Abstract
All organisms contain thousands of proteins and peptides in their body fluids. A deeper insight into the functional relevance of these polypeptides under different physiological and pathophysiological conditions and the discovery of specific peptide biomarkers would greatly enhance diagnosis and therapy of specific diseases. The low-molecular-weight proteome, also termed peptidome, provides a rich source of information. Due to its unique features, the technical challenges differ somewhat from those in “common” proteomics. In this manuscript, we focus on the low-molecular-weight urinary proteome. We review the methodological aspects of sample collection, preparation, analysis, and subsequent data evaluation. In the second part of this review, we summarize the recent progress in the definition and identification of clinically relevant polypeptide markers.
Keywords: Urine, mass spectrometry, clinical proteomics, biomarker, body fluids
Basic considerations
Proteins and peptides (polypeptides) in body fluids play an important role in physiology. A deeper insight into the functional relevance of polypeptides under different physiological and pathophysiological conditions is one of the main challenges in proteome research [1,2,3,4,5,6]. Changes (alterations in concentrations or modifications) may reflect normal and/or pathological processes. Consequently, some polypeptides could serve as biomarkers of specific diseases. These surrogate biomarkers would have the potential to greatly improve diagnostic testing and monitoring the response to therapy, and perhaps also aid drug development.
In contrast to polypeptides in tissues and most types of cells, the polypeptides in body fluids are relatively easily accessible. Among various body fluids, the urine is an especially attractive source of information. One of the first attempts to define the urinary proteome was published by Spahr et al. [7,8]. Using LC-MS, tryptic peptides of pooled urine samples were analyzed and 124 proteins were identified. While this study did not attempt to define any urinary biomarkers for a disease, it clearly highlighted the plethora of information in the urinary proteome and also indicated a possible approach towards its mining. In a very recent study, Adachi et al. identified more than 1,500 proteins (or their fragments) in the urine of healthy individuals, further underlining the complexity of the human urinary proteome [9].
Several advantages make urine a preferred choice for biomarker discovery:
Urine can be obtained in large quantities using non-invasive procedures. Consequently, ample material is available for analysis, as well as assessment of reproducibility or improvement in sample preparation protocols. In addition, repeated sampling of urine from the same individual is usually easy due to the excellent cooperation of the subjects.
Urine generally contains proteins and peptides of mostly lower molecular mass that are highly soluble. These lower-molecular-weight compounds (< 30 kDa) can be analyzed “as is”, avoiding any additional manipulation (e.g., tryptic digests, see below). Furthermore, only modest attention has been paid to solubilization, a process with a major influence on the proteomic analysis of cells or tissues.
In general, peptides and small proteins in the urine are relatively stable, probably due to the fact that urine is “stored” for hours in the bladder, hence proteolytic degradation by endogenous proteases may be essentially complete by the time of voiding. This is in sharp contrast to blood, for which activation of proteases (and consequently generation of an array of proteolytic breakdown products) is inevitably associated with its collection [10,11]. In two independent sets of experiments, Schaub et al. [12] and Theodorescu et al. [13] showed that the urinary proteome did not change significantly when urine was stored up to 3 days at 4°C or up to 6 hours at room temperature. In addition, urine can be stored for several years at −20°C without significant alteration of its proteome. However, these considerations may not apply to specialized applications, such as the recently described urinary exosomes that may be less stable than the rest of the urinary proteome [14].
Changes (both physiological and pathophysiological) in the genitourinary tract and the kidney are reflected by changes in the urinary proteome. Hence, biomarkers in the urine defined by these molecular changes would enable diagnosis of disease as well as assessment of disease progression or response to therapy.
A disadvantage of urine, in contrast to other body fluids, is the wide variation in protein concentration, due, in part, to differences in the daily intake of fluid. However, this shortcoming can be countered by standardization based on creatinine [15] or peptides generally present in urine [16]. Another potential drawback is the inconsistency of the pH that may alter the activity of proteases and consequently lead to greater variability in the concentration or composition of particular peptide fragments.
Definition of disease-specific biomarkers in urine, and most likely in other compartments, is complicated by significant changes in the proteome during the day. These changes are likely caused by common factors such as variations in the diet, metabolic or catabolic processes, circadian rhythms, exercise, as well as circulatory levels of various hormones. [17]. Consequently, the reproducibility of any assay is reduced by these physiological changes, even if the analytical method shows high reproducibility. However, these variations are limited to a part of the urinary proteome; a basal or “housekeeping” portion that remains unaffected by these processes facilitates the analysis.
One of the first reports on specific urinary proteomic biomarkers was published in 1979 by Anderson et al. [18]. Currently, single biomarkers in body fluids are routinely analyzed in hospitals and clinical laboratories and serve as parameters with variable discriminative values. However, for most diseases, highly sensitive and specific single markers have not been defined. Therefore, the focus is shifting from methods to identify a single biomarker to the simultaneous analysis of a set of markers that form a disease-specific pattern. This approach is likely to improve the sensitivity and specificity of diagnosis, thus increasing its reliability. However, such an approach is also more susceptible to errors and bias; hence, special attention must be paid to the statistical analysis.
Technical aspects of proteome analysis in body fluids
As for any multidimensional assay, proteome analysis of an increasing number of variables requires ever more time and effort. In addition, to obtain statistically significant data, expanding the number of analyzed components requires increasing the number of analyzed samples and, consequently, greater computing power, thus rendering the task even more difficult. While it is generally possible to determine the concentration of a single protein within seconds, it would probably require very long time and the parallel use of an array of high-end mass spectrometers, to analyze all proteins in a single urine sample. For practical purposes, it is important to find a balance between the desire for maximal data and the limitations of effort and time for the analysis. In addition, such an approach must ensure a reproducible analysis (including sampling, preparation, and data evaluation) and generation of comparable data for future studies. Collection of data with wide comparability would eliminate the necessity for repeated control measurements otherwise necessary for validation.
Clinical proteome analysis can be seen as an approach to compare a large number of variables in a limited number of datasets. Hence, comparability of the data is an extremely important issue to enable the differential display of a large number of polypeptides in a single, reproducible and time-limited step with high confidence. As outlined in detail elsewhere [11], every manipulatory step increases the possibility of introducing artifacts, reduces reproducibility, and may further increase the complexity of samples. These problems can be illustrated in the analysis of the proteolytic fragments generated by digestion with trypsin. While this step is frequently necessary to allow mass spectrometric analysis of proteins, most investigators have found that this procedure is not entirely reproducible (especially with respect to incomplete cleavage), generates “unexplainable” mass peaks in the spectra, and increases the number of analytes. In contrast, peptides and small proteins can be analyzed directly without proteolytic digestion, thus reducing the time for analysis [16]. By concentrating on peptides and smaller proteins (below ca. 30 kDa), the information held in the larger proteins may be lost. However, the advantage of such an approach is that a sample of reduced complexity can also be analyzed in shorter time, and the loss of information may not be as critical as anticipated.
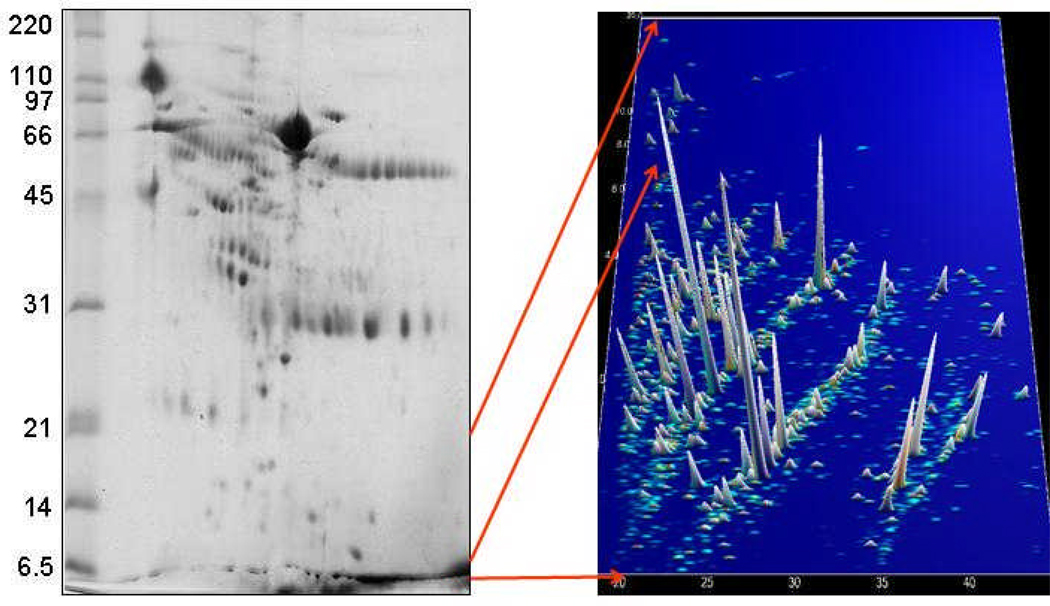
It is presently impossible to analyze the proteome of a complex biological sample by mass spectrometry without prior fractionation. Consequently, pre-MS separation is necessary. Among the methods currently used, 2-DE-MS is the method of choice for the analysis of larger proteins. However, the method is rather time-consuming, technically challenging, and requires special consideration to achieve acceptable comparability and reproducibility [4,19,20,21]. In addition, 2-DE-MS is not suitable for the analysis of smaller polypeptides (<10 kDa). This issue is also illustrated in Figure 1. As this technology is not the focus of this manuscript and has been reviewed by others in this issue, we will not expound upon it in this review. Three different technologies, SELDI, LC-MS and CE-MS, have been used mostly for the analysis of the low-molecular-weight proteome. The advantages and limitations of these three technologies with respect to the application towards peptides and small proteins and to the goal of defining biomarkers are summarized in Table 1.
Figure 1.
Separation of human urine for proteome analysis. The left panel shows a 2-D gel (courtesy of Visith Thongboonkerd). Molecular mass (in kDa) is indicated on the left. Most of the low-molecular-weight proteins remain unseparated in the front. Polypeptides in this mass range can be analyzed by CE-MS, as indicated in the right panel. Mass (in kDa) is plotted against migration time (in min), the intensity of the peaks is indicated by height and color.
Table 1.
Comparison of SELDI, LC-MS, and CE-MS methods.
| Technology | Advantages | Limitations |
|---|---|---|
| SELDI | Easy-to-use system, high throughput, automation, small sample volume, TOF/TOF sequencing possible |
Restricted to selected polypeptides, low-resolution MS, lack of comparability |
| LC-MS | Automation, multidimensional, high sensitivity, any MS/MS sequencing possible |
Time-consuming, sensitive for interfering compounds, restricted mass range |
| CE-MS | Automation, high sensitivity, fast, small sample volumes, multidimensional, low cost, any MS/MS sequencing possible |
Not well suited for larger polypeptides (>30 kDa) |
Liquid chromatography coupled to mass spectrometry (LC-MS)
Liquid chromatography (LC) provides a powerful fractionation method compatible with virtually any mass spectrometer [4,22,23,19]. LC-columns can separate large amounts of analytes with high resolution [22,23]. Therefore, if sensitivity of detection is a consideration, LC may be an excellent choice [24]. A sequential separation using different media in two independent steps provides a multidimensional fractionation that can generate vast amounts of information. Multidimensional protein identification technology (MudPIT) [25,26,27,28] or a 2D liquid-phase fractionation approach [29] is well suited for in-depth analysis of body fluids. However, the massive datasets may be less helpful than anticipated. Limitations include difficulties with comparative analysis, in part due to the variability in multidimensional separations and the substantial time required for analysis of a single sample (generally in days). Furthermore, the method suffers from its sensitivity to interfering compounds (e.g., lipids or detergents, large molecules that may precipitate and/or adsorb to the column) [11]. Therefore, this approach is not well suited for routine clinical analysis. An alternative strategy is thus needed for subsequent validation and development of an assay with clinical application [30].
Surface-enhanced laser desorption ionization (SELDI)
SELDI is an alternative MS-based approach for the proteomic analysis of body fluids used frequently in the discovery phase. It is a simple and user-friendly solution to several obstacles of proteome analysis and has consequently been used in several clinically relevant investigations [31,32,33,34,35,36,37]. SELDI follows the strategy of reducing the complexity of samples by fractionation based on selective interactions of polypeptides with different immobilized matrices. These active surfaces can be reversed-phase or ion-exchange materials, ligands, receptors, antibodies, or DNA, to name just a few. Due to the selectivity and limited capacity of the active surface, only a small fraction of the polypeptides in a sample binds to the surface of the SELDI chip, facilitating the subsequent mass spectrometric analysis of the originally highly complex samples. Numerous reports on biomarkers for a variety of diseases have been published using this strategy [38,39,40,12]. However, the utility of SELDI-MS approaches has been subsequently debated [41,42,43,44]. A drawback of SELDI-MS is certainly the loss of most of the information contained in a biological sample, consequently limiting the significance of the data. Additional problems include lack of comparability of the datasets due to different chip surfaces and conditions, as well as the low capacity and low resolution of the mass spectrometer. The latter can be solved by the use of more appropriate mass spectrometers, such as MALDI-TOF/TOF instruments, as described recently by Orvisky et al. [45]. In this study, the authors enriched for serum peptides by ultrafiltration under denaturing conditions that allowed efficient profiling and identification of peptides up to 5 kDa. Direct TOF/TOF sequencing of the most abundant peptide identified des-Ala-fibrinopeptide A as a serum biomarker of hepatocellular carcinoma.
Capillary electrophoresis coupled with mass spectrometry (CE-MS)
CE-MS is a second alternative MS-based approach for the proteomic analysis of body fluids. It has been used in the discovery and validation phases and subsequent application. This approach is based on CE as a front-end fractionation coupled to a mass spectrometer. CE separates proteins based on migration in the electrical field (300–500 V/cm) with high resolution in a single step. CE-MS offers several advantages: (1) it provides fast separation and high resolution [46], (2) it is quite robust and uses inexpensive capillaries [11], (3) it is compatible with most buffers and analytes [47], and (4) it provides a stable constant flow, thereby avoiding gradients in the buffer that may otherwise hamper detection by MS [48]. As with LC, CE can be interfaced with essentially any mass spectrometer. Several technical considerations that must be taken into account to achieve stable CE-MS coupling have been extensively reviewed [49,50,47,51]. The acidity of the buffer generally used for CE-MS analysis of proteins and peptides prohibits the application of CE for analysis of high-molecular-weight proteins because they tend to precipitate at low pH. However, large proteins can be effectively removed by ultrafiltration (see below). As the urinary proteome contains thousands of different peptides and low-molecular-weight proteins [24,16], this feature of CE-MS does not appear to represent a drawback. Another limitation of CE-MS is the relatively small sample volume that can be loaded onto the capillary, leading to a lower sensitivity of detection in comparison to LC. Improved methods of ionisation by micro- or nano-ion spray have resolved this challenge to a large extent. In addition, improvements in the detection limits of mass spectrometers enable detection in the low- or sub-femtomol range, rendering the issue of sensitivity less important [52,53,54]. Consequently, CE-MS has become a viable alternative to the commonly used proteomic technologies and has recently been successfully applied in several clinical studies [55,56,13].
Ionization and choice of mass spectrometers
The two principal choices for coupling separation with mass spectrometry detection are either off-line coupling with (mostly) MALDI or on-line coupling which is essentially restricted to electrospray ionization (ESI). Off-line coupling comes with the disadvantage of a loss in resolution due to fractionation. However, it is technically less challenging than on-line coupling. MALDI can be easily automated and, in comparison to ESI, generates less complex spectra of mostly singly charged ions. A major disadvantage for the analysis of complex samples is the pronounced signal suppression in MALDI that is observed to a lesser degree in ESI [24]. Online coupling using ESI, while retaining the resolution obtained in fractionation, is technically more challenging and, in addition, results in spectra of higher complexity due to multiply charged ions. However, with the availability of suitable software solutions [57], the latter is not an issue anymore. In light of the technical advancements in the ionspray sources that enable stable ESI (and consequently eliminate much of the original challenge), the benefits of ESI appear to outweigh the associated technical challenges.
In addition to coupling, the choice of the mass spectrometer strongly influences the data. Fourier transform-ion cyclotron resonance (FT-ICR) MS instruments may enthrall investigators with their high resolution and mass accuracy. However, at least in our hands, FT-ICR instruments showed lower sensitivity compared to time-of-flight (TOF) mass spectrometers and are quite expensive. Using TOF, detection limits are in the high-attomol range [48]. The achieved resolution of approximately 10,000 is sufficient to resolve 6- to 7-fold charged ions. Masses of ions with higher charges can be determined using conjugated signals. In comparison, quadrupole or ion-trap mass spectrometers appear to be less well suited for that purpose due to their lower resolution.
Sample preparation
Urine represents a highly complex mixture of molecules varying widely in polarity, hydrophobicity, and size. In addition, clear differences between early-stream and midstream urine samples can be noted ([12] and Mischak et al., unpublished data), further highlighting the importance of standardized protocols for collection of urine. A sample preparation protocol should be reproducible and result in minimal, or at least reproducible, loss of polypeptides. Ideally, a crude unprocessed sample should be analyzed directly, thus avoiding artificial losses or bias arising from sample preparation. However, this approach is frequently not practical, due to the presence of interfering compounds, such as aggregates, salts, lipids, and carbohydrates. Elimination of large-molecular-weight compounds by ultrafiltration has improved the quality of samples and reproducibility of their analysis. If the ultrafiltration step is performed in the presence of a detergent and a chaotropic agent (e.g., urea and SDS), protein-protein interactions (and consequently irreproducible loss of analyte) are avoided [58]. Furthermore, it appears advisable to remove salts and other low-molecular-weight compounds in a single step using, for example, anion-exchange [59] or reversed-phase materials [60], or desalting [58]. In addition, the “human error” in sample handling, especially in pipeting, may affect reproducibility. Robotic handling of the samples improves the reproducibility and should be considered whenever possible [61].
Clearly, different technologies may require different sample preparation procedures. However, it cannot be overemphasized that the reproducibility of sample preparation and comparability of different samples (e.g., from patients showing different degrees of proteinuria) is one of the most important considerations in the analysis. Unfortunately, each additional step in sample preparation is prone to generate new artifacts.
Identification of biomarkers
Current literature indicates that peptidomics enables fast and reliable analysis of polypeptides from several types of highly complex biological samples, such as urine, blood, or cerebrospinal fluid [17,62,63]. Although these polypeptides can serve as excellent biomarkers for diagnostic purposes, their potential physiological role remains unknown as long as their identity, defined by their amino acid sequence, is not determined. The identification of the defined biomarkers presents some unique challenges. Generally, the biomarkers cannot be easily isolated and their sequence analysis must be thus performed from a complex mixture. In addition, potential biomarkers are likely to be small fragments of larger proteins and frequently post-translationally modified. Thus, to identify a small and potentially modified fragment of a protein with a molecular weight greater than 100 kDa requires extensive de novo sequencing.
In “common proteomics”, identification involves separation of intact proteins, enzymatic digestions, MS analysis of the digestion products, and standard methods for sequencing by tandem mass spectrometry (MS/MS). However, the commonly used standard methods for MS/MS sequencing and data evaluation generally do not account for post-translational modifications (PTM) [9]. Identification of any PTM is essential for identification of a specific biomarker and requires further characterization. Furthermore, some PTMs may be disease-specific and can themselves serve as biomarkers (e.g., advanced glycation end-products in diabetes mellitus [64]). In combination, these issues comprise a large source for errors and failures in sequence assignments. Sequencing of unmodified biomarkers, in contrast to assigning a protein’s primary sequence based on the analysis of a tryptic digest, remains a great challenge. As identification of proteins and characterisation of their PTMs is a difficult task, particularly for less abundant proteins, many potential markers identified in peptidomic experiments have been among the abundant proteins [65,66]. Whether these will prove to be the most robust and/or specific needs to be determined.
New fragmentation technologies such as electron capture dissociation (ECD) with FT-ICR MS enable localization of even labile PTMs, such as glycosylation. FT-ICR MS offers two complementary fragmentation techniques for analysis of PTMs by tandem mass spectrometry, infrared multiphoton dissociation (IRMPD) and ECD [67,68]. ECD fragmentation results in complementary cleavage of the backbone N-Cα bond with minimal loss of PTMs. ECD FT-ICR MS has been successfully used to identify urinary polypeptides larger than 8 kDa, owing to the high mass accuracy of FT-ICR MS [69]. Furthermore, localization of glycosylation sites in various glycoproteins, including human IgA1, was accomplished using ECD FT-ICR [70].
Additional technologies using electron-based dissociation techniques, such as electron transfer dissociation (ETD), have shown great promise [71,72], but need to be further developed. Of note, these technologies have certainly shown the best performance for the sequencing of urinary peptides (Mischak, unpublished and Coon et al., manuscript in preparation). Other improvements in the construction of FT mass spectrometers and new software solutions (based on ProSight PTM) also significantly improved top-down proteomics for MS/MS of proteins larger than 10 kDa [73]. Patrie et al. identified 101 unique proteins (5–59 kDa) from whole-cell lysates of Methanosarcina acetivorans using these new approaches. This study also detected several incorrectly predicted start sites [74]. This and other recent work [75,76] suggests that, in the near future, PTMs can be routinely analyzed directly during top-down proteome analysis with high throughput.
Because the termini of the naturally-occurring polypeptides in the urine have not been generated by defined enzymatic cleavage and they frequently harbour PTM, direct identification of these polypeptide is challenging (for more details, see [24]). Among the greater obstacles are limited mass accuracy (especially of the MS/MS spectra), the change in parent-ion mass due to modifications (taking into account all possible modifications results in too many degrees of freedom), and a bias of the search algorithms towards high sequence coverage of an unmodified protein. In addition, MS/MS experiments frequently produce a limited number of preferred fragmentation products (at proline residues, carbohydrate side-chains, etc.).
In general, any of the separation methods can be interfaced with MS/MS instruments. LC- or CE-coupling and the advantages and disadvantages of the two separation methods as well as several different MS/MS instruments have recently been described by Zurbig et al [24]. While LC coupling has the advantage of higher capacity, hence providing more material for MS/MS analysis, CE has the advantage that the number of basic amino acids correlates with the polypeptide migration time at pH 2. This feature facilitates the independent entry of different sequencing platforms for peptide sequencing of C E-MS-defined biomarkers from highly complex mixtures.
Alternatively, fractions during an LC or CE separation can be collected and spotted off-line onto a MALDI target plate. Subsequently, the polypeptides of interest can be analyzed using MALDI-TOF/TOF [77,11]. This approach has the advantage that the signal of interest can be located in MS mode and optimal fragmentation conditions can be determined without repeated separation. However, sequencing of native peptides with MALDI-TOF/TOF frequently seems to produce data of sufficient quality, due to insufficient mass accuracy. In our hands, more than 90% of the spectra obtained using a Bruker MALDI-TOF/TOF did not allow identification of the native peptide of interest. Even so, it represents a simple method and several biomarker candidate peptides have been identified using MALDI-MS/MS, as shown for graft-versus-host disease [78] or diabetic nephropathy [79].
Data evaluation, bioinformatic approaches in proteomics
The information content of a complex proteome analysis requires adequate tools for data analysis. The essential information to be extracted includes the identity and quantity of the polypeptides. A prerequisite for the comparative evaluation of urine (or any other comparative analysis) is the ability to identify identical compounds with high probability in consecutive samples. Hence, resolution and accuracy of the parameters for identification are of major importance. One method to increase the resolution of the MS data is to combine these with the parameters of the separation (e.g., retention or migration time, but every other unique measure may serve as an additional or alternative identifying parameter). Software solutions that automatically select peaks based on parameters such as signal/noise ratio or appearance in several consecutive spectra have been described, such as MSight [80], DeCyder MS (GE Healthcare), or MosaiquesVisu [57,11,58]. It is important that the software is able to perform charge deconvolution with a low error rate and combine peaks (and amplitude) that represent identical compounds at different charge states, as reported for MosaiquesVisu [57].
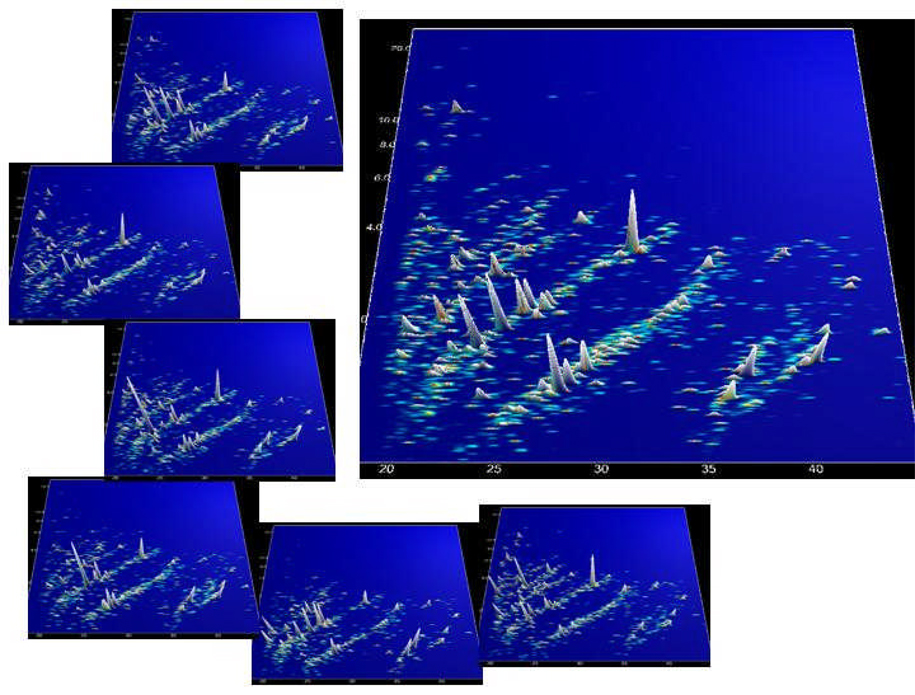
Accuracy (and hence resolution) can be improved by calibrating the identifying parameters. This calibration can be achieved by using external standards or, preferably, internal standards (e.g., peptides that are frequently present in any sample) [81]. The importance of proper calibration is also evident from Figure 2. Definition of biomarkers requires the compilation of datasets to enable comparison and statistical evaluation. If the identifying parameters are not well defined, meaningful comparison of the data is impossible.
Figure 2.
Digital data compilation. Individual datasets from CE-MS analysis of human urine samples were calibrated using internal standards. The left panel displays these data in a 3-dimensional contour plot: mass (in kDa on a logarithmic scale) plotted against normalized migration time (min). The MS signal intensity is represented by the peak height as well as color. The data were digitally compiled to a group-specific polypeptide pattern, shown in the right panel.
Most, if not all, proteomic studies indicated that a single biomarker does not allow reliable diagnosis, staging, or prognosis of a kidney disease. This finding immediately raises the question of how to combine several biomarkers to provide a diagnostic or predictive pattern. While a definitive answer is probably still far away, a number of approaches have emerged.
Hierarchical decision tree-based classification methods, such as CART [82,83], were among the first algorithms to utilize the available information on multiple biomarkers. However, empirical observations suggested that these approaches were not sufficient because the number of incorrect predictions made by the classification algorithm increases with the complexity of the decision tree [84]. The number of datasets available to establish the decision tree is generally low, resulting in a lack of statistical significance beyond the second or third nodes of the tree.
Support Vector Machines (SVM) (for an example, see [85]) provided a tool to overcome some of these limitations due to the theoretical principles upon which they are based. Excellent empirical performance of SVM has been reported in a number of diverse applications [86,87,84]. These approaches provided superior cross-validated predictive performance, but mixed results were obtained with blinded datasets. Reliable results have been obtained when the number of variables was low and substantial differences between the datasets existed. However, when the differences were more subtle, over-fitting (also referred to as “memorizing”, a term often employed in Artificial Intelligence research) to the training set and thus poor classification of blinded datasets was observed (Mischak et al., unpublished data). To avoid such memorizing effects, the number of variables and dimensions must be lowered. One approach is to use a linear combination of several biomarkers (e.g. by addition/subtraction of logarithmic amplitudes). While such an approach does not reflect the complexity of the problem and hence cannot be considered the best possible classifier, it is also not prone to over-fitting, and generally performs well in the blinded dataset.
An important aspect is the indication of the level of confidence in the results. In other words, a classification such as ‘this urine sample has been drawn from an individual with type II diabetes’ should also have a numeric score indicating how likely the classification is correct: i.e. ‘with 90% confidence this urine sample has been drawn from an individual with type II diabetes’. Clearly, 90% confidence is more reliable than 50% confidence, especially if there are only two alternatives to be considered: disease presence versus disease absence (in which case 50% confidence indicates little more than random guessing). While SVMs provide a very encouraging classification performance on a range of difficult problems, they generally cannot assign confidence levels and thus no information is available as to how much the prediction can be trusted.
A promising probabilistic classification method that shares many of the positive characteristics of the SVM, but in addition provides the important levels of confidence with each classification prediction, is based on the Gaussian Process (for a comprehensive, although somewhat technical, explanation of this methodology, see [88]). A general purpose and computationally efficient Gaussian Process-based classification method has recently been successfully applied to the problem of correct prediction of BRCA1 and BRCA2 heterozygous genotypes [89]. The probabilistic nature of Gaussian Process-based classification methods provides a means of inferring optimally weighted combinations and possible selection of biomarkers; a detailed study of this capability is currently ongoing.
No matter which of these approaches is used, two basic considerations apply: 1) the number of independent variables should be kept to a minimum, certainly less than the number of samples investigated, and 2) any such approach must be confirmed with a blinded validation set. It should be imperative to include such a blinded dataset in any report on potential biomarkers.
Urinary biomarkers for renal diseases
One of the first applications of urinary peptidome analysis for a clinically relevant questions was reported by Rogers et al. [90]. With the aim to define renal cell carcinoma (RCC)-specific biomarkers, the authors investigated urine samples from 218 individuals using SELDI analysis. Samples from patients before nephrectomy for RCC (n=48), normal healthy volunteers (n=38), and outpatients with benign diseases of the urogenital tract (n=20) were used as a training set for biomarker definition. The defined markers were subsequently validated in two blinded assessments with an initial "blind" group of 32 samples (12 patients with RCC, 11 healthy controls, and 9 patients as disease controls) and a second group of 80 samples (36 patients with RCC, 31 healthy volunteers, and 13 patients with benign urological conditions). While in the first round sensitivities and specificities of 81.8–83.3% were achieved, the values significantly declined, ranging from 41.0% to 76.6%, for the second set of samples collected 10 months later. The authors analyzed possible contributing factors including sample stability, changing laser performance, and chip variability to assess a long-term robustness of the approach. One of the main conclusions from this study was the evident need for rigorous evaluation of such variables that may influence stability/robustness.
One of the main areas of research has been the evaluation of transplant-associated complications. SELDI has been recently used by Clarke et al. [91] and Schaub et al. [92] to detect potential biomarkers for allograft rejection in kidney transplant patients. Clusters of five and three urinary proteins correctly classified 34 and 50 patients, respectively, with high sensitivity and specificity. Unexpectedly, these two groups defined completely different biomarkers for the same disorder and neither found differences between patients with an allograft without rejection versus patients with only native kidneys. In the context of acute renal allograft rejection, Schaub et al. [65] reported that some of the potentially diagnostic urinary protein SELDI peaks were derived from naturally-occurring proteolytic fragments of beta2-microglobulin. Additional experiments showed that proteolysis of urinary beta2-microglobulin required a pH below 6 and aspartic proteases. Transplant patients with acute tubulointerstitial rejection had a lower urinary pH than did patients with allografts with stable function and healthy individuals. In addition, the patients with rejection had greater amounts of aspartic proteases and intact beta2-microglobulin in the urine.
Wittke et al. [55] used CE-MS to analyze urinary samples from patients with different grades of subclinical or clinical acute rejection, patients with urinary tract infection and patients without evidence of rejection or infection. Substantial differences were found between patients with transplanted kidneys and patients with native kidneys, most likely due to treatment with cyclosporin A, a calcineurin-inhibitor immunosuppressant. In addition, a distinct urinary polypeptide pattern identified 16 of the 17 patients with acute tubulointerstitial rejection; these markers differed from the markers of vascular rejection. Potentially confounding variables, such as acute tubular lesions, tubular atrophy, tubulointerstitial fibrosis, calcineurin inhibitor toxicity, proteinuria, hematuria, allograft function, and different immunosuppressive regimens did not affect the results. However, an additional polypeptide pattern that allowed differentiating between infection and acute rejection was developed. The defined polypeptide patterns were further validated in a blinded assessment of samples from transplant patients potentially exhibiting renal rejection; most samples were correctly classified using these biomarkers.
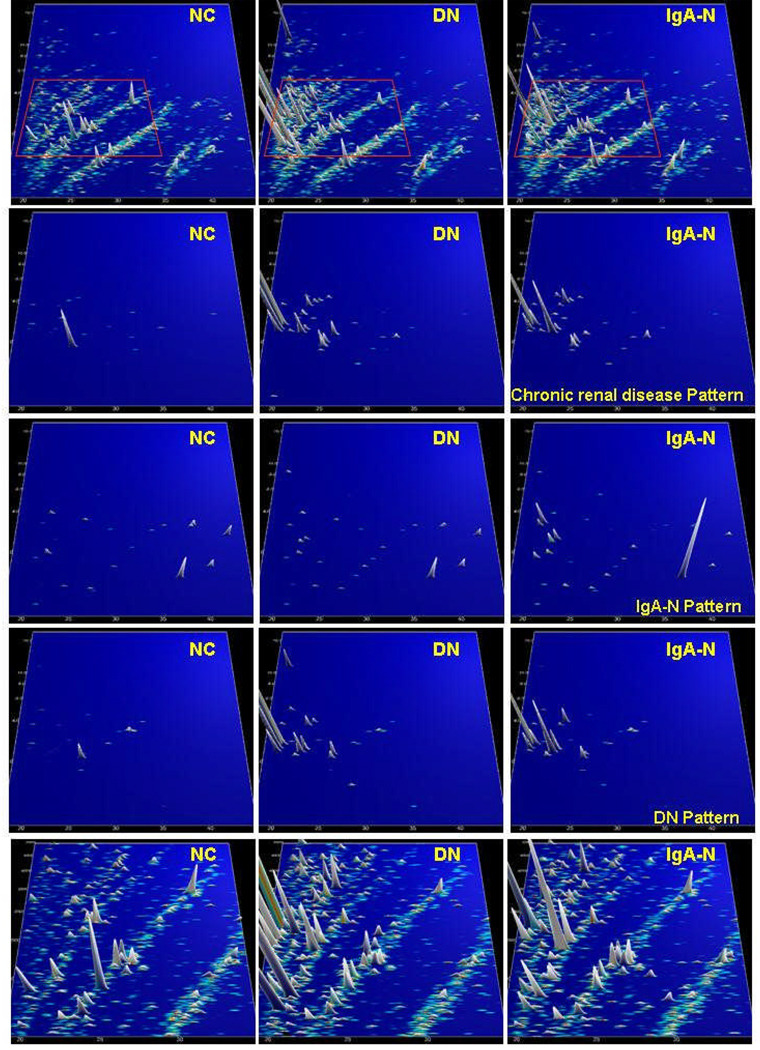
Another area of interest is the definition of urinary polypeptide biomarkers for chronic renal diseases. One of the first reports was the analysis of urinary polypeptide markers of membranous glomerulonephritis by SELDI and CE-MS [93]. Using identical urine samples, three potential biomarkers were defined using SELDI analysis compared to 200 potential biomarkers from the CE-MS analysis. The authors concluded that better results can be obtained using a panel of well-defined biomarker proteins rather than a few not-too-well-defined peaks. CE-MS technology was also used for the analysis of the urinary proteome of patients with type II diabetes mellitus and the frequently observed diabetic nephropathy [79]. The patients exhibited variable degrees of renal damage, as evidenced by different magnitudes of albuminuria. One hundred sixty-eight urinary proteins were present in over 90% of the samples, suggesting a consistent urinary proteome that was subsequently further investigated and used for calibration and standardisation [16]. Additional work on urine samples from patients with other chronic renal diseases suggested that panels of 20 to 50 urinary polypeptide markers allow not only the diagnosis of a specific (primary) kidney disease, but also the discrimination (differential diagnosis) with high sensitivity and specificity between different kidney diseases such as IgA nephropathy, focal-segmental glomerulosclerosis, membranous glomerulonephritis, minimal-change disease, and diabetic nephropathy [84,66,16]. As an example, compiled urinary polypeptide patterns from healthy controls and patients with diabetic nephropathy or IgA nephropathy, as well as the distribution of selected biomarkers, are shown in Figure 3.
Figure 3.
Protein patterns of healthy volunteers (NK), and patients with diabetic nephropathy (DN) and IgA nephropathy (IgA-N), respectively. Upper panel: compiled patterns consisting of 20 to 100 single measurements, molecular mass (0.8–25 kDa, on a logarithmic scale) against normalized migration time (18–45 min), peak height and color encode the signal intensity. Three middle panels: only selected candidate disease-specific biomarkers are displayed on the same scale. An array of general biomarkers for kidney disease present in DN and IgA-N can be defined. In addition, biomarkers that are specific for DN or IgA-N can be identified, as indicated on the right-hand side. Lower panel: zoom of the upper patterns (all peptides analysed, 1.5–5 kDa, 19–35 min). As evident, several additional biomarkers (of mostly lesser statistical value) are present, which can be further exploited.
In a recent study, Decramer et al. [56] utilized CE-MS-based urinary proteome analysis to define specific biomarker patterns for different grades of ureteropelvic junction obstruction, a frequently encountered pathology in newborns. The patients did not have clinical proteinuria. In a blinded prospective study, these patterns predicted with 95% accuracy the clinical outcome of the newborns nine months in advance. These data not only indicated the potential of urinary proteomics to enable the diagnosis of renal disease, but also suggest to the potential to gauge the prognosis.
In addition to the diagnosis and prognosis of disease, urinary proteome analysis may be an excellent tool for fast, non-invasive monitoring of disease progression or response to therapy. The lack of the ability to monitor these parameters has greatly hampered development of specific therapeutics in the past. While renal diseases represent a major clinical problem, only a few disease-specific drugs have been developed so far, due in large extent to the absence of good monitoring capability.
In a randomized double-blind study, Rossing et al. [94] evaluated the treatment of macroalbuminuric patients with daily doses of 8 mg, 16 mg, and 32 mg candesartan or placebo for two months. Examination of the urine samples from these patients with CE-MS revealed a significant change in 15 of 113 proteins characteristic for diabetic renal damage. Similar data have been obtained for patients with vasculits (Haubitz et al., manuscript in preparation and [16]), for whom the vasculitis-specific protein pattern reverted towards normal after treatment.
In addition to the definition of disease-specific polypeptide patterns, stage-specific polypeptide markers can be defined. Mischak et al. [79] and Meier et al. [95] defined stage-specific biomarkers for diabetic nephropathy in patients with diabetes mellitus type I or type II. In both studies, the individual data sets of healthy volunteers (9 and 39, respectively), patients with diabetes type I or II without marcoalbuminuria (28 and 46, respectively), and with intermittent or persistent macroalbuminuria (16 and 66, respectively) were combined to create typical polypeptide patterns. In patients with type II diabetes mellitus and a normal albumin excretion rate, the detected polypeptide pattern differed significantly from that in patients with greater albuminuria. Comparable results were obtained for patients with diabetes type I, suggesting that the urinary proteome contains a much greater variety of polypeptides than previously demonstrated.
Urinary biomarkers for urological disorders
As urine is in direct contact with the bladder, it is to be expected that urinary biomarkers will display (pathological) changes in the bladder as well as the urinary tract. Vlahou et al. have sougth to define biomarkers for bladder cancer [96,2,97,98], with moderate success. Subsequently, several other groups have reported preliminary data on the use of SELDI-MS for detection of urothelial cancer [99,100]. Although the findings were generated by the same technology, they differed and were not comparable, most likely due to different chip surfaces and conditions. In a more thorough investigation that also included assessment of blinded datasets, Munro et al. [101] employed SELDI technology to define biomarkers for recurrent transitional cell carcinoma (TCC) of the bladder. In this well performed study with extensive quality control measures and robotic sample handling, the authors established a biomarker pattern that enabled classification of blinded datasets with 65% specificity and 75% sensitivity. Theodorescu et al. [13] described the detection and validation of biomarkers of urothelial carcinoma using CE-MS. In a study of 46 patients with urothelial carcinoma and 33 healthy volunteers, a bladder cancer-specific urinary proteomic pattern was identified. The model was refined by an analysis of 366 urine samples from healthy volunteers and patients with malignant and non-malignant genitourinary diseases. In a blinded assessment, the prediction model based on 22 polypeptides correctly classified all patients with urothelial carcinoma and all healthy volunteers (100% sensitivity and specificity). In addition, the differentiation between bladder cancer from other malignant and non-malignant diseases, such as renal nephrolithiasis, ranged in sensitivity from 86% to 100%. Additional experiments in a multicenter blinded study confirmed these results and showed that superficial cancer can be distinguished from muscle-invasive disease with high accuracy (Theodorescu et al., in preparation).
The analysis of urine as a diagnostic tool was also applied to patients with prostate cancer. The heterogeneity of progressive prostate cancer (PCa) has hampered development of an effective early detection assay. Existing prostate cancer screening has relatively poor specificity. M’koma et al. [102] reported analysis of urine samples from 407 patients using MALDI-TOF analysis of eluates from reversed-phase material over a mass range of 1,000–5,000. The results distinguished between PCa and other pathological alterations of the prostate with 70% to 80% sensitivity and specificity.
In a pilot study [58], CE-MS techniques defined potential urinary markers of prostate cancer. Forty-seven urine samples from patients who underwent prostate biopsy were analyzed; 26 patients had PCa and 21 had benign prostatic disease. The data indicated several polypeptides as potential biomarkers for PCa patients, with 92% sensitivity and 96% specificity. Additional data suggested that the early-stream urine was the best sample for the definition of PCa-specific biomarkers, indicating that these biomarkers likely originated from prostatic secretions. Based on these results, the same group refined the prostate-specific pattern with 116 urine samples from 54 patients with PCa and 62 patients with benign pathology. A pattern of 26 potential biomarkers was validated in a blinded assessment of urine samples from 36 patients with PCa and 24 patients with benign prostatic conditions (Theodorescu et al., in preparation). The prediction model correctly classified 32 of the 36 patients with PCa and 16 of the 24 patients with benign pathology.
Application of urinary proteome analysis to other diseases
While the main focus of urinary biomarker discovery using CE-MS has been genitourinary diseases, other diseases may also produce urinary polypeptide patterns of diagnostic significance. A recent report by Nemirovskiy et al. [103] indicated that analysis of urinary polypeptides improved the assessment of patients with osteoarthritis. Accentuated MMP activity increases the amount of a 45-mer collagen type II peptide. The authors found that this specific fragment in the urine and proposed that the activity of matrix metalloproteases could be monitored in vivo by measuring the urinary excretion of particular collagen fragments. Interestingly, several of the urinary proteome biomarkers reported recently were also collagen fragments [56,13,55]; therefore, it is tempting to speculate that they indirectly indicate activity of disease-specific proteases.
Other examples are the clinical follow-up of patients after allogeneic hematopoietic stem cell transplantation (HSCT) [78,104]. Urine samples from 40 patients after HSCT (35 allogeneic, 5 autologous) and 5 patients with sepsis were collected during a period of 100 days (a maximum of 10 samples per patient) and analyzed. A pattern consisting of 16 differentially-excreted polypeptides indicated early graft versus host disease, a severe life-threatening complication of allogenic HSCT. The pattern of markers discriminated patients with early graft versus host disease from patients without complications with 82% specificity and 100% sensitivity. A subsequent blinded multicenter validation study of 100 patients with more than 600 samples collected prospectively confirmed the results, although with reduced specificity and sensitivity (Weissinger et al., submitted).
In two independent sets of experiments, Dominiczak et al. and Peter et al. (manuscripts submitted) examined patients undergoing coronary artery bypass grafting or patients after acute coronary infarction. Urine samples from patients and controls were analyzed using CE-MS to identify biomarkers for coronary artery disease. In a blinded assessment based on more than 200 samples, specific urinary biomarkers identified the patients with greater than 90% sensitivity and specificity. These findings confirmed the association between arteriosclerosis risk factors and renal dysfunction [105].
Identification of uremic toxins using proteomics
Another application of proteomics that has gained considerable interest is the examination and definition of potential uremic toxins. Spent dialysate and hemofiltrate fluid is an excellent source for proteomic analysis, as it contains little albumin and other interfering large proteins. In 1994, Forssmann et al. [106] used an advanced LC-MS approach to identify proteins from hemofiltrate using a “peptide bank” with up to 300 different chromatographic fractions prepared from 10,000 liters of human hemofiltration fluid [107]. With this approach, several peptides with various biochemical functions were isolated [108,109].
Li et al. [110] examined urine and serum samples from uremic patients and healthy subjects using LC and MALDI-TOF MS as well as LC/ESI-MS/MS to define uremic toxins. One of the identified molecules, an octapeptide with molecular weight 1,007.94 Da (Val-Val-Arg-Gly-Cys-Thr-Trp-Trp), was biologically active; it accelerated the death of rabbits with chronic renal failure.
Using CE-MS, the effect of different dialysis membranes (low-flux vs. high-flux) on the number of polypeptides from 1 kDa to 10 kDa (“middle molecules” of uremia) in the dialysate was investigated [59]. Larger polypeptides (above 10 kDa) were present in only the dialysates obtained with high-flux membranes, while most of polypeptides in dialysates obtained with low-flux membranes were smaller than 10 kDa. In a pilot study the potential of CE-MS and CE-MS/MS to identify uremic retention molecules in dialysis fluids obtained with low-flux and high-flux membranes was assessed [111]. The results again indicated higher efficiency of removal of larger peptides using high flux membranes. In an unrelated study, the same technology was used to identify polypeptides in the plasma of dialysis patients that are generally absent in the plasma of normal controls [62]. A combination of data from the study of human plasma and hemodialysate should identify potential uremic toxins. These findings may lead to improvement of the efficacy of dialysis.
Pathophysiological role of biomarkers
Although the majority of the potential urinary biomarkers described to date have not been sequenced, sequences are available for more than 100 such peptides (e.g., [65,58,16,94,66]. Not unexpectedly, most of these peptides are derived from the most abundant proteins in the blood and urine: albumin, beta 2-macroglobulin, uromodulin, and collagen. Consequently, a valid question is whether peptidomics is not just another way to measure glomerular injury, that could probably be assessed with similar precision, but less effort, by meauring albuminuria [112]. This question cannot be answered with absolute certainty. However, the fact that differential diagnosis based on urinary proteome analysis is possible [84,66,113] and that patients in complete remission without albuminuria still exhibit apparently disease-specific changes in urinary polypeptides [84] strongly suggests that these peptides contain clues about the pathogenesis and are not merely degradation products. It is tempting to speculate that the disease-specific peptides may be indirect indicators of the activity of disease-specific proteases, as recently suggested by Haubitz [66]. This hypothesis is further strengthened by work recently published by Nemirovsky et al. [103], in which the presence of specific collagen fragments correlated with the disease-specific activity of matrix metalloproteases.
While the evidence is still scarce, it is an attractive hypothesis that urinary peptides of diagnostics value are not merely degradation products of abundant larger proteins, but a result of distinct, disease-specific processes, in many cases due to significant changes in the activity of proteases A similar scenario may be applicable to albuminuria. Consequently, an albumin-derived biomarker is not simply “an albumin fragment”, but rather a specific fragment, defined by its specific C- and N-terminus. Unfortunately, such essential detailed information is frequently absent (e.g, see the recently published database of urinary proteins [9]). A thorough examination of the sequences of the urinary peptides and comparison with protease specificities may strengthen the above hypothesis and lead to better insight into the regulation and pathophysiological role of specific proteases in many diseases.
Limitations of proteome analysis
Currently, the lack of standards and comparability among different methods appears to be one of the major limitation of proteome analysis. The vast majority of the published reports cannot be compared, thereby greatly reducing their relevance. Development of universally accepted protocols for collection, storage, and preparation of samples, as well as required analytical performance (e.g., mass resolution and accuracy), will improve the situation considerably. A first step in this direction may be the recently published suggestions for mandatory standards and guidelines [30]. The establishment of reliable 2-DE-, LC-, and CE-MS databases based on data derived from standard protocols would benefit the field. The reports based on different technologies, albeit promising, clearly indicate a need for standardization and show that a “common platform” that allows comparison of datasets from different laboratories is urgently required. Otherwise, these bits of information will never generate a “big picture” that is vital for proteomics to be applied with its full potential. Given the complexity of the task, it is crucial that thousands of comparable datasets be available for data evaluation and validation. As this task cannot be accomplished by individual laboratories, it is also essential to establish standards for quality control (e.g., minimal requirements for mass accuracy and resolution of the choice of the mass spectrometers, [30]). An excellent first step in that direction is the “Human Kidney and Urine Proteome Project” (HKUPP, http://hkupp.kir.jp) that aims to combine the efforts of leading scientists in the field.
Lack of appropriate and user-friendly bioinformatics software for data analysis also hinders progress toward clinical applications. So far, no standard has been developed for this, resulting in a set of different solutions that may work well for particular problems. However, as the different groups use highly divergent approaches, the data are generally not comparable. A repository of all data in a common format, together with specific software solutions generally available, would be an excellent step towards establishing comparable data and results.
While some analyses of the urinary proteome are quite promising, it is evident that the data should be further validated in other laboratories. This process may be more difficult than anticipated because certain technological advances are currently available only in single laboratories.
Another limitation is the current lack of sequence data for many potential biomarkers. This may be due to a variety of reasons, such as the above-outlined shortcomings in the sequencing of naturally occurring peptides (especially if they contain PTMs), and also the scarce quantity of these potential biomarkers. Peptides present in with small amounts may be detected by MS, but multiple fragmentation products may escape detection in MS/MS. Improvements in software solutions for sequence assignment as well as in detection limits of MS/MS instruments will hopefully shrink these shortcomings in the near future.
Summary and Outlook
From the very first clinical observations of kidney diseases, it became apparent that urinary proteins reflect renal pathology. In the past, personal skills (simple observation, smelling or even tasting of urine) were required in renal medicine, and were skillfully performed by our predecessors. Presently, advanced technologies are available to improve the analytical description of the protein content of urine. The contribution of proteomics to the understanding of the pathogenesis, diagnosis, and assessment of response to treatment of disease has been significant. However, its impact is modest in comparison to the expectations generated by the more than 25 years of technological progress with proteomics.
Proteome analysis is still far from displaying its full potential as a routine tool for clinical application. However, the first studies, sometimes with several hundred patients, have clearly revealed its potential for clinical diagnosis [13,56]. While it may be years or even decades until the entire urinary (or any other) proteome is explored, these results unmistakably indicate that urinary proteome analysis can be utilized today to deliver clinically important information. Certainly, the current technologies can and will be improved. However, application rather than improvement of the technology should be the primary goal of clinical proteomics. We should take full advantage of the subset of the proteome that is accessible and contains highly valuable information for medical assessment, and put its analysis to good use in the clinic.
Acknowledgements
We are grateful to Eric Schiffer and Visith Thongboonkerd for supplying unpublished data and for critically reading this manuscript. This work was supported in part by a grant from the lower Saxony Ministry of Economics (HM) and by grants from the National Institutes of Health (DK61525, DK64400, DK47322, and DK71802) and General Clinical Research Center of the University of Alabama at Birmingham (M01 RR00032) (BAJ and JN).
References
- 1.Ma Y, Liu G, Du M, Stayton I. Electrophoresis. 2004;25:1473–1484. doi: 10.1002/elps.200405895. [DOI] [PubMed] [Google Scholar]
- 2.Vlahou A, Fountoulakis M. J.Chromatogr.B Analyt.Technol.Biomed.Life Sci. 2005;814:11–19. doi: 10.1016/j.jchromb.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 3.Kolch W, Mischak H, Pitt AR. Clin.Sci.(Lond) 2005;108:369–383. doi: 10.1042/CS20050006. [DOI] [PubMed] [Google Scholar]
- 4.Issaq HJ. Electrophoresis. 2001;22:3629–3638. doi: 10.1002/1522-2683(200109)22:17<3629::AID-ELPS3629>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 5.Thongboonkerd V, Malasit P. Proteomics. 2005;5:1033–1042. doi: 10.1002/pmic.200401012. [DOI] [PubMed] [Google Scholar]
- 6.Fliser D, Novak J, Thongboonkerd V, Argiles A, et al. J Am Soc Nephrol. 2007 doi: 10.1681/ASN.2006090956. in press. [DOI] [PubMed] [Google Scholar]
- 7.Davis MT, Spahr CS, McGinley MD, Robinson JH, et al. Proteomics. 2001;1:108–117. doi: 10.1002/1615-9861(200101)1:1<108::AID-PROT108>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 8.Spahr CS, Davis MT, McGinley MD, Robinson JH, et al. Proteomics. 2001;1:93–107. doi: 10.1002/1615-9861(200101)1:1<93::AID-PROT93>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 9.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omenn GS, States DJ, Adamski M, Blackwell TW, et al. Proteomics. 2005 May;:3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 11.Kolch W, Neususs C, Pelzing M, Mischak H. Mass Spectrom Rev. 2005;24:959–977. doi: 10.1002/mas.20051. [DOI] [PubMed] [Google Scholar]
- 12.Schaub S, Wilkins J, Weiler T, Sangster K, et al. Kidney Int. 2004;65:323–332. doi: 10.1111/j.1523-1755.2004.00352.x. [DOI] [PubMed] [Google Scholar]
- 13.Theodorescu D, Wittke S, Ross MM, Walden M, et al. Lancet Oncol. 2006;7:230–240. doi: 10.1016/S1470-2045(06)70584-8. [DOI] [PubMed] [Google Scholar]
- 14.Zhou H, Yuen PS, Pisitkun T, Gonzales PA, et al. Kidney Int. 2006;69:1471–1476. doi: 10.1038/sj.ki.5000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vestergaard P, Leverett R. J.Lab Clin.Med. 1958;51:211–218. [PubMed] [Google Scholar]
- 16.Schiffer E, Mischak H, Novak J. Proteomics. 2006;6:5615–5627. doi: 10.1002/pmic.200600230. [DOI] [PubMed] [Google Scholar]
- 17.Fliser D, Wittke S, Mischak H. Electrophoresis. 2005;26:2708–2716. doi: 10.1002/elps.200500187. [DOI] [PubMed] [Google Scholar]
- 18.Anderson NG, Anderson NL, Tollaksen SL. Clin.Chem. 1979;25:1199–1210. [PubMed] [Google Scholar]
- 19.Aebersold R, Goodlett DR. Chem.Rev. 2001;101:269–295. doi: 10.1021/cr990076h. [DOI] [PubMed] [Google Scholar]
- 20.Yanagida M. J Chromatogr.B Analyt.Technol.Biomed.Life Sci. 2002;771:89–106. doi: 10.1016/s1570-0232(02)00074-0. [DOI] [PubMed] [Google Scholar]
- 21.Morrison RS, Kinoshita Y, Johnson MD, Uo T, et al. Mol.Cell Proteomics. 2002;1:553–560. doi: 10.1074/mcp.r200004-mcp200. [DOI] [PubMed] [Google Scholar]
- 22.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 23.Issaq HJ, Conrads TP, Janini GM, Veenstra TD. Electrophoresis. 2002;23:3048–3061. doi: 10.1002/1522-2683(200209)23:17<3048::AID-ELPS3048>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 24.Zurbig P, Renfrow MB, Schiffer E, Novak J, et al. Electrophoresis. 2006;27:2111–2125. doi: 10.1002/elps.200500827. [DOI] [PubMed] [Google Scholar]
- 25.Chen EI, Hewel J, Felding-Habermann B, Yates JR., III Mol.Cell Proteomics. 2006;5:53–56. doi: 10.1074/mcp.T500013-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Cagney G, Park S, Chung C, Tong B, et al. J.Proteome.Res. 2005;4:1757–1767. doi: 10.1021/pr0500354. [DOI] [PubMed] [Google Scholar]
- 27.Ru QC, Katenhusen RA, Zhu LA, Silberman J, et al. J.Chromatogr.A. 2006;1111:166–174. doi: 10.1016/j.chroma.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 28.Kislinger T, Gramolini AO, Maclennan DH, Emili A. J.Am.Soc.Mass Spectrom. 2005;16:1207–1220. doi: 10.1016/j.jasms.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 29.Soldi M, Sarto C, Valsecchi C, Magni F, et al. Proteomics. 2005;5:2641–2647. doi: 10.1002/pmic.200401269. [DOI] [PubMed] [Google Scholar]
- 30.Mischak H, Apweiler R, Banks RE, Conaway M, et al. PROTEOMICS - Clinical Applications. 2007;1:148–156. doi: 10.1002/prca.200600771. [DOI] [PubMed] [Google Scholar]
- 31.Shiwa M, Nishimura Y, Wakatabe R, Fukawa A, et al. Biochem Biophys Res Commun. 2003;309:18–25. doi: 10.1016/s0006-291x(03)01520-1. [DOI] [PubMed] [Google Scholar]
- 32.Issaq HJ, Veenstra TD, Conrads TP, Felschow D. Biochem.Biophys.Res.Commun. 2002;292:587–592. doi: 10.1006/bbrc.2002.6678. [DOI] [PubMed] [Google Scholar]
- 33.Forde CE, Gonzales AD, Smessaert JM, Murphy GA, et al. Biochem.Biophys.Res.Commun. 2002;290:1328–1335. doi: 10.1006/bbrc.2002.6352. [DOI] [PubMed] [Google Scholar]
- 34.Yip TT, Lomas L. Technol.Cancer Res.Treat. 2002;1:273–280. doi: 10.1177/153303460200100408. [DOI] [PubMed] [Google Scholar]
- 35.Merchant M, Weinberger SR. Electrophoresis. 2000;21:1164–1177. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1164::AID-ELPS1164>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 36.von Eggeling F, Junker K, Fiedle W, Wollscheid V, et al. Electrophoresis. 2001;22:2898–2902. doi: 10.1002/1522-2683(200108)22:14<2898::AID-ELPS2898>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 37.Weinberger SR, Viner RI, Ho P. Electrophoresis. 2002;23:3182–3192. doi: 10.1002/1522-2683(200209)23:18<3182::AID-ELPS3182>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 38.Tang N, Tornatore P, Weinberger SR. Mass Spectrom Rev. 2004;23:34–44. doi: 10.1002/mas.10066. [DOI] [PubMed] [Google Scholar]
- 39.Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, et al. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 40.Rosenblatt KP, Bryant-Greenwood P, Killian JK, Mehta A, et al. Annu.Rev.Med. 2004;55:97–112. doi: 10.1146/annurev.med.55.091902.105237. [DOI] [PubMed] [Google Scholar]
- 41.Kolch W, Mischak H, Chalmers MJ, Pitt A, Marshall AG. Rapid Commun.Mass Spectrom. 2004;18:2365. doi: 10.1002/rcm.1633. [DOI] [PubMed] [Google Scholar]
- 42.Check E. Nature. 2004;429:496–497. doi: 10.1038/429496a. [DOI] [PubMed] [Google Scholar]
- 43.Baggerly KA, Morris JS, Edmonson SR, Coombes KR. J.Natl.Cancer Inst. 2005;97:307–309. doi: 10.1093/jnci/dji008. [DOI] [PubMed] [Google Scholar]
- 44.Baggerly KA, Morris JS, Coombes KR. Bioinformatics. 2002;20:777–785. doi: 10.1093/bioinformatics/btg484. [DOI] [PubMed] [Google Scholar]
- 45.Orvisky E, Drake SK, Martin BM, bdel-Hamid M, et al. Proteomics. 2006;6:2895–2902. doi: 10.1002/pmic.200500443. [DOI] [PubMed] [Google Scholar]
- 46.Johannesson N, Wetterhall M, Markides KE, Bergquist J. Electrophoresis. 2004;25:809–816. doi: 10.1002/elps.200305719. [DOI] [PubMed] [Google Scholar]
- 47.Hernandez-Borges J, Neususs C, Cifuentes A, Pelzing M. Electrophoresis. 2004;25:2257–2281. doi: 10.1002/elps.200405954. [DOI] [PubMed] [Google Scholar]
- 48.Neususs C, Pelzing M, Macht M. Electrophoresis. 2002;23:3149–3159. doi: 10.1002/1522-2683(200209)23:18<3149::AID-ELPS3149>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 49.Gelpi E. J Mass Spectrom. 2002;37:241–253. doi: 10.1002/jms.297. [DOI] [PubMed] [Google Scholar]
- 50.Schmitt-Kopplin P, Frommberger M. Electrophoresis. 2003;24:3837–3867. doi: 10.1002/elps.200305659. [DOI] [PubMed] [Google Scholar]
- 51.Schmitt-Kopplin P, Matthias Englmann. Electrophoresis. 2005;26:1209–1220. doi: 10.1002/elps.200410355. [DOI] [PubMed] [Google Scholar]
- 52.Sassi AP, Andel F, III, Bitter HM, Brown MP, et al. Electrophoresis. 2005;26:1500–1512. doi: 10.1002/elps.200410127. [DOI] [PubMed] [Google Scholar]
- 53.Ullsten S, Danielsson R, Backstrom D, Sjoberg P, Bergquist J. J.Chromatogr.A. 2006;1117:87–93. doi: 10.1016/j.chroma.2006.03.048. [DOI] [PubMed] [Google Scholar]
- 54.Klampfl CW. Electrophoresis. 2006;27:3–34. doi: 10.1002/elps.200500523. [DOI] [PubMed] [Google Scholar]
- 55.Wittke S, Haubitz M, Walden M, Rohde F, et al. Am.J.Transplant. 2005;5:2479–2488. doi: 10.1111/j.1600-6143.2005.01053.x. [DOI] [PubMed] [Google Scholar]
- 56.Decramer S, Wittke S, Mischak H, Zurbig P, et al. Nat.Med. 2006;12:398–400. doi: 10.1038/nm1384. [DOI] [PubMed] [Google Scholar]
- 57.Neuhoff N, Kaiser T, Wittke S, Krebs R, et al. Rapid Commun.Mass Spectrom. 2004;18:149–156. doi: 10.1002/rcm.1294. [DOI] [PubMed] [Google Scholar]
- 58.Theodorescu D, Fliser D, Wittke S, Mischak H, et al. Electrophoresis. 2005;26:2797–2808. doi: 10.1002/elps.200400208. [DOI] [PubMed] [Google Scholar]
- 59.Kaiser T, Hermann A, Kielstein JT, Wittke S, et al. J Chromatogr.A. 2003;1013:157–171. doi: 10.1016/s0021-9673(03)00712-x. [DOI] [PubMed] [Google Scholar]
- 60.Wittke S, Fliser D, Haubitz M, Bartel S, et al. J Chromatogr.A. 2003;1013:173–181. doi: 10.1016/s0021-9673(03)00713-1. [DOI] [PubMed] [Google Scholar]
- 61.Bons JA, de BD, van Dieijen-Visser MP, Wodzig WK. Clin.Chim.Acta. 2006;366:249–256. doi: 10.1016/j.cca.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 62.Weissinger EM, Nguyen-Khoa T, Fumeron C, Saltiel C, et al. Proteomics. 2006;6:993–1000. doi: 10.1002/pmic.200500210. [DOI] [PubMed] [Google Scholar]
- 63.Adermann K, John H, Standker L, Forssmann WG. Curr.Opin.Biotechnol. 2004;15:599–606. doi: 10.1016/j.copbio.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 64.Lapolla A, Tubaro M, Fedele D, Reitano R, et al. Rapid Commun.Mass Spectrom. 2005;19:162–168. doi: 10.1002/rcm.1759. [DOI] [PubMed] [Google Scholar]
- 65.Schaub S, Wilkins JA, Antonovici M, Krokhin O, et al. Am.J.Transplant. 2005;5:729–738. doi: 10.1111/j.1600-6143.2005.00766.x. [DOI] [PubMed] [Google Scholar]
- 66.Haubitz M, Wittke S, Weissinger EM, Walden M, et al. Kidney Int. 2005;67:2313–2320. doi: 10.1111/j.1523-1755.2005.00335.x. [DOI] [PubMed] [Google Scholar]
- 67.Marshall AG, Hendrickson CL, Jackson GS. Mass Spectrom.Rev. 1998;17:1–35. doi: 10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 68.Zubarev RA. Mass Spectrom.Rev. 2003;22:57–77. doi: 10.1002/mas.10042. [DOI] [PubMed] [Google Scholar]
- 69.Chalmers MJ, Mackay CL, Hendrickson CL, Wittke S, et al. Anal.Chem. 2005;77:7163–7171. doi: 10.1021/ac050983o. [DOI] [PubMed] [Google Scholar]
- 70.Renfrow MB, Cooper HJ, Tomana M, Kulhavy R, et al. J.Biol.Chem. 2005;280:19136–19145. doi: 10.1074/jbc.M411368200. [DOI] [PubMed] [Google Scholar]
- 71.Coon JJ, Ueberheide B, Syka JE, Dryhurst DD, et al. Proc.Natl.Acad.Sci.U.S.A. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Good DM, Coon JJ. Biotechniques. 2006;40:783–789. doi: 10.2144/000112194. [DOI] [PubMed] [Google Scholar]
- 73.Patrie SM, Charlebois JP, Whipple D, Kelleher NL, et al. J.Am.Soc.Mass Spectrom. 2004;15:1099–1108. doi: 10.1016/j.jasms.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 74.Patrie SM, Ferguson JT, Robinson DE, Whipple D, et al. Mol.Cell Proteomics. 2006;5:14–25. doi: 10.1074/mcp.M500219-MCP200. [DOI] [PubMed] [Google Scholar]
- 75.Meng F, Forbes AJ, Miller LM, Kelleher NL. Mass Spectrom.Rev. 2005;24:126–134. doi: 10.1002/mas.20009. [DOI] [PubMed] [Google Scholar]
- 76.Roth MJ, Forbes AJ, Boyne MT, Kim YB, et al. Mol.Cell Proteomics. 2005;4:1002–1008. doi: 10.1074/mcp.M500064-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rejtar T, Hu P, Juhasz P, Campbell JM, et al. J.Proteome.Res. 2002;1:171–179. doi: 10.1021/pr015519o. [DOI] [PubMed] [Google Scholar]
- 78.Kaiser T, Kamal H, Rank A, Kolb HJ, et al. Blood. 2004;104:340–349. doi: 10.1182/blood-2004-02-0518. [DOI] [PubMed] [Google Scholar]
- 79.Mischak H, Kaiser T, Walden M, Hillmann M, et al. Clin.Sci.(Lond) 2004;107:485–495. doi: 10.1042/CS20040103. [DOI] [PubMed] [Google Scholar]
- 80.Palagi Patricia M, Daniel Walther, Manfredo Quadroni, Sébastien Catherinet, et al. Proteomics. 2005;5:2381–2384. [Google Scholar]
- 81.Weissinger EM, Wittke S, Kaiser T, Haller H, et al. Kidney Int. 2004;65:2426–2434. doi: 10.1111/j.1523-1755.2004.00659.x. [DOI] [PubMed] [Google Scholar]
- 82.Steinberg D, Colla P. CART - Classification and Regression Trees. San Diego: Salford Systems; 1997. [Google Scholar]
- 83.Breimann L, Friemann J, Olshen RA, Stone JC. Classification and regression trees. Belmont: Wadsworth; 1984. [Google Scholar]
- 84.Weissinger EM, Wittke S, Kaiser T, Haller H, et al. Kidney Int. 2004;65:2426–2434. doi: 10.1111/j.1523-1755.2004.00659.x. [DOI] [PubMed] [Google Scholar]
- 85.Peng S, Xu Q, Ling XB, Peng X, et al. FEBS Lett. 2003;555:358–362. doi: 10.1016/s0014-5793(03)01275-4. [DOI] [PubMed] [Google Scholar]
- 86.Willingale R, Jones DJ, Lamb JH, Quinn P, et al. Proteomics. 2006;6:5903–5914. doi: 10.1002/pmic.200600375. [DOI] [PubMed] [Google Scholar]
- 87.Thukral SK, Nordone PJ, Hu R, Sullivan L, et al. Toxicol.Pathol. 2005;33:343–355. doi: 10.1080/01926230590927230. [DOI] [PubMed] [Google Scholar]
- 88.Girolami M, Rogers S. Neural Computation. 2006;18:1790–1817. [Google Scholar]
- 89.Kote-Jarai Z, Matthews L, Osorio A, Shanley S, et al. Clin.Cancer Res. 2006;12:3896–3901. doi: 10.1158/1078-0432.CCR-05-2805. [DOI] [PubMed] [Google Scholar]
- 90.Rogers MA, Clarke P, Noble J, Munro NP, et al. Cancer Res. 2003;63:6971–6983. [PubMed] [Google Scholar]
- 91.Clarke W, Silverman BC, Zhang Z, Chan DW, et al. Ann Surg. 2003;237:660–664. doi: 10.1097/01.SLA.0000064293.57770.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schaub S, Rush D, Wilkins J, Gibson IW, et al. J Am.Soc.Nephrol. 2004;15:219–227. doi: 10.1097/01.asn.0000101031.52826.be. [DOI] [PubMed] [Google Scholar]
- 93.Neuhoff N, Kaiser T, Wittke S, Krebs R, et al. Rapid Commun.Mass Spectrom. 2004;18:149–156. doi: 10.1002/rcm.1294. [DOI] [PubMed] [Google Scholar]
- 94.Rossing K, Mischak H, Parving HH, Christensen PK, et al. Kidney Int. 2005;68:193–205. doi: 10.1111/j.1523-1755.2005.00394.x. [DOI] [PubMed] [Google Scholar]
- 95.Meier M, Kaiser T, Herrmann A, Knueppel S, et al. J.Diabetes Complications. 2005;19:223–232. doi: 10.1016/j.jdiacomp.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 96.Vlahou A, Schellhammer PF, Wright GL., Jr Adv.Exp.Med.Biol. 2003;539:47–60. doi: 10.1007/978-1-4419-8889-8_4. [DOI] [PubMed] [Google Scholar]
- 97.Vlahou A, Schellhammer PF, Mendrinos S, Patel K, et al. Am.J.Pathol. 2001;158:1491–1502. doi: 10.1016/S0002-9440(10)64100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vlahou A, Giannopoulos A, Gregory BW, Manousakas T, et al. Clin.Chem. 2004;50:1438–1441. doi: 10.1373/clinchem.2003.028035. [DOI] [PubMed] [Google Scholar]
- 99.Liu W, Guan M, Wu D, Zhang Y, et al. Eur.Urol. 2005;47:456–462. doi: 10.1016/j.eururo.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 100.Langbein S, Lehmann J, Harder A, Steidler A, et al. Technol.Cancer Res.Treat. 2006;5:67–72. doi: 10.1177/153303460600500109. [DOI] [PubMed] [Google Scholar]
- 101.Munro NP, Cairns DA, Clarke P, Rogers M, et al. Int.J.Cancer. 2006;119:2642–2650. doi: 10.1002/ijc.22238. [DOI] [PubMed] [Google Scholar]
- 102.M'koma AE, Blum DL, Norris JL, Koyama T, et al. Biochem.Biophys.Res.Commun. 2006 [Google Scholar]
- 103.Nemirovskiy OV, Dufield DR, Sunyer T, Aggarwal P, et al. Anal.Biochem. 2006 doi: 10.1016/j.ab.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 104.Weissinger EM, Hertenstein B, Mischak H, Ganser A. Expert.Rev.Proteomics. 2005;2:639–647. doi: 10.1586/14789450.2.5.639. [DOI] [PubMed] [Google Scholar]
- 105.Verhave JC, Hillege HL, Burgerhof JG, Gansevoort RT, et al. Kidney Int. 2005;67:1967–1973. doi: 10.1111/j.1523-1755.2005.00296.x. [DOI] [PubMed] [Google Scholar]
- 106.Schepky AG, Bensch KW, Schulz-Knappe P, Forssmann WG. Biomed.Chromatogr. 1994;8:90–94. doi: 10.1002/bmc.1130080209. [DOI] [PubMed] [Google Scholar]
- 107.Schulz-Knappe P, Schrader M, Standker L, Richter R, et al. J Chromatogr.A. 1997;776:125–132. doi: 10.1016/s0021-9673(97)00152-0. [DOI] [PubMed] [Google Scholar]
- 108.Fricke K, Schulz A, John H, Forssmann WG, Maronde E. Endocrinology. 2005;146:2060–2068. doi: 10.1210/en.2004-1097. [DOI] [PubMed] [Google Scholar]
- 109.John H, Radtke K, Standker L, Forssmann WG. Biochim Biophys Acta. 2005;1747:161–170. doi: 10.1016/j.bbapap.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 110.Li G, Chu J, Liu X, Yuan Z. Clin.Chim.Acta. 2004;350:89–98. doi: 10.1016/j.cccn.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 111.Weissinger EM, Kaiser T, Meert N, De Smet R, et al. Nephrol.Dial.Transplant. 2004;19:3068–3077. doi: 10.1093/ndt/gfh509. [DOI] [PubMed] [Google Scholar]
- 112.Comper WD, Osicka TM, Clark M, MacIsaac RJ, Jerums G. Kidney Int. 2004;65:1850–1855. doi: 10.1111/j.1523-1755.2004.00585.x. [DOI] [PubMed] [Google Scholar]
- 113.Haubitz M, Fliser D, Rupprecht H, Floege J, et al. Nephrology Dialysis Transplantation. 2005;20:V20–V20. [Google Scholar]