Abstract
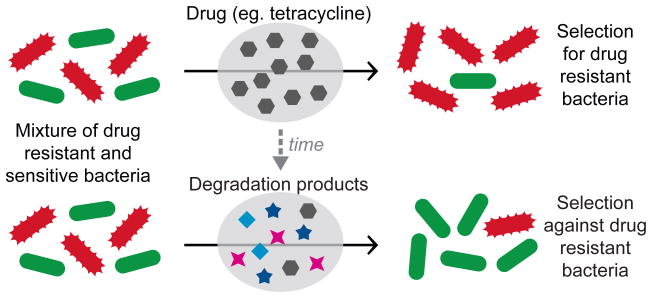
Antibiotics are often unstable, decaying into various compounds with potential biological activities. We found that as tetracycline degrades, the competitive advantage conferred to bacteria by resistance not only diminishes, but reverses to become a prolonged disadvantage due to the activities of more stable degradation products. Tetracycline decay can lead to net selection against resistance, which may help explain the puzzling coexistence of sensitive and resistant strains in natural environments.
More than half of all known antibiotics are secreted by soil bacteria1, mediating communication2,3, metabolism4,5 and warfare6. While resistance to these naturally-occurring antibiotics is prevalent in the soil environment, the genes conferring resistance do not seem to take over and fixate in these natural populations; instead resistant and sensitive bacterial strains coexist7. It is therefore likely that while antibiotics select for resistant strains, other natural mechanisms might exist which select against resistance. Indeed, several natural chemicals are known to specifically inhibit growth of strains resistant to certain antibiotics8,9. The ability of any compound to select for or against resistance depends not only on the selective pressure it exerts, but also on the duration of its activity, determined by its chemical stability. Many antibiotics are short-lived in the natural environment; they decay to an assortment of chemical species which may be more stable than the precursor drug, and may therefore have significant ecological impacts. Thus, competition between antibiotic resistant and sensitive strains may be influenced both by the short-term effect of an antibiotic and by the potential long-term effects of its degradation products (Fig. 1a). Here we ask how the chemical decay of tetracycline influences selection for resistant strains. Tetracycline is widely used clinically10 and agriculturally11, its major degradation pathway is well characterized12, and its decay products are found in soil and wastewater11,13. One of its decay products, anhydrotetracycline, is known to preferentially inhibit the growth of bacteria carrying the Tn10 tetracycline resistance determinant, by binding the tetR regulator to induce expression of the costly tetA efflux pump14–17. We investigated the selective advantage/disadvantage of resistance throughout the degradation process, by directly competing fluorescently labeled tetracycline resistant and sensitive strains of Escherichia coli.
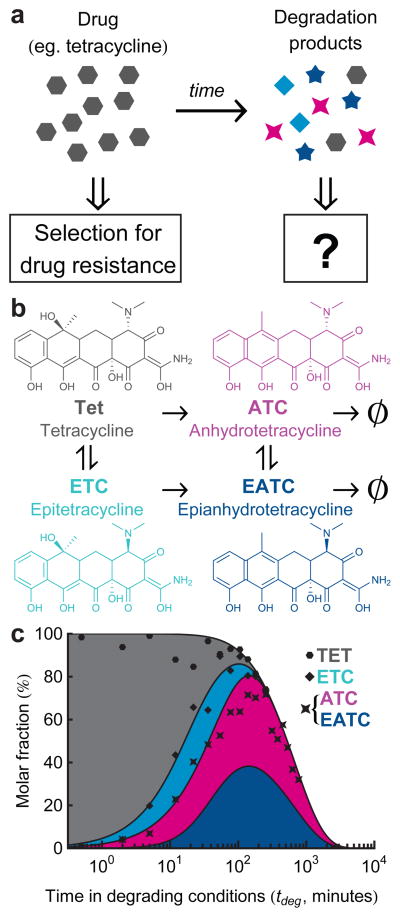
Figure 1. Tetracycline degrades into a range of longer lived compounds, with potential ecological impacts on selection for resistance.
a, While an antibiotic selects for strains resistant to it, it is not clear what selective pressure is imposed by its soup of degradation products. b, Tetracycline degrades into a range of bioactive compounds, which themselves slowly decay further. c, Tetracycline decay products have different concentration profiles through time. Degradation is accelerated by pH of 1.5 and temperature of 75°C [12]. Shaded areas in this stacked plot represent the kinetic model of Ref. 12 with a correction for long-term decay (Supplementary Methods). Points are estimated fractions of Tet and its degradation products, obtained by fitting the spectra of pure compounds to a spectrum of the degraded Tet solution at each individual timepoint (Supplementary Methods). ATC and EATC are not well distinguished spectrally, and so are plotted as their sum. These fitted points confirm the consistency of our samples with the kinetic model of Ref. 12.
Tetracycline (Tet, 1) undergoes reversible epimerization to epitetracycline (ETC, 2) and also irreversible dehydration to anhydrotetracycline (ATC, 3), with both epimerization and dehydration yielding epianhydrotetracycline (EATC, 4)12 (Fig. 1b). To accelerate the degradation process to convenient timescales we exposed tetracycline to phosphoric acid and high temperatures (Ref. 12; Supplementary Supplementary Methods). At different time-points of exposure to these degrading conditions (tdeg), samples of the chemical reaction were taken and the reaction was stopped by shifting to neutral pH and freezing. To track the abundance of tetracycline and its degradation products over time, we measured the absorbance spectrum of each sample and compared it to the spectra of the individual compounds (Supplementary Fig. 1). A previously established kinetic model12, extended to account for the loss of the degradation products at very long timescales, was fully consistent with the spectral data (Fig. 1c; Supplementary Methods and Supplementary Figs. 2–4).
To measure the selective pressure for tetracycline resistance imposed by samples of Tet that had been exposed to degrading conditions for different times, we used a fluorescence-based competition assay between resistant and sensitive E. coli18,19. Matching Tet resistant (TetR) and sensitive (TetS) strains were generated by supplying MG1655 with a plasmid carrying the tetR-tetA genes from the Tn10 transposon, or with the non-resistant parental plasmid, respectively20. These TetS and TetR strains were differentially labeled with chromosomally encoded cyan and yellow fluorescent proteins; pairs of strains were constructed in both dye permutations. Direct competition between the strains, as well as high-resolution measurements of their individual growth rates21, showed equal fitness of the TetS and TetR strains in the absence of Tet (Supplementary Fig. 5). To measure selection for or against resistance, TetS and TetR strains were mixed 1:1 in fresh media, a sample of untreated or degraded Tet was added, and the cultures were grown overnight to stationary phase; the final ratio of sensitive cell count (NTetS) to resistant cell count (NTetR) was measured by flow cytometry (Fig. 2a).
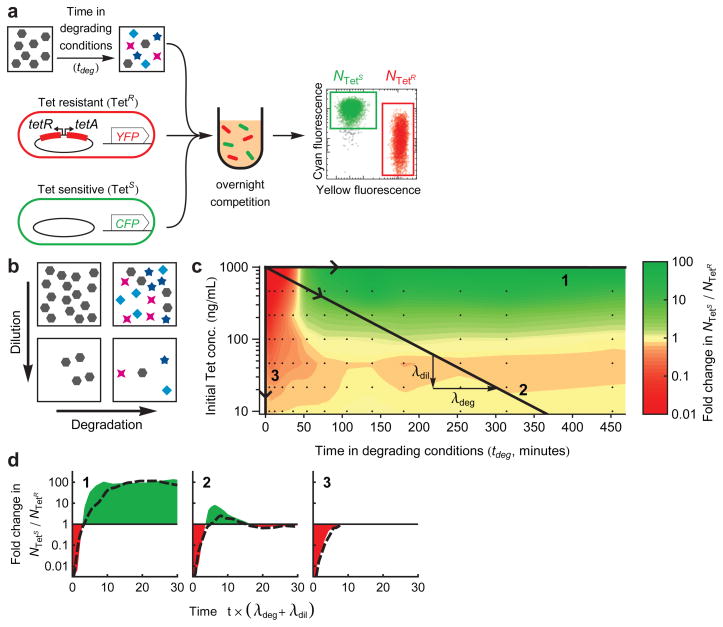
Figure 2. Tetracycline degradation inverts the overall selective advantage of resistant strains.
a, To measure selection for/against resistance by degraded tetracycline solution, a sample of the degradation reaction is taken at timepoint tdeg and is added to a 1:1 mixture of resistant (TetR) and sensitive (TetS) cells inoculated into fresh media. Fluorescent labels (YFP or CFP) allow changes in the ratio NTetS/NTetR to be measured by flow cytometry, after overnight competition. b, Loss of the initial drug can occur by either degradation to alternate compounds (across x-axis), or by dilution (down y-axis). c, Selective pressure in favor (red) or against (green) resistance as a function of the degradation time tdeg and dilution (axes definitions match panel b). Black points mark measurements, between which the color map is interpolated. Numbered black lines are trajectories representing Tet loss by degradation alone (1), dilution alone (3), or a combination of both with respective rates λdeg and λdil (2). d, Selective pressure changes over time as Tet is lost along the three trajectories of panel c. Shaded areas represent the integrated selective pressure in favor of (red) or against (green) resistance. The time axis is normalized by net rate of Tet loss (λdeg + λdil). Dotted black lines are an additive model of selective pressure, constructed by summing the changes in log(NTetS/NTetR) produced by each of the individual compounds (Supplementary Fig. 8), given their concentrations from the kinetic model of Tet decay (Fig. 1c).
While Tet strongly selects in favor of resistance, we found that its cocktail of degradation products actually selects in favor of sensitivity (Fig. 2c,d, trajectory 1). Solutions of Tet with little or no exposure to degrading conditions (tdeg < ~50 min), applied at high concentrations (1000 ng/mL), strongly favored the growth of resistant bacteria. However, following substantial degradation (tdeg > ~50 min), not only did the loss of Tet abolish selection for resistance, but the accumulation of its degradation products caused strong selection against resistance (Fig. 2c,d, trajectory 1). Importantly, while the initial selection in favor of resistance was short lived, the subsequent selection in favor of sensitivity lasted for much longer.
We consider the full ecological impact of an antibiotic as the selective pressure of the antibiotic and its degradation products integrated over time. This integrated selective pressure is represented by the area between the curve of the fold changes in log(NTetS/NTetR) and the line log(NTetS/NTetR) =0 (no selection) (Fig. 2d). When degradation is the primary means of loss (trajectory 1), the initial selection for resistance by Tet is greatly outweighed by the subsequent longer selection against resistance by more stable degradation products (Fig. 1c).
In a natural scenario, an initial drug dosage may not only be lost due to degradation, but also due to dilution, or diffusion (Fig. 2b). To account for dilution in addition to degradation, we applied our TetR-TetS competition assay across a 2D gradient created by serial dilution of each of the time samples of the degradation reaction (Fig. 2c). Dilution of the drug and its degradation products can profoundly affect the overall selection pressure: if Tet loss is dominated by dilution, degradation products do not appear at substantial concentration and so the selective pressure of the initial compound, Tet, is dominant, favoring resistance (Fig. 2c,d, trajectory 3). Tet clearance in the clinical setting and in treatment of farm animals is dominated by dilution rather than degradation22, consistent with the selective advantage of resistance in these settings23.
More generally, we envision that in the environment a drug may be lost simultaneously by both degradation and dilution. An environment initially containing a fixed dose of Tet will then move through different chemical environments along a linear trajectory across Figure 2c, with a slope defined by the ratio between the dilution rate and the degradation rate (−λdil/λdeg, where λdil, λdeg are the reciprocals of the drug’s half-life due to dilution and degradation, respectively). We find that when both degradation and dilution are active, selective pressure can vary over time in a complex non-monotonic manner (Figure 2c, d, trajectory 2). Since degradation products can select in opposing directions depending upon their concentration, net selection depends non-trivially upon both the means of loss and the initial concentration of antibiotic (Supplementary Fig. 6).
We next asked to what extent these complex trajectories of selective effects, exerted throughout Tet decay, can be rationalized in terms of the individual selective pressures exerted by the drug and each of its degradation products. We measured the effect on the TetS-TetR competition imposed by Tet, ETC, ATC and EATC as a function of concentration, and found that each compound has a different selective impact, with ATC selecting against resistance (Supplementary Fig. 7). This observation is consistent with the known action of ATC as a strong inducer of the costly tet operon15,16 and adds to the growing evidence of signaling roles for antibiotics2,3. Drug combinations may produce effects not explicable by the sum of individual drug effects. We adopt the Bliss definition of additivity, where the effect of drugs in combination is equal to the multiplication of their individual effects24. To test for non-additive drug interactions, we measured selective pressures across a 2D gradient of Tet vs. a 1:1 mixture of ATC and EATC, chosen to approximately represent the chemical environments encountered following Tet decay (epimerization leads to nearly equal amounts of ATC and EATC at late times, Fig. 1c). We found that Bliss additivity reproduced all features of the measured 2D gradient, suggesting that interactions amongst Tet and the decay products ATC and EATC are primarily additive (Supplementary Fig. 8).
An additive model of the selective pressures throughout Tet degradation and dilution was then constructed from the kinetic model of chemical composition (Fig. 1c, Supplementary Fig. 3) and the selective effects of each of the individual compounds (Supplementary Fig. 7). This additive model shows very good qualitative and quantitative agreement with the measured selective pressures along trajectories 1, 2 and 3 (Fig. 2d). Selection by Tet and its degradation products can therefore be understood as the additive sum of the effects of each of the compounds.
In the natural environment, antibiotics are not static - a single drug can decay into a range of compounds, each accumulating and degrading with different kinetics and displaying different selective effects. The simple additive sum of the effects of each of the degradation products can lead to complex, non-monotonic, patterns of selections for and against resistance. Consequently, the net evolutionary impact of a drug depends upon the manner of its eventual loss from the environment. When Tet loss is dominated by degradation, the initial selection for resistance by tetracycline can be substantially outbalanced by the prolonged selection against resistance imposed by its longer lived degradation products. While these results were demonstrated for accelerated degradation of tetracycline, they depend on relative, rather than absolute, stabilities of the drug and its degradation products, and therefore may be of relevance also to the natural environment. Interestingly, ATC is a biosynthetic precursor to Tet in the drug producing microbes25, and induces Tet efflux pump expression prior to the imminent production of the drug. This provides an evolutionary rationale for non-toxic drug derivatives to be potent inducers of resistance genes. It will be interesting to test the selective effect of decay of other drugs on various resistance mechanisms and via multiple decay pathways; different decay pathways will produce different metabolites, which could be affected by the environment and even by other surrounding microbes5. Selection against resistance by antibiotic decay may help explain the puzzling coexistence of antibiotic resistant and sensitive microbial strains in the natural soil environment.
Supplementary Material
Figure 3.
Acknowledgments
We thank R. Lenski for gift of plasmids, R. Chait, D. Kahne for helpful insights and R. Ward and M. Elowitz for comments on the manuscript. This work was supported in part by National Institutes of Health Grant R01 GM081617 (to R.K.) and a George Murray Scholarship (to A.C.P.).
Footnotes
Author Contributions
A.C.P., E.A. and R.K designed research; A.C.P. performed research and analyzed data; A.C.P. and R.K. wrote the manuscript.
Competing Financial Interests Statement
The authors declare that they have no competing interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and/or discussion reported in this article.
References
- 1.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. John Innes Foundation; Norwich, UK: 2000. [Google Scholar]
- 2.Yim G, Wang HH, Davies J. Philos Trans R Soc Lond B Biol Sci. 2007;362:1195–200. doi: 10.1098/rstb.2007.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Linares JF, Gustafsson I, Baquero F, Martinez JL. Proc Natl Acad Sci U S A. 2006;103:19484–9. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price-Whelan A, Dietrich LE, Newman DK. Nat Chem Biol. 2006;2:71–8. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 5.Dantas G, Sommer MO, Oluwasegun RD, Church GM. Science. 2008;320:100–3. doi: 10.1126/science.1155157. [DOI] [PubMed] [Google Scholar]
- 6.Walsh C. Antibiotics: actions, origins, resistance. ASM Press; Washington, D.C: 2003. p. 335. [Google Scholar]
- 7.D’Costa VM, McGrann KM, Hughes DW, Wright GD. Science. 2006;311:374–377. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 8.Halling-Sorensen B, Sengelov G, Tjornelund J. Arch Environ Contam Toxicol. 2002;42:263–71. doi: 10.1007/s00244-001-0017-2. [DOI] [PubMed] [Google Scholar]
- 9.Bochner BR, Huang HC, Schieven GL, Ames BN. J Bacteriol. 1980;143:926–33. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chopra I, Hawkey PM, Hinton M. J Antimicrob Chemother. 1992;29:245–77. doi: 10.1093/jac/29.3.245. [DOI] [PubMed] [Google Scholar]
- 11.Sarmah AK, Meyer MT, Boxall AB. Chemosphere. 2006;65:725–59. doi: 10.1016/j.chemosphere.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 12.Yuen PH, Sokoloski TD. J Pharm Sci. 1977;66:1648–1650. doi: 10.1002/jps.2600661143. [DOI] [PubMed] [Google Scholar]
- 13.Jia A, Xiao Y, Hu J, Asami M, Kunikane S. J Chromatogr A. 2009;1216:4655–62. doi: 10.1016/j.chroma.2009.03.073. [DOI] [PubMed] [Google Scholar]
- 14.Moyed HS, Nguyen TT, Bertrand KP. J Bacteriol. 1983;155:549–556. doi: 10.1128/jb.155.2.549-556.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckert B, Beck CF. J Bacteriol. 1989;171:3557–3559. doi: 10.1128/jb.171.6.3557-3559.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lederer T, et al. Biochemistry. 1996;35:7439–7446. doi: 10.1021/bi952683e. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen TN, Phan QG, Duong LP, Bertrand KP, Lenski RE. Mol Biol Evol. 1989;6:213–225. doi: 10.1093/oxfordjournals.molbev.a040545. [DOI] [PubMed] [Google Scholar]
- 18.Hegreness M, Shoresh N, Hartl D, Kishony R. Science. 2006;311:1615–1617. doi: 10.1126/science.1122469. [DOI] [PubMed] [Google Scholar]
- 19.Chait R, Craney A, Kishony R. Nature. 2007;446:668–671. doi: 10.1038/nature05685. [DOI] [PubMed] [Google Scholar]
- 20.Lenski RE, et al. Mol Ecol. 1994;3:127–135. doi: 10.1111/j.1365-294x.1994.tb00113.x. [DOI] [PubMed] [Google Scholar]
- 21.Kishony R, Leibler S. J Biol. 2003;2:14. doi: 10.1186/1475-4924-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly RG, Buyske DA. J Pharmacol Exp Ther. 1960;130:144–9. [PubMed] [Google Scholar]
- 23.Levy SB, FitzGerald GB, Macone AB. N Engl J Med. 1976;295:583–588. doi: 10.1056/NEJM197609092951103. [DOI] [PubMed] [Google Scholar]
- 24.Bliss CI. Annals of Applied Biology. 1939;26:585–615. [Google Scholar]
- 25.McCormick JR, Jensen ER, Johnson S, Sjolander NO. J Am Chem Soc. 1968;90:2201–2. doi: 10.1021/ja01010a063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.