with PPP3R1 I/D, significantly influence muscle phenotypic responses. Although the results from IGF1 CA repeat × PPP3R1 I/D interactions should be interpreted with caution because of the limited sample size for some of the combined genotype groups, they do provide support for the generation of new hypotheses involving IGF1 CA repeat × PPP3R1 I/D interactions that should be tested in future studies. Such studies will provide a better understanding of the role of gene polymorphisms on the responses to ST.
Acknowledgments
Grants: This study was supported by National Institute on Aging Research Contract AG-4-2148 and National Institute on Aging Grants AG-1620501, AG-021500, AG-00268 (training grant), and AG-022791.
References
- 1.Baxter RC, Martin JL, Beniac VA. High molecular weight insulin-like growth factor binding protein complex. J Biol Chem. 1989;264:11843–11848. [PubMed] [Google Scholar]
- 2.Clarkson PM, Devaney JM, Gordish-Dressman H, Thompson PD, Hubal MJ, Urso M, Price TB, Angelopoulos TJ, Gordon PM, Moyna NM, Pescatello LS, Visich PS, Zoeller RF, Seip RL, Hoffman EP. ACTN3 genotype is associated with increases in muscle strength in response to resistance training in women. J Appl Physiol. 2005;99:154–163. doi: 10.1152/japplphysiol.01139.2004. [DOI] [PubMed] [Google Scholar]
- 3.Deal C, Ma J, Wilkin F, Paquette J, Rozen F, Ge B, Hudson T, Stampfer M, Pollak M. Novel promoter polymorphism in insulin-like growth factor-binding protein-3: correlation with serum levels and interaction with known regulators. J Clin Endocrinol Metab. 2001;86:1274–1280. doi: 10.1210/jcem.86.3.7280. [DOI] [PubMed] [Google Scholar]
- 4.DeLellis K, Ingles S, Kolonel L, McKean-Cowdin R, Henderson B, Stanczyk F, Probst-Hensch NM. IGF1 genotype, mean plasma level and breast cancer risk in the Hawaii/Los Angeles multiethnic cohort. Br J Cancer. 2003;88:277–282. doi: 10.1038/sj.bjc.6600728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delmonico MJ, Kostek MC, Doldo NA, Hand BD, Bailey JA, Rabon-Stith KM, Carignan C, Hurley BF. Effects of moderate velocity strength training on peak muscle power and movement velocity: do women respond differently from men? J Appl Physiol. 2005;99:1712–1718. doi: 10.1152/japplphysiol.01204.2004. [DOI] [PubMed] [Google Scholar]
- 6.Dunn SE, Burns JL, Michel RN. Calcineurin is required for skeletal muscle hypertrophy. J Biol Chem. 1999;274:21908–21912. doi: 10.1074/jbc.274.31.21908. [DOI] [PubMed] [Google Scholar]
- 7.Dunn SE, Chin ER, Michel RN. Matching of calcineurin activity to upstream effectors is critical for skeletal muscle growth. J Cell Biol. 2000;151:663–672. doi: 10.1083/jcb.151.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiatarone Singh MA, Ding M, Manfredi TJ, Solares GS, O'Neill EF, Clements KM, Ryan ND, Kehayias JJ, Fielding RA, Evans WJ. Insulin-like growth factor I in skeletal muscle after weight-lifting exercise in frail elders. Am J Physiol Endocrinol Metab. 1999;277:E135–E143. doi: 10.1152/ajpendo.1999.277.1.E135. [DOI] [PubMed] [Google Scholar]
- 9.Florini JR, Ewton DZ, Coolican SA. Growth hormone and insulin-like growth factor system in myogenesis. Endocr Rev. 1996;17:481–517. doi: 10.1210/edrv-17-5-481. [DOI] [PubMed] [Google Scholar]
- 10.Folland J, Leach B, Little T, Hawker K, Myerson S, Montgomery H, Jones D. Angiotensin-converting enzyme genotype affects the response of human skeletal muscle to functional overload. Exp Physiol. 2000;85:575–579. [PubMed] [Google Scholar]
- 11.Foulstone EJ, Savage PB, Crown AL, Holly JM, Stewart CE. Role of insulin-like growth factor binding protein-3 (IGFBP-3) in the differentiation of primary human adult skeletal myoblasts. J Cell Physiol. 2003;195:70–79. doi: 10.1002/jcp.10227. [DOI] [PubMed] [Google Scholar]
- 12.Hameed M, Lange KH, Andersen JL, Schjerling P, Kjaer M, Harridge SD. The effect of recombinant human growth hormone and resistance training on IGF-I mRNA expression in the muscles of elderly men. J Physiol. 2004;555:231–240. doi: 10.1113/jphysiol.2003.051722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hameed M, Orrell RW, Cobbold M, Goldspink G, Harridge SD. Expression of IGF-I splice variants in young and old human skeletal muscle after high resistance exercise. J Physiol. 2003;547:247–254. doi: 10.1113/jphysiol.2002.032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrix LJ, Carter MW, Scott DT. Covariance analyses with heterogeneity of slopes in fixed models. Biometrics. 1982;38:641–650. [PubMed] [Google Scholar]
- 15.Hill M, Goldspink G. Expression and splicing of the insulin-like growth factor gene in rodent muscle is associated with muscle satellite (stem) cell activation following local tissue damage. J Physiol. 2003;549:409–418. doi: 10.1113/jphysiol.2002.035832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill M, Wernig A, Goldspink G. Muscle satellite (stem) cell activation during local tissue injury and repair. J Anat. 2003;203:89–99. doi: 10.1046/j.1469-7580.2003.00195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurley BF, Redmond RA, Pratley RE, Treuth MS, Rogers MA, Goldberg AP. Effects of strength training on muscle hypertrophy and muscle cell disruption in older men. Int J Sports Med. 1995;16:378–384. doi: 10.1055/s-2007-973024. [DOI] [PubMed] [Google Scholar]
- 18.Hurley BF, Roth SM. Strength training in the elderly: effects on risk factors for age-related diseases. Sports Med. 2000;30:249–268. doi: 10.2165/00007256-200030040-00002. [DOI] [PubMed] [Google Scholar]
- 19.Huygens W, Thomis MA, Peeters MW, Vlietinck RF, Beunen GP. Determinants and upper-limit heritabilities of skeletal muscle mass and strength. Can J Appl Physiol. 2004;29:186–200. doi: 10.1139/h04-014. [DOI] [PubMed] [Google Scholar]
- 20.Ivey FM, Roth SM, Ferrell RE, Tracy BL, Lemmer JT, Hurlbut DE, Martel GF, Siegel EL, Fozard JL, Metter EJ, Fleg JL, Hurley BF. The effects of age, gender, and myostatin genotype on the hypertrophic response to strength training and detraining. J Gerontol A Biol Sci Med Sci. 2000;55:M641–M648. doi: 10.1093/gerona/55.11.m641. [DOI] [PubMed] [Google Scholar]
- 21.Jernstrom H, Chu W, Vesprini D, Tao Y, Majeed N, Deal C, Pollak M, Narod SA. Genetic factors related to racial variation in plasma levels of insulin-like growth factor-1: implications for premenopausal breast cancer risk. Mol Genet Metab. 2001;72:144–154. doi: 10.1006/mgme.2000.3130. [DOI] [PubMed] [Google Scholar]
- 22.Jernstrom H, Deal C, Wilkin F, Chu W, Tao Y, Majeed N, Hudson T, Narod SA, Pollak M. Genetic and nongenetic factors associated with variation of plasma levels of insulin-like growth factor-1 and insulin-like growth factor-binding protein-3 in healthy premenopausal women. Cancer Epidemiol Biomarkers Prev. 2001;10:377–384. [PubMed] [Google Scholar]
- 23.Kato I, Eastham J, Li B, Smith M, Yu H. Genotype-phenotype analysis for the polymorphic CA repeat in the insulin-like growth factor-I (IGF-I) gene. Genet Epidemiol. 2003;18:203–209. doi: 10.1023/A:1023379100539. [DOI] [PubMed] [Google Scholar]
- 24.Kostek MC, Delmonico MJ, Reichel JB, Roth SM, Douglass L, Ferrell RE, Hurley BF. Muscle strength response to strength training is influenced by insulin-like growth factor 1 (IGF-1) genotype in older adults. J Appl Physiol. 2005;98:2147–2154. doi: 10.1152/japplphysiol.00817.2004. [DOI] [PubMed] [Google Scholar]
- 25.Larsson L, Grimby G, Karlsson J. Muscle strength and speed of movement in relation to age and muscle morphology. J Appl Physiol. 1979;46:451–456. doi: 10.1152/jappl.1979.46.3.451. [DOI] [PubMed] [Google Scholar]
- 26.Lemmer JT, Hurlbut DE, Martel GF, Tracy BL, Ivey FM, Metter EJ. Age and gender responses to strength training and detraining. Med Sci Sports Exerc. 2000;32:1505–1512. doi: 10.1097/00005768-200008000-00021. [DOI] [PubMed] [Google Scholar]
- 27.Lindle RS, Metter EJ, Lynch NA, Fleg JL, Fozard JL, Tobin J, Roy TA, Hurley BF. Age and gender comparisons of muscle strength in 654 women and men aged 20–93 yr. J Appl Physiol. 1997;83:1581–1587. doi: 10.1152/jappl.1997.83.5.1581. [DOI] [PubMed] [Google Scholar]
- 28.Lynch NA, Metter EJ, Lindle RS, Fozard JL, Tobin JD, Roy TA, Fleg JL, Hurley BF. Muscle quality I. Age-associated differences between arm and leg muscle groups. J Appl Physiol. 1999;86:188–194. doi: 10.1152/jappl.1999.86.1.188. [DOI] [PubMed] [Google Scholar]
- 29.Metter EJ, Talbot LA, Schrager M, Conwit R. Skeletal muscle strength as a predictor of all-cause mortality in healthy men. J Gerontol A Biol Sci Med Sci. 2002;57:B359–B365. doi: 10.1093/gerona/57.10.b359. [DOI] [PubMed] [Google Scholar]
- 30.Metter EJ, Talbot LA, Schrager M, Conwit R. Arm-cranking muscle power and arm isometric muscle strength are independent predictors of all-cause mortality in men. J Appl Physiol. 2004;96:814–821. doi: 10.1152/japplphysiol.00370.2003. [DOI] [PubMed] [Google Scholar]
- 31.Miller MD, Crotty M, Giles LC, Bannerman E, Whitehead C, Cobiac L, Daniels LA, Andrews G. Corrected arm muscle area: an independent predictor of long-term mortality in community-dwelling older adults? J Am Geriatr Soc. 2002;50:1272–1277. doi: 10.1046/j.1532-5415.2002.50316.x. [DOI] [PubMed] [Google Scholar]
- 32.Mitsiopoulos N, Baumgartner RN, Heymsfield SB, Lyons W, Gallagher D, Ross R. Cadaver validation of skeletal muscle measurement by magnetic resonance imaging and computerized tomography. J Appl Physiol. 1998;85:115–122. doi: 10.1152/jappl.1998.85.1.115. [DOI] [PubMed] [Google Scholar]
- 33.Miyazaki M, Hitomi Y, Kizaki T, Ohno H, Haga S, Takemasa T. Contribution of the calcineurin signaling pathway to overload-induced skeletal muscle fiber-type transition. J Physiol Pharmacol. 2004;55:751–764. [PubMed] [Google Scholar]
- 34.Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musaro A, McCullagh KJ, Naya FJ, Olson EN, Rosenthal N. IGF-1 induces skeletal muscle myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature. 1999;400:581–585. doi: 10.1038/23060. [DOI] [PubMed] [Google Scholar]
- 36.Olson EN, Williams RS. Remodeling muscles with calcineurin. Bioassays. 2000;22:510–519. doi: 10.1002/1521-1878(200011)22:11<1049::AID-BIES14>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 37.Pallafacchina G, Calabria E, Serrano AL, Kalhovde JM, Schiaffino S. A protein kinase B-dependent and rapamycin-sensitive pathway controls skeletal muscle growth but not fiber type specification. Proc Natl Acad Sci USA. 2002;99:9213–9218. doi: 10.1073/pnas.142166599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parsons SA, Millay DP, Wilkins BJ, Bueno OF, Tsika GL, Neilsen JR, Liberatore CM, Yutzey KE, Crabtree GR, Tsika RW, Molkentin JD. Genetic loss of calcineurin blocks mechanical overload-induced skeletal muscle fiber type switching but not hypertrophy. J Biol Chem. 2004;279:26192–26200. doi: 10.1074/jbc.M313800200. [DOI] [PubMed] [Google Scholar]
- 39.Riechman SE, Balasekaran G, Roth SM, Ferrell RE. Association of interleukin-15 and interleukin-15 receptor genetic variation with resistance exercise training responses. J Appl Physiol. 2004;97:2214–2219. doi: 10.1152/japplphysiol.00491.2004. [DOI] [PubMed] [Google Scholar]
- 40.Ritter O, Hack S, Schuh K, Rothlein N, Perrot A, Osterziel KJ, Schulte HD, Neyses L. Calcineurin in human heart hypertrophy. Circulation. 2002;105:2265–2269. doi: 10.1161/01.cir.0000016044.19527.96. [DOI] [PubMed] [Google Scholar]
- 41.Rosen CJ, Kurland ES, Vereault D, Adler RA, Rackoff PJ, Craig WY, Witte S, Rogers J, Bilezikian JP. Association between serum insulin growth factor-I (IGF-I) and a simple sequence repeat in IGF-1 gene: implications for genetic studies of bone mineral density. J Clin Endocrinol Metab. 1998;83:2286–2290. doi: 10.1210/jcem.83.7.4964. [DOI] [PubMed] [Google Scholar]
- 42.Roth SM, Ferrell RE, Hurley BF. Strength training for the prevention and treatment of sarcopenia. J Nutr Health Aging. 2000;4:143–155. [PubMed] [Google Scholar]
- 43.Schuller AG, Groffen C, van Neck JW, Zwarthoff EC, Drop SL. cDNA cloning and mRNA expression of the six mouse insulin-like growth factor binding proteins. Mol Cell Endocrinol. 1994;104:57–66. doi: 10.1016/0303-7207(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 44.Semsarian C, Wu MJ, Ju YK, Marciniec T, Yeoh T, Aleen DG, Harvey RP, Graham RM. Skeletal muscle hypertrophy is mediated by a Ca2+ -dependent calcineurin signaling pathway. Nature. 1999;400:576–581. doi: 10.1038/23054. [DOI] [PubMed] [Google Scholar]
- 45.Serrano AL, Murgia M, Pallafacchina G, Calabria E, Coniglio P, Lomo T, Schiaffino S. Calcineurin controls nerve activity-dependent specification of slow skeletal muscle fibers but not muscle growth. Proc Natl Acad Sci USA. 2001;98:13108–13113. doi: 10.1073/pnas.231148598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slattery ML, Baumgartner KB, Byers T, Guiliano A, Sweeney C, Herrick J, Curtin K, Murtaugh M, Wolff R. Genetic, anthropometric, and lifestyle factors associated with IGF-1 and IGFBP-3 levels in Hispanic and non-Hispanic white women. Cancer Causes Control. 2005;16:1147–1157. doi: 10.1007/s10552-005-0318-2. [DOI] [PubMed] [Google Scholar]
- 47.Spangenburg EE, Abraha T, Childs TE, Pattison JS, Booth FW. Skeletal muscle IGF-binding protein-3 and -5 expressions are age, muscle, and load dependent. Am J Physiol Endocrinol Metab. 2003;284:E340–E350. doi: 10.1152/ajpendo.00253.2002. [DOI] [PubMed] [Google Scholar]
- 48.Stewart CE, Rotwein P. Growth, differentiation, and survival: multiple physiological functions for insulin-like growth factors. Physiol Rev. 1996;76:1005–1026. doi: 10.1152/physrev.1996.76.4.1005. [DOI] [PubMed] [Google Scholar]
- 49.Talmadge RJ, Otis JS, Rittler MR, Garcia ND, Spencer SR, Lees SJ, Naya FJ. Calcineurin activation influences muscle phenotype in a muscle-specific fashion. BMC Cell Biol. 2004;5:1–12. doi: 10.1186/1471-2121-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang W, Arnett DK, Devereux RB, Panagiotou D, Province MA, Miller MB, de Simone G, Gu C, Ferrell RE. Identification of a novel 5-base pair deletion in calcineurin B (PPP3R1) promoter region and its association with left ventricular hypertrophy. Am Heart J. 2005;150:845–851. doi: 10.1016/j.ahj.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 51.Thomis MA, Beunen GP, Maes HH, Blimkie CJ, Leemputte MV, Claessens AL, Marchal G, Willems E, Vlietinek RF. Strength training: importance of genetic factors. Med Sci Sports Exerc. 1998;30:724–731. doi: 10.1097/00005768-199805000-00013. [DOI] [PubMed] [Google Scholar]
- 52.Toogood AA, Shalet SM. Ageing and growth hormone status. Bailleres Clin Endocrinol Metab. 1998;12:281–296. doi: 10.1016/s0950-351x(98)80023-2. [DOI] [PubMed] [Google Scholar]
- 53.Tracy BL, Ivey FM, Hurlbut D, Martel GF, Lemmer JT, Siegel EL, Metter EJ, Fozard JL, Fleg JL, Hurley BF. Muscle quality. II. Effects of strength training in 65- to 75-yr-old men and women. J Appl Physiol. 1998;86:195–201. doi: 10.1152/jappl.1999.86.1.195. [DOI] [PubMed] [Google Scholar]
- 54.Villafuerte BC, Zhang WN, Phillips LS. Insulin and insulin-like growth factor-I regulate hepatic insulin-like growth factor binding protein-3 by different mechanisms. Mol Endocrinol. 1996;10:622–630. doi: 10.1210/mend.10.6.8776722. [DOI] [PubMed] [Google Scholar]
- 55.Wang MG, Yi H, Guerini D, Klee CB, McBride OW. Calcineurin A alpha (PPP3CA), calcineurin A beta (PPP3CB) and calcineurin B (PPP3R1) are located on human chromosomes 4, 10q21→q22 and 2p16→p15, respectively. Cytogenet Cell Genet. 1996;72:236–241. doi: 10.1159/000134198. [DOI] [PubMed] [Google Scholar]