Abstract
Integrative biomedical research projects query, analyze, and integrate many different data types and make use of datasets obtained from measurements or simulations of structure and function at multiple biological scales. With the increasing availability of high-throughput and high-resolution instruments, the integrative biomedical research imposes many challenging requirements on software middleware systems. In this paper, we look at some of these requirements using example research pattern templates. We then discuss how middleware systems, which incorporate Grid and high-performance computing, could be employed to address the requirements.
Keywords: integrative biomedical research, system level integrative analysis, multi-scale integrative investigation
1 Introduction
Integrative biomedical research projects study complex interrelationships between different biological entities and across different biological scales (e.g., molecular, cellular, and organ) in order to understand the function and structure of biological processes in normal and disease conditions. A better understanding of the relationships can result in a better understanding of the mechanisms of complex diseases such as cancer and can lead to improved disease diagnosis and treatment strategies.
Integrative research studies carry out a wide range of experiments and capture a wide variety of data types. Datasets may be captured from high-throughput molecular analyzers (such as data from microarray analysis, mass spectroscopy measurements, and measurements done using real-time polymerase chain reaction (PCR) platforms), from high-resolution imaging of tissues and organs (using high-power light and confocal microscopy scanners, high-resolution magnetic resonance (MR) and positron emission tomography (PET) scanners), and from phenotypic observations such as electrocardiogram measurements. The datasets are integrated, analyzed, and mined using computational biology, image analysis, and bioinformatics techniques in order to look for patterns in data that can predict responses to treatment and provide insight into mechanisms of disease.
Biomedical researchers increasingly leverage high-throughput instruments capable of generating semantically complex terabyte-level molecular datasets and to scanners capable of rapidly capturing multi-terabyte high-resolution images of tissue and organs. As a result, researchers have a great opportunity to study biological processes at unprecedented resolutions and scales. While advanced data-capture technologies provide highly detailed views of structure and function of biological and physical processes, the researcher is faced with the need to efficiently store, manage, and analyze large volumes of semantically complex data in order to synthesize meaningful information. In addition, studies involving complex diseases often require collaboration between researchers with complementary expertise areas. In such studies, information can be captured and stored in disparate, heterogeneous databases and may need to be analyzed through applications developed and hosted by different groups.
In this paper, we examine and discuss the informatics requirements imposed on systems software by large-scale, multi-institutional integrative biomedical research studies. We describe these requirements using example research pattern templates (Saltz et al., 2008a, b, c). We discuss how Grid and high-performance computing techniques and middleware components could be used to address some of these requirements. We give examples of software middleware systems, which our group has been involved in the development of and which make use of Grid and high-performance computing techniques.
2 Example Integrative Biomedical Research Patterns
Specific research questions investigated in a study will determine the particular approach employed, the types of experiments (and some case simulations) carried out, the types and sizes of datasets collected, and the types of analyses executed. Nevertheless, research projects targeting similar problems employ common principles and processes. These principles and processes can be classified into broad groups of common patterns, referred to here as pattern templates (Saltz et al., 2008a, b, c). The concept of pattern templates is inspired by the work on pattern languages (Alexander, 1977) that capture common aspects of architectural design patterns and by the principles of software design patterns for software development (Gamma et al., 1994). In this work, we use pattern templates to capture, classify, and describe requirements, best practices, and constraints on families of projects and applications. In this section, we present examples of two pattern templates: system-level integrative analysis and multi-scale integrative investigation.
2.1 System-Level Integrative Analysis
The system-level integrative analysis template represents research studies that have the following characteristics: (1) a set of focused biological system questions are targeted in each study; (2) a closely coordinated set of experimental measurements are carried out; and (3) results from these experiments are integrated in order to answer the biomedical questions.
A good example of an application described by this pattern template is the effort on the part of the CardioVascular Research Grid (CVRG; http://www.cvrgrid.org) and the Reynolds project to answer the following question: “Who should receive implantable cardioverter defibrillators (ICDs)?”. This question has great practical significance since high-risk patients may receive ICDs. This study collects data from a set of patients with and without ICDs. The datasets gathered from the patients include gene expression, single nucleotide polymorphism (SNP), microarray data, ECG measurements, recorded firings of ICDs, and image data. These datasets are analyzed and integrated to predict the likelihood of potentially lethal arrhythmias.
Another example of this pattern template is the effort on the part of the Ohio State Center for Integrative Cancer Biology (http://icbp.med.ohio-state.edu/). One of the focused questions targeted in this project is “which ovarian cancer patients are best suited for a given therapy?”. The project carries out a coordinated set of measurements from Chromatin-immunoprecipitation microarray (or ChIP-chip), differential methylation hybridization (DMH), and gene expression profiling experiments (Han et al., 2008). Epigenetics, gene sequence, microarray, and proteomics datasets collected from these experiments are integrated in order to understand the impact of epigenetic changes on particular genomic pathways. A deep understanding of this biological system can be used to develop new drugs and to evaluate which patients are best suited for a given therapy.
2.2 Multi-Scale Integrative Investigation
The multi-scale integrative investigation pattern template models research studies that have the following characteristics: (1) the goal in these studies is to measure and quantify biomedical phenomena; (2) data is obtained from experimental measurements (in some cases simulations) of multiple biological scales (e.g., molecular, cellular, and macro-anatomic scales); (3) these datasets are analyzed and integrated to understand the morphology and processes of the biomedical phenomena in space and time.
An example of the multi-scale integrative investigation is the study of the tumor microenvironment (TME) in order to understand the mechanisms of cancer development. Many research studies have shown that cancer development occurs in space and time; interactions among multiple different cell types, regulation, protein expression, signaling, and blood vessel recruitment happen in time and space. The TME consists of different types of cells, including fibroblasts, glial cells, vascular and immune cells, and the extra cellular matrix (ECM) that holds them together. The cellular organization of tissues is possible via cellular signal interchange in the TME. In a TME project, the investigation may focus on how alternations in intercellular signaling could happen and how morphology and cellular-level processes are associated with genetics, genomics, and protein expression. Understanding cellular signal interchange can lead to a better understanding of malignant cancer development and progression.
Image acquisition, processing, classification, and analysis play a central role in the multi-scale integrative investigation template. Datasets may arise from high-resolution microscopy images obtained from tissue samples. Hundreds of images can be obtained from one tissue specimen, thus generating both two- and three-dimensional morphological information. In addition, image sets can be captured at multiple time points to form a temporal view of morphological changes. The images are processed through a series of simple and complex operations expressed as a data analysis workflow. The workflow may include steps such as cropping, correction of various data acquisition artifacts, segmentation, registration, classification of image regions and cell types, as well as interactive inspection and annotation of images. The analysis workflow annotates image regions with cell types and the spatial characteristics of the cells. This information is then combined with molecular information to investigate correlations and associations between molecular and morphological information. Genetic and cellular information can further be integrated with biological pathway information to study the impact of genetic, epigenetic, and cellular changes on major pathways.
Simulation also plays a crucial role in the multi-scale integrative investigation template. As knowledge of basic biomedical phenomena increases, the ability to carry out meaningful detailed simulations increases dramatically. Some researchers are now carrying out TME simulations and we expect the prevalence of this to increase dramatically with the improved quality of detailed multi-scale data.
3 Informatics Requirements of Pattern Template Examples
Both example templates involve the integration of many types of information from a variety of data resources in order to synthesize information. In the case of the system-level integrative analysis template, for example, multi-institutional studies access data from resources hosted at different institutions. Information is drawn from commercial/enterprise systems, e.g., health information records, laboratory information management systems, the Picture Archiving and Communications System (PACS) (Huang et al., 1991), as well as genetic, genomic, epigenetic, and microscopy databases. As a result, successful implementation of the example templates is influenced by (1) how effectively the researcher can discover information that is available and relevant to the research project and (2) how efficiently they can query, analyze, and integrate information from different resources. This is a challenging issue since disparate data sources are heterogeneous and cannot readily interoperate.
The lack of interoperability among data sources is more apparent in multi-institutional settings; however, interoperability of resources even within a large institution is often limited. The same type of data may be expressed in different formats. Naming schemes and semantic metadata associated with data types are heterogeneous and not compatible: they are often developed and managed in silos. Middleware systems need to provide tools and infrastructure support to overcome these barriers to syntactic and semantic interoperability. With syntactic interoperability, the functionality of a data or analytical resource can be accessed via programmatic interfaces and data structures can be exchanged between resources programmatically. With semantic interoperability, systems can consistently exchange information and can perform reasoning on conceptual knowledge types. This enables semantically correct and unambiguous use of resources and their content.
Handling of semantic information plays a key role in the two templates, but more so in the multi-scale integrative investigation template. Image annotations and molecular information in the study of the TME, for example, can be associated with concepts defined in domain-specific ontologies. Annotated image and molecular data form a multi-dimensional model of the TME represented in a semantic knowledge base. It is desirable for a researcher to be able to query these complex and hierarchical micro-anatomic structures and molecular compositions using both semantic annotations spatial predicates in order to explore interactions across different biological scales. As an example, consider Figure 1, which illustrates a microscopy image annotated with different types of cells and regions. The ontology in this example may have the following concepts and relationships: “Endothelial cells touch blood vessel lumen” and “Protein C is expressed only in endothelial cells”. The data gathered from experiments and analysis may have the following information: “Region A is a cell (from image analysis)”, “Region A expresses protein C (from molecular assay)”, and “Region B (from expert markup)”. Analysis of the image also shows that Region B touches Region A. Given the ontology and the spatial characteristics of the dataset, it can be inferred that “Region A is an endothelial cell”, “Region B is a blood vessel”. Thus, a query searching for endothelial cells or blood vessel should return Region A and Region B, respectively. With a more comprehensive version of this example ontology, other examples of questions researchers may ask include “What is the morphological/molecular effect on cell type 1 if we make a genetic change in cell type 2?” or “Are the modifications of the gene expression more evident in fibroblasts, macrophages or endothelial cells?”.
Fig. 1.
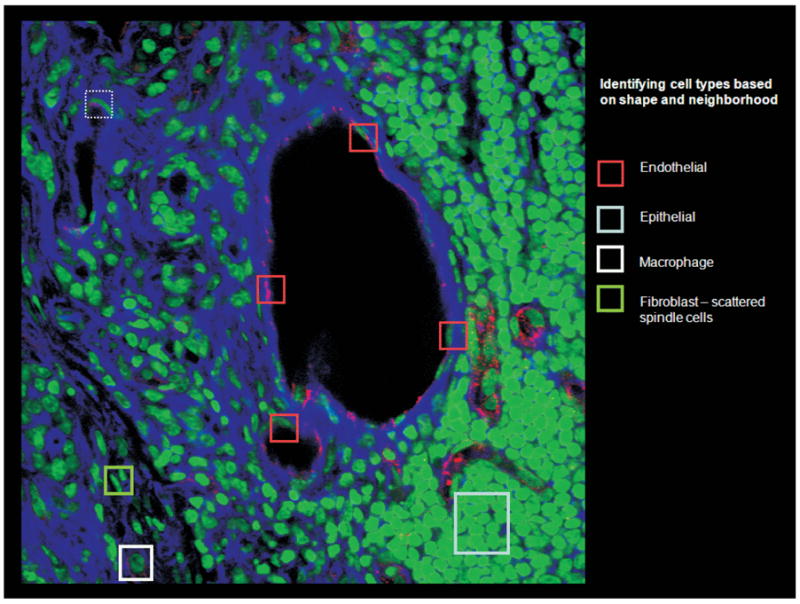
Microscopy image marked with cell types and regions annotated automatically and manually.
It is also desirable to support the definition of new concepts and classifications based on already existing annotations and spatial information. The following rule, for example, creates a new concept, “foreign body”, using terms from ontology and spatial relationships. It states that any entity that is classified as “unknown” and is within 10 units of distance from a macrophage is a foreign body (a macrophage is a first line of defense against foreign bodies in animals). Here, the term “macrophage” may come from a cell ontology:
High-performance computing requirements in these example templates arise from many sources. Clinical outcome information in the system-level integrative analysis template may be captured in unstructured text documents. Structured information for data integration can be extracted from the text documents using natural language processing (NLP) techniques. NLP methods are computationally expensive, and a study may need to process hundreds or thousands of text documents. Whole genome analyses and coordinated analyses of molecular and imaging data are other sources of compute- and data-intensive operations. High-throughput instruments are capable of generating multi-gigabyte genomic datasets. Processing and mining of these datasets on a desktop machine may take several hours, even days. Similarly, microscopy images obtained from advanced scanners may reach multiple gigabytes in size. In studies involving three-dimensional reconstructions and time-dependent acquisition of images, storage and processing of the image datasets can be prohibitively expensive on desktop machines.
Exploration of terabyte-scale datasets require support for a researcher to create a global view of all datasets in a study and efficiently drill down to localized, higher-resolution representations. Systems software should be able to support efficient computation of various summaries and representative samples from datasets to speed up the knowledge extraction process. These systems should implement architecture-conscious caching techniques, descriptive metadata structures, and index schemes to manage the summary products so that they can be retrieved and processed quickly. Mechanisms are also desired that can incorporate trading accuracy of query/analysis results for performance, while meeting user-defined accuracy or performance requirements.
High-performance computing (HPC) support is also needed to enable efficient management and querying of very large semantic and spatial information. A high-resolution image may contain thousands of cells and hundreds of slides may be obtained from a specimen. The researcher has to manage and interact with large volumes of image data, large numbers of objects (corresponding to segmented and classified regions), and semantic information. The size of the semantic information component can easily exceed millions of explicit semantic annotations (about cell information and genomic data).
Compelling workflow and federated query use cases, which involve multiple data sources and analytical services, arise from the example templates described here. Studies in the system-level integrative analysis template, for instance, query information associated with multiple groups of subjects, compare and correlate the information about the subject under study with this information, and classify the analysis results. The multi-scale template, and the system-level analysis template to an extent, can involve processing large volumes of image data, including three-dimensional reconstruction, segmentation, and feature detection and classification. These types of analyses require high-performance analytical services, the backend of which should leverage distributed memory clusters, filter/stream-based high-performance computing, multi-core systems, symmetric multiprocessor (SMP) systems, and parallel file systems. In addition, workflow execution engines capable of efficient inter-resource (inter-service) large-scale data transfers are needed. One of the challenges is to be able to enable composition of hierarchical workflows, in which a workflow step itself can be another workflow, and coordinate execution of the interactions between services as well as fine-grained dataflow operations within a service.
4 Grid and HPC for Integrative Biomedical Research
This section presents examples of software support that employ Grid and HPC to address the requirements of integrative biomedical research. We describe the caGrid infrastructure to present an implementation choice for system-level integrative analysis studies in multi-institutional settings. Next we describe a proof-of-concept implementation to illustrate the use of parallel computing for semantic query support in the multi-scale integrative investigation pattern template.
4.1 Cagrid Infrastructure
A more detailed description of caGrid can be found in earlier publications (Saltz et al., 2006; Oster et al., 2008). Here we provide a brief overview of the main characteristics of caGrid and how it can be used to support the system-level integrative analysis template as well as potential future improvements to caGrid in order to more efficiently support the pattern templates.
caGrid is a Grid middleware infrastructure designed (1) to facilitate collaborative research studies, (2) to allow researchers to both contribute data and analytical resources and discover resources in the environment, and (3) to enable federated queries across disparate databases and execution of workflows encompassing distributed databases and analytical tools. caGrid implements an infrastructure wherein the structure and semantics of data can be programmatically determined and by which distributed data and analytical resources can be programmatically discovered and accessed.
caGrid is built upon the Grid Services standards as a service-oriented architecture. Each data and analytical resource in caGrid is implemented as a Grid Service. caGrid services are standard Web Services Resource Framework (WSRF) v1.2 services (Foster et al., 2005) and can be accessed by any specification-compliant client. A key difference of caGrid from other Grid systems is the emphasis on interoperability of resources in the caGrid environment. To this end, caGrid draws from the basic principles of the model-driven architecture. Data and analytical resources are made available to the environment through object-oriented service application programming interfaces (APIs). These APIs operate on registered data models, expressed as object classes and relationships between the classes in UML. caGrid leverages the existing NCI data modeling infrastructure, the Cancer Data Standards Repository (caDSR) and the Enterprise Vocabulary Services (EVS) (Covitz et al., 2003; Phillips et al., 2006), to manage, curate, semantically annotate, and employ these data models. At the Grid level, services produce and consume objects serialized into XML documents that conform to XML schemas registered in the Mobius GME (generic modeling environment) service (Hastings et al., 2007). In effect, properties and semantics of data types are defined in caDSR and EVS and the structure of their XML materialization in the Mobius GME.
The caGrid infrastructure also consists of coordination services, a runtime environment to support the deployment, execution, and invocation of data and analytical services, and tools for easier development of services, management of security, and composition of services into workflows. The coordination services provide support for common Grid-wide operations required by clients and other services. These operations include metadata management, advertisement and discovery, federated query, workflow management, and security. The coordination services can be replicated and distributed to achieve better performance and scalability to large numbers of clients.
caGrid has been used in the implementation of applications and tools to support informatics needs of a number of biomedical research projects. caGrid is the Grid architecture of the cancer Biomedical Informatics Grid (caBIG, https://cabig.nci.nih.gov) program. It is also used as the core middleware infrastructure in the CVRG (http://cvrgrid.org).
Data integration from multiple and distributed resources is a common requirement arising in the example templates. In the system-level integrative analysis template, researchers may create molecular, proteomic, clinical, and image databases using in-house tools or existing public or commercial applications. The service-oriented and model-driven architecture of caGrid can provide support for interoperability of these databases and for federated and secure access to these resources.
The first step in producing interoperable resources is for resource providers to agree on some common data structures and their semantics. These agreed-upon data structures could be published in the environment so that other researchers can reuse them. In the caBIG environment, for example, community accepted data models are registered in caDSR and their semantics are drawn from vocabularies managed in the EVS. XML schemas corresponding to the registered data models are stored in the GME so they are available at the Grid level. Using the caGrid service development tools (Hastings et al., 2007), a research group can then wrap their databases and analysis methods as caGrid data and analytical services, respectively. Data managed or manipulated in these services is exposed via service interfaces to the environment using the published data models and XML schemas. In this way, a client application can interact with these databases via well-defined interfaces and data models without needing to know how the data is stored in respective database systems.
The security infrastructure of caGrid (Langella et al., 2008) can be employed to ensure that only users with appropriate privileges can access controlled and sensitive information or interact with the services.
A cooperative group, for example, could expose their data collection repositories as secure caGrid services, allowing collaborating sites to securely access data and analytical tools, which are wrapped as services, without needing to install any special software. Using the federated query mechanism in caGrid, a researcher can compose and execute a query across multiple data services to, for example, retrieve microarray data, SNP data, image data, and clinical outcome data on a group of patients. The results of the query can then be processed through an analysis workflow involving analysis services at multiple locations.
caGrid provides support for federated querying of multiple data services to enable distributed aggregation and joins on object classes and object associations defined in domain object models. The current support for federated query is aimed at the basic functionality required for data subsetting and integration. Extensions to this basic support are needed to provide more comprehensive support. Scalability of federated query support is important when there are large numbers of clients and queries span large volumes of data and a large number of services. Middleware components need to be developed that will enable distributed execution of queries by using HPC systems available in the environment as well as by carefully creating sub-queries, pushing them to services or groups of services for execution, and coordinating data exchange between services to minimize communication overheads.
caGrid provides a workflow management service that supports the execution and monitoring of workflows expressed in the Business Process Execution Language (BPEL) (http://www.ibm.com/developerworks/library/specification/ws-bpel/). The use of BPEL in caGrid facilitates easier sharing and exchange of workflows. Although a powerful language, BPEL is difficult for users to use. There are frameworks such as WEEP (http://weep.grid-miner.org/index.php/Main_Page) that are designed to provide high-level API and runtime support for management and execution of BPEL-based workflows. Higher-level user interfaces and support are necessary to more easily integrate complex analysis workflows in the biomedical research process. To this end, the caGrid development effort has implemented support to use the Taverna Work-flow Management System (Hull et al., 2006) in version 1.2 of caGrid core infrastructure. The Taverna environment provides a graphical development graphical user interface (GUI) and support for shims to enable composition of incompatible services.
Researchers are capable of collecting or generating large volumes of data thanks to advanced scanners and analysis instruments. The data- and compute-intensive nature of scientific data analysis applications demands scalable solutions. Workflow support should implement several optimization mechanisms to reduce execution times. First, the workflow system should take advantage of distributed computing resources on the Grid. The Grid environment provides computation and storage resources. The workflow system should support execution of workflow components at different sites (Grid nodes) and reliable, efficient staging of data across the Grid nodes. A Grid node may itself be a cluster system or a potentially heterogeneous and dynamic collection of machines. The workflow system should enable scheduling of tasks onto such machines and efficient execution of the portions of the workflow, which are mapped to that Grid node, on cluster systems. Second, performance of workflows is affected by application-specific parameters. Performance optimization of a workflow requires finding a set of optimal values for these performance parameters. Workflow-level performance parameters include grouping of data processing components comprising the workflow into meta-components, distribution of components across sites and machines within a site, and the number of copies of a component to be executed. These parameters have an impact on the computation, input/output (I/O), and communication overheads, and as a result, the total execution time. Another means of improving performance is by adjusting component-level performance parameters in a workflow. An example is the data chunk size in applications, which analyze spatial datasets. Another example is the version of the algorithm employed by a component to process the data. The workflow system should provide support to improve performance through manipulation of such parameters along multiple dimensions of the parameter space.
In recent work we have developed a workflow middleware that supports data analysis workflows in a Grid environment with cluster-based Grid nodes and that allows for performance improvement via manipulation of workflow-level and component-level performance parameters (Kumar et al., 2009). We are in the early stages of investigating how this system can be integrated with caGrid. The workflow middleware integrates four systems: WINGS (Gil et al., 2007), Pegasus (Deelman et al., 2004), Condor (Thain et al., 2005), and DataCutter (Beynon et al., 2001). The WINGS system is used to facilitate high-level semantic representation of workflows. In the WINGS system, the building blocks of workflows are components and data types. An application- or domain-specific library defines the input and output data types of each component and how metadata for input types are associated with that for output types. The data types are defined in an application-specific ontology. A user can describe a workflow using semantic properties associated with components and data types using the Web Ontology Language (OWL). Using this workflow template, the user can specify a dataset instance (e.g., an image or group of images) as input to the workflow. A specification of the workflow is then generated by the workflow system in the form of a directed acyclic graph. Once a workflow has been specified, the user can adjust workflow-level and component-level parameters and quality-of-service requirements to enable performance optimizations. The Pegasus Workflow Management System is used to reliably map and execute application workflows onto diverse computing resources in the Grid. Condor is used to schedule tasks across machines. Pegasus submits tasks in the form of a directed acyclic graph (DAG) to Condor instances running locally at each Grid site. We have extended Condor’s default job-scheduling mechanism to support performance optimizations stemming from quality–performance tradeoffs. DataCutter is employed for pipelined dataflow style execution of portions of a workflow mapped to a Grid site consisting of cluster-style systems. A task mapped by Pegasus to a site and scheduled for execution by Condor may correspond to a meta-component. In that case, the execution of the meta-component is carried out by DataCutter in order to enable the combined use of task and data parallelism and data streaming among components of the meta-component.
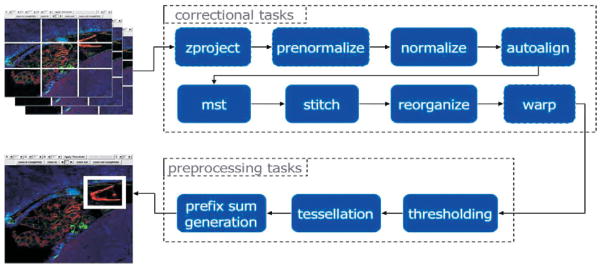
We have performed an experimental evaluation of this workflow system using a pixel intensity quantification (PIQ) workflow across two cluster systems hosted in different departments at the Ohio State University. The first consists of 64 dual-processor nodes equipped with 2.4 GHz AMD Opteron processors and 8 GB of memory, interconnected by a Gigabit Ethernet network. The storage system consists of two 250 GB SATA disks installed locally on each compute node, joined into a 437 GB RAID0 volume. The second cluster is a 32-node cluster consisting of faster dual-processor 3.6 GHz Intel Xeon nodes each with 2 GB of memory and only 10 GB of local disk space. This cluster is equipped with both an InfiniBand interconnect as well as a 1 Gbps Ethernet network. The two clusters are connected by a 10-Gigabit wide-area network connection: each node is connected to the network via a Gigabit card. The PIQ workflow is shown in Figure 2.
Fig. 2.
The PIQ workflow.
In our experiments, we have created a grouping of multiple components of the workflow into meta-components to generate a workflow with a coarser task granularity. The zproject and prenormalize steps formed one meta-component, the normalize, autoalign, and mst steps formed another meta-component, and the stitch, reorganize, and warp steps formed the third meta-component. Preprocess is the fourth meta-component in the workflow template and includes the threshold, tessellate, and prefix sum components. When component collections are embedded within a meta-component, they are not explicitly unrolled at the time of workflow instance generation. Instead, they are implicitly unrolled within a DataCutter instance. DataCutter will create multiple copies of filters that handle the processing of component collection tasks. Also using the DataCutter system, data processing within the preprocess meta-component is pipelined and streamed between the components without any disk I/O during execution. Our experimental results show that by combining components into meta-components and employing high-performance dataflow techniques within a meta-component mapped to a cluster system, we can improve the performance of the workflow by over 50%. The workflow system enables this performance improvement by allowing the user to adjust performance parameters, in this case the granularity of the workflow steps.
4.2 Parallel Materialization in a Spatial Knowledge Base
The multi-scale investigation template requires support for querying of data using semantic annotations as well as spatial predicates. It is also desirable to enable creation of new concepts and semantic information using existing annotations and spatial relationships. Ontology languages such as RDFS and OWL, in their current form, support explicit role relationships between individuals (instance data) and do not offer any special support for spatial data types and predicates. Rule-based programming offers a convenient abstraction to link spatial and non-spatial information. Existing semantic stores, such as SwiftOWLIM (Kiryakov et al., 2005), are optimized for materializing RDFS/OWL ontologies. Rule systems such as JESS (Hill, 2003) are suitable for forward chaining on rules. They support user-defined predicates as black-boxes. The introduction of spatial predicates in rules, however, may result in a large number of computations.
We describe a parallel approach that combines a semantic storage engine with a rule engine to carry out materialization in a spatial knowledge base (SKB), which consists of an ontology, explicit assertions, and rules involving spatial predicates. Materialization is the process of computing all inferred assertions as per the semantics of the ontology and rules with spatial predicates in the knowledge base. Materialization can speed up the execution of subsequent queries into the database: materialized assertions can be loaded into a database and optimization techniques including indexing can be utilized.
The parallel approach employs spatial partitioning techniques along with a demand-driven strategy for execution. It makes use of spatial predicates in rules to partition space and achieve data parallelism. The demand-driven strategy is used to achieve load-balanced execution among processing nodes.
We define a SKB as consisting of four parts: TBox, ABox, spatial object mapping, and rule set. A TBox consists of concepts and roles and expresses relationships between them in an ontology language such as RDFS or OWL. An ABox consists of individuals and property assertions. Individuals may belong to one or more concepts and may be related to each other via roles. The individuals may be associated with a spatial object that covers a certain portion of a multi-dimensional space. The third part contains this mapping. The final piece is a set of rules that capture higher-level application semantics using rules that involve ontology and spatial constraints. A rule is of the form left-hand side (LHS) ? right-hand side (RHS), which is interpreted as when the conditions of LHS are true for some binding of variables. The RHS is asserted for the set of bindings. The LHS is a conjunction of conditional elements (CEs). A CE may be an ontology constraint or a spatial predicate. The RHS is a conjunction of ontology constraints.
The first step in materialization is the calculation of inferred assertions due to the semantics of the ontology. Rule-based inference is applied on the inferred information, which may generate additional assertions. These two steps may be repeated until no new assertions are generated.
The parallel approach employs a master node and worker nodes. The master node is responsible for partitioning the space and assigning partitions to workers. Workers report to the master when they are idle and are assigned a partition by the master node. Execution proceeds in a demand-driven fashion.
4.2.1 Master Node: Partitioning Space
Each spatial individual in the dataset is assigned a minimum bounding sphere (MBS), which reflects the boundaries of the individual. If the individual is a point, the MBS of the individual will be zero. The master node parses the rule set and expands the MBS associated with spatial individuals to form spheres of influence (SOIs). The SOI is the indication of how two spatial individual might affect a rule in the rule set. For example, if there is a rule
This rule states that if the distance between two individuals x and y is less than 10 units, y is an element of concept C. If the two individuals are points, their MBS will be zero. However, if they are within 10 units of each other, the rule should return true. In this example, the SOIs of individuals in the dataset because of this rule will be 5 units.
Initially, each spatial individual is assigned a SOI, which is the same as its MBS. For a given rule set, the master node computes the minimum radius of the sphere of influence (R) over all objects in the dataset. To compute R for a rule set, the master node computes a minimum radius value per rule, R1, R2, … in the rule set. It then computes the maximum of these values. If the SOI of an individual is smaller than the minimum value, it is expanded to be the minimum value. Since the parallel algorithm partitions the space, the computation of SOIs is necessary to ensure correctness when materialization is carried out. Without SOIs, it is possible that two individuals that may be required for a rule’s evaluation may be incorrectly assigned to two different partitions.
The master node maintains a stack of partitions. A partition consists of a bounding rectangle and a set of individuals. Initially, there is a single partition that holds all individuals. The top partition is retrieved, and if the size of the partition (i.e. number of individuals) is less than the target size, it is treated as a partition ready to be processed by a worker node. Otherwise, it is partitioned into two smaller partitions and the two smaller partitions are pushed onto the stack. We employ a variant of the kd-tree (Ooi et al., 1987), node-splitting algorithm to partition the space. Given a partition, the algorithm chooses the longer dimension of the containing space and finds a splitting plane that creates two balanced partitions. That is, the number of individuals in partition is approximately equal to the number of individuals in the other partition. Individuals are assigned to one of the partitions based on which partition contains their SOIs. Note that some individuals’ SOIs may transcend the splitting plane. These individuals are assigned to both partitions. When a master generates a small enough partition, it signals an available worker and passes on the partition information, i.e., the bounds of the partition.
4.2.2 Worker Node: Processing a Partition
Upon receiving a partition, a worker node extracts the individuals relevant to that partition and the rules and pushes them into the rule engine. This involves the following steps:
Identify concepts and roles in the LHS of the rule set.
Extract individuals (or pairs of individuals) corresponding to the concepts (or roles).
Filter individuals (or pairs of individuals) if the concept (or role) is spatially restricted. A concept is said to be spatially restricted if the concerned variables are involved in spatial predicates. Individuals that fall outside the partition are not considered.
Add filtered assertions to the rule engine and allow the rule engine to process information. This may result in new assertions, which are fed back to the rule engine.
The spatial predicate is never computed for a binding of individuals that are not mapped to the same partition. The worker node records the new assertions generated by the rule engine and signals availability to master and waits for the next partition. This finishes the processing of a partition.
4.2.3 Combining Results from Partitions
When the master is done sending all of the partitions, it signals the end of the first iteration to all of the workers. Workers combine the new assertions from all partitions and perform ontological materialization. In most situations, this would be the end of processing and a trivial union of results from all workers would correspond to the materialization of the dataset. However, if the rule set is recursive, further processing may be required. A rule set is recursive if a fact asserted by the RHS of a rule may cause additional matches in the LHS of some rule. Recall that a spatial individual may be mapped to more than one partition because of its SOI overlapping a splitting plane. It is possible that processing of one partition may cause inferred assertions on the individual, which may in turn affect the processing of the second partition. It is possible that a spatial individual is assigned to two partitions and is inferred as being member of a concept in one of them. This new assertion must be propagated to the other partition. The execution proceeds in iterations to solve this problem. At the end of the first iteration, worker nodes exchange newly generated assertions, which may satisfy the LHS of some rule. To optimize communication, the workers do not exchange all inferred assertions. They only exchange inferred assertions corresponding to concepts (and roles) that appear in the LHS of some rule in the ruleset. Each worker then revisits each partition it processed in the previous iteration and checks for a change in the number of input individuals. If a worker detects a difference, then the partition is processed as in the first iteration. If no worker detects any change in input size for any partition, then execution is halted as the fixed point as been reached. Thus, the parallel approach handles recursive rule sets over multiple iterations.
4.2.4 Proof-of-Concept Implementation
We have developed a proof-of-concept implementation of the presented approach using DataCutter (Beynon et al., 2001), a component-based middleware framework. In DataCutter, an application processing structure is implemented as a set of components, referred to as filters, that exchange data through a stream abstraction. We used a version of DataCutter that employs MPI as the message-passing substrate. The master and worker processes are DataCutter filters. The DataCutter subsystem is responsible for launching the master and worker processes on nodes in a cluster and setting up communication links. We used SwiftOWLIM library (Kiryakov et al., 2005), which is a very efficient semantic repository. We also utilize JESS (Hill, 1987), a rule engine system written in Java, for execution of rules. We implemented the spatial predicates as user-defined functions and a ruleset parser using the ANTLR tool (Parr and Quong, 1995).
We performed an experimental evaluation using a distributed memory parallel machine and synthetically generated confocal microscopy dataset. Each compute node has a dual AMD 250 Opteron processor running at 2.8 GHz with 8 GB of main memory. The nodes were connected through an Infiniband Switch.
Confocal microscopy datasets were synthetically generated in the spirit of real images obtained from confocal microscope in a TME setting. Scientists are primarily interested in the relative locations of epithelial cells, endothelial cells, and macrophages. Typically epithelial cells are found to be lining ducts or blood vessels. Endothelial cells are usually found embedded in the extra-cellular matrix. We generated a dataset with 100,000 cells in a single three-dimensional image. Epithelial cells are the most numerous (about 85% of the total number) while endothelial cells and macrophages, together, form about 5% of the total. The rest of the cells are marked as unknown to simulate the situation when an image analysis algorithm cannot classify a cell with the required confidence level.
Table 1 shows a set of sample rules. The first rule states that an unknown cell that is in close proximity to an epithelial cell may be marked as an epithelial cell. This is a recursive rule. The second rule states that if an endothelial cell is very close to an epithelial cell, it may in fact be a special type of endothelial cell that lies within the blood vessel. Macrophages are the body’s first line of defense against foreign bodies. The third rule states that any unidentified entity that is very close to a macrophage is a foreign body.
Table 1.
Example rules for confocal microscopy datasets in a TME study
| Rule 1 | EpithelialCell(?x) ? UnknownCell(?y) → EpithelialCell(?y) ? d(?x,?y) < 20 |
| Rule 2 | EndothelialCell(?x) ? EpithelialCell(?y) → BloodVesselEndothelialCell(?x) ? d(?x,?y) < 20 |
| Rule 3 | UnknownCell(?x) ? Macrophage(?y) → ForeignBody(?x) ? d(?x,?y) < 20 |
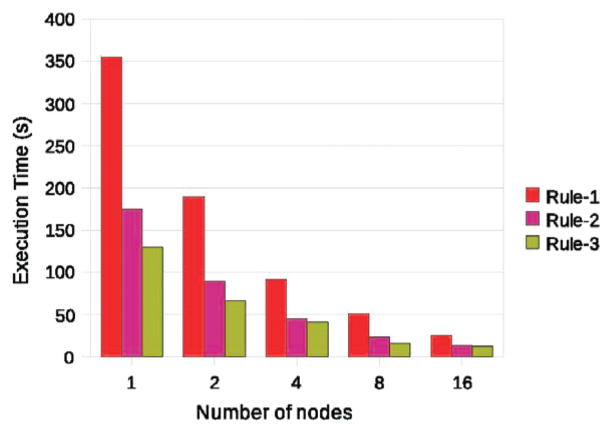
Figure 3 shows the scalability of the system for the three rules. The first rule is the most expensive since it is recursive and requires multiple iterations. The second rule involves testing more combinations of individuals than the third rule and is more computationally expensive. We can see good speedup in the case of the recursive rule as well. The number of unknown cells is relatively small (8%) and they are distributed in the extra-cellular matrix. The computational work is well distributed among partitions and execution stops in two iterations.
Fig. 3.
Execution times on 1–16 processors.
5 Conclusions
Integrative biomedical research is a challenging field for information technology developers. The requirements of integrative research projects span a wide range of challenging problems, from semantic information management to processing of large data volumes to integration of information in distributed environments. Individual middleware systems and software components can address some of these requirements, as described in this paper. However, there is still substantial work to be done in order to fully support large-scale, multi-institutional integrative biomedical research studies. It is necessary to implement integrated environments that combine model-driven architectures, service-oriented architectures, Grid computing systems, HPC techniques, and techniques in semantic information management.
Acknowledgments
This research was supported in part by the National Science Foundation under Grants #CNS-0403342, #CNS-0426241, #CSR-0509517, #CSR-0615412, #ANI-0330612, #CCF-0342615, #CNS-0203846, #ACI-0130437, #CNS-0615155, #CNS-0406386, the Ohio Board of Regents through grants BRTTC #BRTT02-0003 and AGMT TECH-04049, and the NIH through grants #R24 HL085343, #R01 LM009239, and #79077CBS10.
Biographies
Tahsin Kurc is Chief Software Architect in the Center for Comprehensive Informatics at Emory University. He is also an acting research associate professor at the Department of Biomedical Engineering. He received his PhD in computer science from Bilkent University, Turkey, in 1997 and his BSc in electrical and electronics engineering from Middle East Technical University, Turkey, in 1989. He was a Postdoctoral Research Associate at the University of Maryland from 1997 to 2000. Before coming to Emory, he was a Research Assistant Professor in the Biomedical Informatics Department at the Ohio State University. His research areas include high-performance data-intensive computing, runtime systems for efficient storage and processing of very large scientific datasets, domain decomposition methods for unstructured domains, parallel algorithms for scientific and engineering applications, and scientific visualization on high-performance machines.
Shannon Hastings received his MSc in Computer Science from Rensselaer Polytechnic Institute and his BSc in Computer Science from the Ohio University. He is currently a co-director of the Software Research Institute at the Ohio State University and the lead architect of the Introduce toolkit in the caGrid infrastructure. His research areas include high-performance and distributed computing, Grid computing, middleware systems for large-scale distributed scientific data management, distributed image management and analysis frameworks, large-scale service-based software systems designs, and software quality and testing concepts and frameworks.
Vijay S. Kumar is pursuing his PhD in computer science at The Ohio State University. He received a BEng in computer science from the Birla Institute of Technology and Science, Pilani, India in 2003. His research interests include enabling intelligent performance optimizations for data-intensive scientific application workflows, supporting performance–quality trade-offs in data analysis, parallel algorithms for spatial data analysis, and knowledge representation for the performance domain.
Stephen Langella is currently co-Director of the Software Research Institute (SRI) in the Department of Biomedical Informatics at the Ohio State University where he investigates his current interests in the areas of security, Grid computing, high-performance distributed computing and large-scale data management. He is a director of the caGrid Knowledge Center, a community resource to groups and communities wishing to use and contribute to caGrid. He is the lead security architect for the caGrid project, a multi-institutional effort and the core middle-ware for the Cancer Biomedical Informatics Grid. He received his BSc in Computer Science from the University of Buffalo and his MSc in Computer Science from Rensselaer Polytechnic Institute. Previously he worked as a Research Scientist at General Electric’s Global Research Center in the Visualization and Computer Vision and Advanced Computing Technologies Laboratories.
Ashish Sharma is a Senior Systems Architect in the Center for Comprehensive Informatics and an Assistant Professor at the Department of Biomedical Engineering at Emory University. He received his PhD in Computer Science from the University of Southern California in 2005 and his BEng in Electrical Engineering from Punjab University, India, in 1999. His research interests include medical imaging, massive data processing, scientific visualization, and Grid computing. Some of his recent research topics include image analysis of microscopy data, mul-tiresolution image processing, and scientific visualization of massive datasets.
Tony Pan has an MSc from Rensselaer Polytechnic Institute. His current research interests include Grid-enabled storage, management, processing, and analysis of large-scale microscopy and radiology images using distributed middleware. More recently, he has been involved with developing Grid-based image and data analysis applications and infrastructure for both the National Cancer Institute’s Cancer Bioinformatics Grid as well as the National Heart, Lung and Blood Institute’s CardioVascular Research Network.
Scott Oster is a co-Director of the Software Research Institute (SRI) in the Department of Biomedical Informatics at the Ohio State University, wherein he helps to coordinate the department’s software development projects towards the aim of high-quality reusable infrastructure. For the past several years he has acted as the Lead Architect for the caGrid project: a multi-institutional effort and the core Grid middleware for the Cancer Biomedical Informatics Grid. Prior to coming to Ohio State in 2003, he worked at Cougaar Software Inc. as a Senior Computer Engineer working on several DARPA software research projects focusing on distributed intelligent agent technology. He received his MSc in Computer Science from Rensselaer Polytechnic Institute in 2002, and his BSc in Computer Science from the University of Iowa in 2000. His research areas include high-performance, data-intensive computing, middleware frameworks for efficient storage and processing of very large scientific datasets, Grid and distributed computing, agent frameworks, software quality and testing frameworks, and efficient and elegant designs for layered, componentized, and service-based architectures.
David Ervin pursued his undergraduate studies at Capital University and received his BA in Computer Science in the Spring of 2005. While attending Capital, he was active in the fledgling Computational Science Across the Curriculum Program, and in 2004 presented his research involving Genetic Algorithms at the National Conference on Undergraduate Research in Indianapolis. His current research focus is on the caGrid project; the core middleware implementation of the National Cancer Center’s caBIG project. He is currently the caGrid Data Services architect.
Justin Permar is a Technical Manager in the Software Research Institute (SRI) and Department of Biomedical Informatics (BMI). He is the Operations Manager for the caGrid Knowledge Center, a resource funded by the National Cancer Institute (NCI). The Knowledge Center’s mission is to provide comprehensive information resources and services to facilitate collaborative scientific research using caGrid. Justin received a BSc degree in Computer Engineering, with Honors, from Brown University in 2002. His honors thesis focused on capturing and analyzing neural signals in the motor cortex as part of an engineering effort in support of neuroscience research at Brown. He joined Ohio State after more than four years of research and development at the Massachusetts Institute of Technology Lincoln Laboratory (MIT LL) in Lexington, MA. His research at MIT LL included distributed systems and high-performance messaging-oriented middleware applied to a radar control center modernization project. His research areas include middleware, high-performance messaging, Grid infrastructure and applications support, and system-of-systems integration and interoperability.
Sivaramakrishnan Narayanan received his BEng (hons) from Birla Institute of Technology and Science, Pilani, India in 2002 and his PhD from The Ohio State University (OSU) under the guidance of Dr Joel Saltz in 2008. His research at OSU involved optimizing access to large-scale structural and semantic scientific data on the Grid. He currently works in query optimization for parallel databases at Greenplum, Inc.
Yolanda Gil is Associate Division Director at the Information Sciences Institute of the University of Southern California, and Research Associate Professor in the Computer Science Department. Her research interests include intelligent user interfaces, knowledge-rich problem solving, scientific and Grid computing, and the semantic Web. An area of recent interest is large-scale distributed data analysis through knowledge-rich computational workflows. She received her MSc and PhD degrees in Computer Science from Carnegie Mellon University. She was elected to the Council of the Association for the Advancement of Artificial Intelligence (AAAI) in 2003.
Ewa Deelman is a Research Assistant Professor at the USC Computer Science Department and a Project Leader at the USC Information Sciences Institute (ISI). Her research interests include the design and exploration of collaborative, distributed scientific environments, with particular emphasis on workflow management as well as the management of large amounts of data and metadata. At ISI, she is leading the Pegasus project, which designs and implements workflow mapping techniques for large-scale workflows running in distributed environments. She received her PhD from Rensselaer Polytechnic Institute in Computer Science in 1997 in the area of parallel discrete event simulation.
Mary Hall received her PhD in 1991 from Rice University and joined the School of Computing at the University of Utah as an associate professor in 2008. Her research focuses on compiler-based autotuning technology to exploit performance-enhancing features of a variety of computer architectures, investigating application domains that include biomedical imaging, molecular dynamics and signal processing. Prior to joining University of Utah, she held positions at the University of Southern California, Caltech, Stanford and Rice University.
Joel H. Saltz is Director of the Center for Comprehensive Informatics, Chief Medical Information Officer of Emory Healthcare, and Professor of Department of Pathology, Emory School of Medicine. Prior to coming to Emory, he was Professor and Chair of the Department of Biomedical Informatics at The Ohio State University (OSU) and Davis Endowed Chair of Cancer at OSU. He is trained both as a computer scientist and as a medical scientist. He received his MD and PhD degree in Computer Science at Duke University. He completed a residency in Clinical Pathology at Johns Hopkins University and is a board certified clinical pathologist. His research interests are in the development of systems software, databases, and compilers for the management, processing, and exploration of very large datasets, Grid computing, biomedical computing applications, medical informatics systems, and Grid and high-performance computing in biomedicine.
References
- Alexander C. A Pattern Language: Towns, Buildings, Construction (Center for Environmental Structure Series) Oxford University Press; Oxford: 1977. [Google Scholar]
- Beynon MD, Kurc T, Catalyurek U, et al. Distributed processing of very large datasets with DataCutter. Parallel Computing. 2001;27(11):1457–1478. [Google Scholar]
- Covitz PA, Hartel F, Schaefer C, et al. caCORE: a common infrastructure for cancer informatics. Bioinformatics. 2003;19(18):2404–2412. doi: 10.1093/bioinformatics/btg335. [DOI] [PubMed] [Google Scholar]
- Deelman E, Blythe J, Gil Y, et al. Grid Computing Lecture Notes in Computer Science. XXXX. Springer; Berlin: 2004. Pegasus: mapping scientific workflows onto the Grid; pp. 11–20. [Google Scholar]
- Foster I, Czajkowski K, Ferguson D, et al. Modeling and managing state in distributed systems: the role of OGSI and WSRF. Proceedings of the IEEE. 2005;93(3):604–612. [Google Scholar]
- Gamma E, Helm R, Johnson R, et al. Design Patterns: Elements of Reusable Object-Oriented Software (Addison-Wesley Professional Computing Series) Addison-Wesley; Reading, MA: 1994. [Google Scholar]
- Gil Y, Ratnakar V, Deelman E, et al. Wings for Pegasus: creating large-scale scientific applications using semantic representations of computational workflows. The 19th Annual Conference on Innovative Applications of Artificial Intelligence (IAAI); 2007.2007. [Google Scholar]
- Han J, Potter D, Kurc T, et al. An application of a service-oriented system to support arrayannotation in custom chip design for epigenomic analysis. Cancer Informatics. 2008;6:111–125. [PMC free article] [PubMed] [Google Scholar]
- Hastings S, Langella S, Oster S, et al. Distributed data management and integration: the Mobius project. Proceedings of the Global Grid Forum 11 (GGF11) Semantic Grid Applications Workshop; Honolulu, HI. 2004. pp. 20–38. [Google Scholar]
- Hastings S, Oster S, Langella S, et al. Introduce: an open source toolkit for rapid development of strongly typed Grid services. Journal of Grid Computing. 2007;5(4):407–427. [Google Scholar]
- Hill EF. Jess in Action: Java Rule-Based Systems. Manning Publications Co; Greenwich, CT: 2003. [Google Scholar]
- Huang HKK, Witte G, Ratib O, et al. Picture Archiving and Communication Systems (PACS) in Medicine. Springer; Berlin: 1991. [Google Scholar]
- Hull D, Wolstencroft K, Stevens R, et al. Taverna: a tool for building and running workflows of services. Nucleic Acids Research. 2006;34:729–732. doi: 10.1093/nar/gkl320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiryakov A, Ognyanov D, Manov D. WISE Workshops Lecture Notes in Computer Science. Vol. 3807. Springer; Berlin: OWLIM––a pragmatic semantic repository for OWL; pp. 182–192. [Google Scholar]
- Kumar V, Kurc T, Hall M, et al. An integrated framework for parameter-based optimization of scientific workflows. The 18th International Symposium on High Performance and Distributed Computing (HPDC 2009); Germany. 2009; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langella S, Hastings S, Oster S, et al. Sharing data and analytical resources securely in a biomedical research Grid environment. Journal of the American Medical Informatics Association. 2008;15(3):363–373. doi: 10.1197/jamia.M2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi B, McDonell K, Sacks-Davis R. Spatial kd-tree: an indexing mechanism for spatial databases. The IEEE COMPSAC Conference; Tokyo, Japan. 1987.1987. [Google Scholar]
- Oster S, Langella S, Hastings S, et al. caGrid 1.0: an enterprise Grid infrastructure for biomedical research. Journal of the American Medical Informatics Association. 2008;15(2):138–149. doi: 10.1197/jamia.M2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr TJ, Quong RW. ANTLR: a predicated-LL(k) parser generator. Software—Practice and Experience. 1995;25(7):789–810. [Google Scholar]
- Phillips J, Chilukuri R, Fragoso G, et al. The caCORE Software Development Kit: streamlining construction of interoperable biomedical information services. BMC Medical Informatics and Decision Making. 2006;6(2) doi: 10.1186/1472-6947-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltz J, Hastings S, Langella S, et al. A roadmap for caGrid, an enterprise Grid architecture for biomedical research. Studies in Health Technolology and Informatics. 2008a;138:224–237. [PMC free article] [PubMed] [Google Scholar]
- Saltz J, Kurc T, Hastings S, et al. e-Science, caGrid, and translational biomedical research. IEEE Computer. 2008b;41(11):58–66. doi: 10.1109/MC.2008.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltz J, Oster S, Hastings S, et al. caGrid: design and implementation of the core architecture of the cancer biomedical informatics Grid. Bioinformatics. 2006;22(15):1910–1916. doi: 10.1093/bioinformatics/btl272. [DOI] [PubMed] [Google Scholar]
- Saltz J, Oster S, Hastings S, et al. Translational research design templates, Grid computing, and HPC. The 22nd IEEE International Parallel & Distributed Processing Symposium (IPDPS’08); 2008c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thain D, Tannenbaum T, Livny M. Distributed computing in practice: the Condor experience. Concurrency and Computation: Practice and Experience. 2005;17(2):323–356. [Google Scholar]