Summary
Although many methods exist to study the recognition and signaling properties of proteins in isolation, it remains a challenge to perform these investigations on a system-wide or proteome-wide scale and within the context of biological complexity. Protein microarray technology provides a powerful tool to assess the selectivity of protein–protein interactions in high-throughput and to quantify the abundances and post-translational modification states of many different proteins in complex mixtures. Here, we provide an overview of the various applications of protein microarray technology and compare the strengths and technical challenges associated with each approach. Overall, we conclude that if this technology is to have a substantial impact on our understanding of cell biology and physiology, increased emphasis must be placed on obtaining rigorously controlled, quantitative data from protein function microarrays and on assessing the selectivity of reagents used in conjunction with protein-detecting microarrays.
Introduction
Over the past two decades, new tools have been developed that allow researchers to study the genome in high-throughput and with high fidelity. Fewer tools exist, however, to study the proteome. This discrepancy stems from fundamental differences between nucleic acids and proteins. Nucleic acids are relatively uniform in their physicochemical properties and can be recognized with exquisite selectivity through the process of complementary base pairing. This mechanism forms the basis for all current techniques in genome analysis, including DNA sequencing and expression profiling. Proteins, on the other hand, are biochemically diverse and their functions are inextricably linked to their three-dimensional conformations. At present, we lack the ability to predict the recognition properties of proteins using primary sequences alone and to design protein-detecting reagents that recognize individual proteins in complex mixtures. As neither DNA sequence nor mRNA abundance fully informs protein function or activity, our understanding of the proteome lags far behind our understanding of the genome. In this review, we outline how protein microarray technology is currently being used to bridge this gap and what challenges must be overcome before it is established as a routine tool to dissect the proteome.
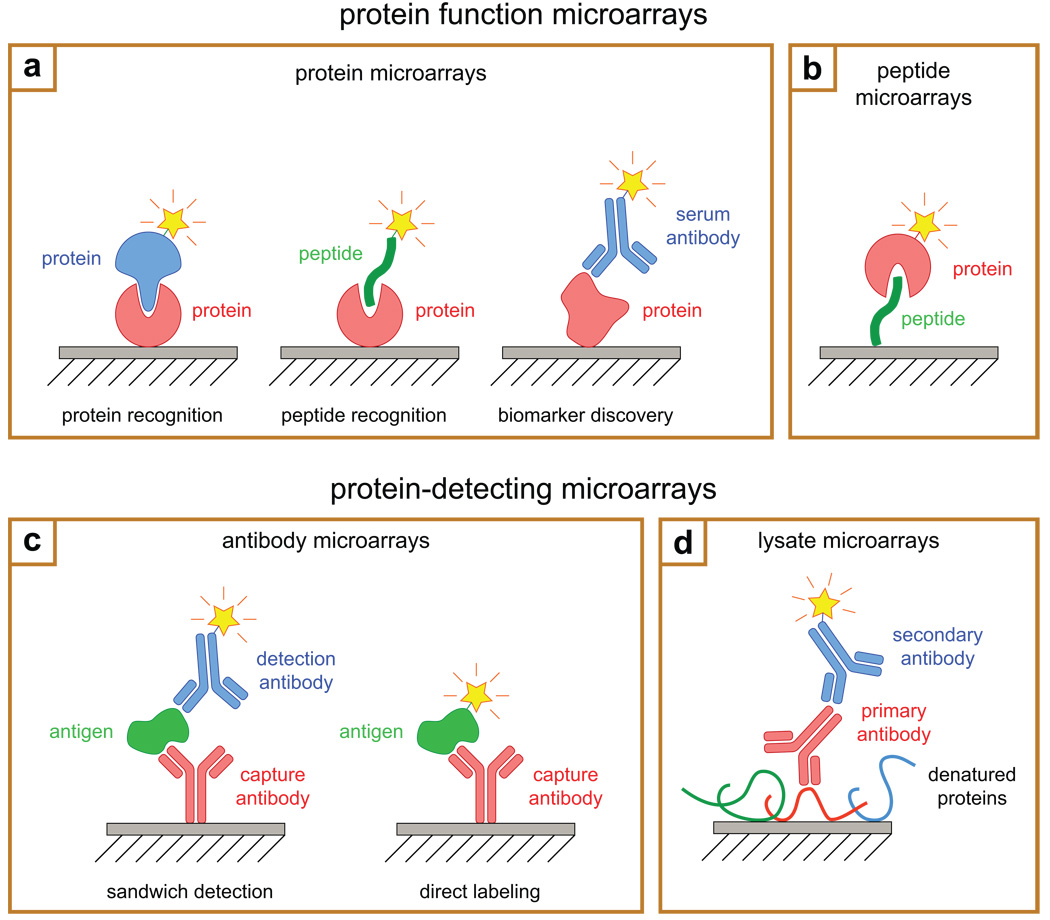
Protein microarrays can be divided into two general categories: ‘protein function microarrays’ and ‘protein-detecting microarrays’ (Fig. 1) [1]. Protein function microarrays comprise purified proteins, protein domains, or peptides, and are generally used to study molecular recognition or to screen for putative interaction partners. Protein-detecting microarrays, on the other hand, rely on reagents that recognize proteins in a selective fashion (e.g., antibodies) and are used to quantify the abundances and post-translational modification states of proteins in complex mixtures (e.g., cell lysates, tumor biopsies, and serum). The following discussion will address each of these categories separately.
Figure 1.
Common formats for protein microarrays. Protein function microarrays (a,b) are used to study molecular recognition and to identify protein–protein interactions, whereas protein-detecting microarrays (c,d) are used to detect and quantify proteins in biological samples. a, Microarrays of full-length proteins or protein domains are used to screen for protein–protein interactions (left); to study protein–peptide interactions (middle); or to identify antibodies in patient serum (right). b, Peptide microarrays are used to investigate protein–peptide interactions; to study substrate selectivity; and to profile enzyme activity in biological samples. c, Antibody microarrays are used to isolate proteins from complex mixtures. In the sandwich format (left), captured proteins are detected using a second, solution-phase antibody. In the direct-detection format (right), relative quantification is achieved by chemically labeling biological samples before applying them to the array. d, In contrast to antibody microarrays, lysate microarrays comprise the biological samples themselves. Complex mixtures of proteins are immobilized on nitrocellulose-coated glass substrates and detected using solution-phase antibodies. A labeled secondary antibody is often used to generate the signal.
Protein Function Microarrays
One of the primary goals of functional proteomics is to understand molecular recognition within the context of the proteome. Protein function microarrays provide a powerful way to assess binding selectivity across entire families of related proteins and, in the limit, across entire proteomes. In 2000, MacBeath and Schreiber showed that purified, recombinant proteins could be microarrayed on chemically derivatized glass substrates in a way that preserves their function [2]. Since then, variations of this technology have been used to study large collections of recombinant proteins. One approach that has been particularly successful is to focus on families of interaction domains.
(1) Microarrays of Protein Interaction Domains
Protein interaction domains mediate the formation of multi-protein complexes that confine signaling proteins to appropriate subcellular locations and help determine the selectivity of enzyme–substrate interactions. In 2002, Espejo et al. showed that a variety of interaction domain families (WW, SH3, SH2, 14.3.3, FHA, PDZ, PH and FF) retain their ability to mediate selective interactions when abstracted from their full-length parent proteins and printed in a microarray format [3]. Shortly afterwards, Newman and Keating used microarrays of coiled-coil domains to study the selectivity of pairing between human basic region leucine zipper (bZIP) transcription factors [4]. This latter study provided an unbiased view of protein–protein interactions and revealed a surprisingly high level of binding-partner selectivity. More recently, Keating and coworkers have used their arrays to assess the selectivity of computationally designed peptide ligands [5]. In a similar fashion, protein microarrays have been used to generate interaction maps for WW domains in yeast [6]; to investigate the association of the Kaposi Sarcoma-associated herpes virus protein K15 with the human endocytic machinery [7]; and to assess interactions between myelin basic proteins and SH3 domains [8]. In each case, the information gained from screening a panel of potential binding partners was used to guide more in-depth investigations into protein function.
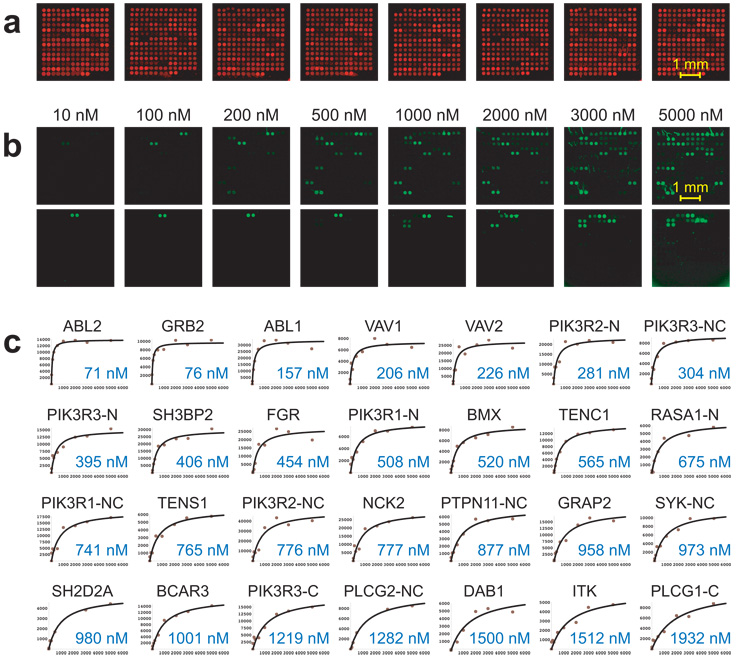
While these studies all uncovered valuable information regarding biophysical interactions, the data they generated were binary in nature: proteins were reported either to ‘interact’ or ‘not interact’. A complete understanding of molecular recognition, however, requires quantitative information. Additionally, by measuring binding affinities, researchers can gauge the physiological relevance of observed interactions and gain insight into the interplay of proteins in the complex environment of a living cell, where proteins often compete for the same targets. Quantitative protein microarrays were initially described in 2004, when Boutell et al. prepared microarrays comprising 50 allelic variants of p53 and probed the arrays with different concentrations of labeled GADD45-DNA [9]. From the resulting saturation binding curves, they were able to obtain equilibrium dissociation constants (KD’s) for each binding event. In a similar fashion, Jones et al. prepared microarrays of human SH2 and PTB domains and probed the arrays with different concentrations of phosphotyrosine-containing peptides derived from the four human ErbB receptors (Fig. 2) [10]. The resulting quantitative interaction maps revealed a previously unrecognized property of receptor tyrosine kinases (RTKs): they differ in the extent to which they become more promiscuous when overexpressed. Intriguingly, this property correlates with oncogenicity, suggesting that some RTKs may promote cancer by activating signaling pathways that are not turned on under normal conditions.
Figure 2.
Microarrays of protein domains for the quantitative study of molecular recognition. a, 159 SH2 and PTB domains were microarrayed in separate wells of a 96-well microtiter plate. The fluorescence arises from a trace amount of Cy5-labeled BSA that was added to each protein before arraying. b, SH2/PTB domain microarrays were probed with eight different concentrations of a fluorescently labeled phosphopeptide derived from ErbB2 (pTyr1139). c, Binding curves showing fluorescence as a function of peptide concentration for 28 high-affinity interactions. The data were fit to an equilibrium binding function to determine dissociation constants (KD’s; blue text). This figure was adapted, with permission, from Jones et al. [10].
Protein microarrays have also been used in a quantitative fashion to characterize enzymes. For example, Funeriu et al. prepared microarrays of six members of the cathepsin family of cysteine proteases [11]. Their work demonstrated three basic applications for enzyme microarrays; they can be used to determine enzyme kinetics, to screen for enzyme inhibitors in high-throughput, and to obtain dose-response curves for selected compounds.
In some cases, protein domains mediate interactions that are too weak to quantify directly using protein microarrays. For example, most PDZ domains bind their peptide ligands with dissociation constants between 2 µM and 50 µM. Stiffler et al. showed, however, that microarrays can be used as a high-fidelity, high-throughput screening tool to identify putative domain–peptide interactions, which can then be retested and quantified in a more targeted fashion [12]. By choosing an appropriate intensity threshold, they showed that protein microarrays highlight biophysical interactions with a false positive rate of 19% and a false negative rate of 6%.
One of the key advantages of protein domain microarrays is that they provide a way to assess binding selectivity in an unbiased fashion and on a family-wide scale. When performed in a quantitative fashion, these data can be used to train predictive models of molecular recognition. MacBeath and coworkers recently prepared microarrays of 157 mouse PDZ domains and used these arrays to survey interactions with over 200 peptides derived from the C-termini of mouse proteins [13]. The resulting interaction dataset was used to construct statistical models that capture the binding preferences not only of mouse PDZ domains, but of PDZ domains derived from other organisms as well [13,14]. Interestingly, their models showed that PDZ domains do not fall into discrete classes, as had previously been assumed. Instead, they lie on a functional continuum and it appears that their binding selectivity has been optimized across the proteome in order to minimize cross-reactivity [13].
(2) Peptide Microarrays
Many of the above studies focused on interactions between immobilized domains and solution-phase peptides. As an alternative approach, microarrays of short peptides can be prepared and interrogated with solution-phase proteins (Fig. 1b). Standardized methods now exist to synthesize peptides in situ in a microarray format (‘SPOT’ synthesis) [15,16]. Peptide microarrays are particularly useful when the objective is to screen one or a few proteins against a much larger number of potential binding partners. In one application of SPOT synthesis, Katz et al. prepared a microarray of 59 overlapping peptides spanning the length of three anti-apoptotic Bcl-2 family proteins [17]. The array was then probed with a fragment of the pro-apoptotic protein ASPP2. The authors identified two homologous binding sites on the Bcl-2 proteins, which were found to be associated with pro- and anti-apoptotic functions, respectively. In an interesting variation of SPOT synthesis, Boisguerin et al. developed an efficient way to prepare microarrays of inverted peptides displaying their C-termini [18]. They then used this method to study interactions mediated by the PDZ domains of α-1-syntrophin and Erbin.
Peptide microarrays also provide a convenient way to study the recognition properties of enzymes. For example, Hilhorst et al. used commercially available microarrays of 140 peptides to characterize the substrate selectivity of protein kinase A (PKA) [19]. By measuring the levels of phosphorylated peptides at different times of incubation with PKA, they were able to determine reaction kinetics for each peptide in a multiplex fashion.
Microarrays featuring more than 1,100 peptide substrates of human kinases are now available and have been used to study glucocorticoid-mediated effects on T-cell receptor-initiated signal transduction [20]; to identify kinases involved in angiotensin II–dependent hypertensive renal fibrosis [21]; to characterize the kinome in Barret’s esophagus [22]; and to study the effects of COX-2 inhibition on kinase signaling pathways in colorectal cancer cells [23]. In each case, the arrays were used as a screening tool to identify active kinases. Traditional biochemical techniques were then used to corroborate the microarray findings and to investigate the individual roles of the kinases. Overall, these studies show how peptide microarray technology can be used as a discovery tool to generate new biological hypotheses.
(3) Whole Proteome Microarrays
The ultimate extension of protein microarray technology is to prepare arrays comprising entire proteomes. In a groundbreaking study in 2001, Snyder and coworkers expressed, purified, and arrayed 5,800 yeast proteins. They then probed the resulting microarrays with six different phospholipids to reveal lipid-binding proteins [24]. In a subsequent study, they used their yeast proteome microarrays to highlight in vitro substrates of 87 protein kinases [25]. By integrating their kinase–substrate data with large-scale protein–protein interaction data and transcription factor binding data, they assembled a global regulatory network for yeast that uncovered several frequently used regulatory modules. The insight they gained into the logic of protein regulation could only have been obtained using this proteome-wide approach. In a similar fashion, Popescu et al. constructed microarrays comprising 2,158 unique Arabidopsis thaliana proteins and used these arrays to identify 570 phosphorylation substrates of mitogen-activated protein kinases [26]. The majority of substrates highlighted in their screens were not previously known and overall the list of substrates was enriched in transcription factors involved in the regulation of development, defense, and stress response. Several substrates were validated by subsequent reconstitution experiments, showing that whole proteome arrays provide a powerful way to identify biologically relevant interactions.
Proteome-wide microarrays also enable surprising biological discoveries. Lin et al. recently incubated yeast proteome microarrays with the essential nucleosome acetyl transferase of H4 (NuA4) complex [27]. Unexpectedly, they identified many nonchromatin substrates. Most notably, they found that the metabolic enzyme phosphoenolpyruvate carboxykinase is a physiological substrate of NuA4 and that acetylation of this enzyme plays a role in regulating the chronological life span of yeast.
Investigations of this nature are now much more accessible, as human proteome arrays are commercially available. These arrays are particularly useful for identifying biomarkers of disease. For example, human proteome arrays were used to detect autoimmune response markers in ovarian [28] and breast cancer [29], and Escherichia coli proteome arrays were used to identify markers of inflammatory bowel disease [30]. Whole proteome arrays also serve as a convenient discovery tool that can be interfaced with sophisticated biochemistry. Merbl et al. recently incubated human proteome microarrays with cell extracts that replicate the mitotic checkpoint and anaphase release [31]. By developing their arrays with antibodies that recognize polyubiquitination, they were able to identify novel substrates of the anaphase-promoting complex. While proteome chips have not yet been used in a quantitative fashion to characterize and model binding selectivity, they are proving extremely useful as unbiased and normalized tools for biological and clinical discovery.
Protein-Detecting Microarrays
As detailed above, protein function microarrays enable broad and unbiased investigations of molecular recognition. Information gained from these studies can be used to map out biophysical and biochemical connections between proteins. To determine how information flows through these networks in a dynamic fashion, however, requires methods to measure the abundances and post-translational modification states of many different proteins in biological samples in high throughput. Protein-detecting microarrays provide this capability (Fig. 1c,d). Since a large number of selective antibodies are commercially available, most studies in this area have relied on antibodies, in conjunction with microarray technology, to profile cellular lysates, tumor biopsies, and human serum.
(1) Antibody Microarrays
Perhaps the most frequently used strategy is to prepare microarrays of immobilized antibodies on solid supports, where they function as ‘capture’ reagents (Fig. 1c). Antibody microarrays enable multiplexed analyses of non-denatured proteins with very low reagent and sample consumption. To detect and quantify the captured analytes, a second antibody is often used that recognizes a different epitope on the captured protein. This procedure mimics a sandwich ELISA, but in a multiplexed microarray format. This strategy has been used to track the phosphorylation status of receptor tyrosine kinases following growth factor-stimulation [32]; to assess changes in the tyrosine phosphorylation state of selected proteins following drug treatment [33]; to discover cancer-associated glycan variations on specific proteins in the serum of pancreatic cancer patients [34]; and to identify prognostic markers of early mortality in the serum of patients with end-stage renal disease [35].
One limitation of the sandwich assay is that it is often difficult to identify matched pairs of capture and detection antibodies. In addition, the detection step requires a cocktail of labeled antibodies. As more antibodies are included in this cocktail, the level of background binding increases, as does the risk of cross-reactive binding. To circumvent these problems, the proteins in the biological samples can be directly labeled using one or more fluorescent dyes or small molecule haptens (Fig. 1c) [36,37]. An advantage of this strategy is that pairs of samples can be compared in a ratiometric fashion using spectrally distinct fluorophores. This approach has been used to discover biomarkers of pancreatic cancer [38], bladder cancer [39], and mantle-cell lymphoma [40]. One caveat of this method, however, is that it relies on the uniform labeling of proteins across different samples, which cannot be assured. In addition, direct chemical labeling can modify antigenic epitopes, thereby blocking antibody recognition.
In principle, one can avoid these problems using label-free detection. To date, however, label-free methods are too insensitive for most biological investigations. For example, proof-of-concept experiments using surface plasmon resonance [41,42] and optical interferometry [43] reported limits of detection in the range of 20–30 ng/mL of target protein. In contrast, fluorescence-based detection methods enable quantification down to 1 pg/mL [35,44]. Future development of label-free detection methods must focus on increasing assay sensitivity.
Although antibody microarrays offer great potential as a tool for quantitative proteomics, proteome-wide analyses are still far from reach. Currently, only a small number of signaling events can be followed reliably using well-validated antibodies. One solution is to combine a variety of experimental approaches. Gaudet et al. used antibody microarrays, in conjunction with kinase activity assays, western blotting, and flow cytometry, to assemble a compendium of ~10,000 measurements of signals and responses in HT-29 cells treated with different combinations of TNFα, EGF, and insulin [45]. This disparate dataset was then used to derive a predictive model of apoptotic signaling [46]. Their integrated approach shows how the high-throughput technology of antibody microarrays can be used when suitable detection reagents are available, and how additional measurements can be added using lower-throughput methods.
(2) Lysate Microarrays
As an alternative to preparing microarrays of antibodies, the biological samples themselves can be spotted on nitrocellulose-coated glass substrates and the immobilized proteins detected and quantified using solution-phase antibodies (Fig. 1d). This strategy was initially described by Paweletz et al. in 2001 [47] and is referred to either as a ‘reverse phase protein array’ or as a ‘lysate microarray’. Each microarray contains the entire set of biological samples and reports on the relative levels of one or two proteins. By preparing multiple copies of the array, however, it is possible to follow the abundances and post-translational modification states of many different proteins. One of the advantages of this approach is that the proteins being quantified are denatured and so do not have to remain folded and functional on the solid support. Additionally, protein complexes are disrupted and so do not complicate subsequent analyses.
To date, this method has been used in a clinical setting to follow pro-survival checkpoint proteins as a function of cancer progression [47] and to discover and validate specific biomarkers for disease diagnosis and patient stratification [48,49]. In addition, lysate microarrays have been used to study the kinetics of intracellular signaling in a system-wide fashion. By tracking 62 phosphorylation sites in stimulated Jurkat T-cells, Chan et al. uncovered a previously unrecognized link between T-cell receptor activation and Raf-1 activity [50].
Lysate microarray technology has also been adapted to a high-throughput screening format. By treating cells grown in microtiter plates with cell-permeable small molecules, and by arraying the resulting lysates, Sevecka and MacBeath showed that compounds could be screened on the basis of how they perturb the ‘state’ of the cell [51]. Interestingly, they showed that compounds with the same target induced similar states, suggesting that this technology could be used to help identify the targets of bioactive compounds with unknown mechanisms of action.
Although lysate microarray technology holds great promise for quantitative proteomics, its use remains limited due to a lack of well-validated antibodies. In our experience, we have found that the majority of commercially available antibodies exhibit a substantial amount of cross-reactivity when used in this assay, even when the antibody produces a single discernable band of the correct molecular weight on a western blot [51]. Other groups have found that the precise blocking and detection protocol [52], as well as the composition of the lysis buffer [53], substantially affects antibody performance. Further development is therefore necessary before this approach is widely adopted.
Conclusion
Protein microarray technology offers a powerful way to dissect the complexity of the proteome. The diversity of applications and the economy of scale make this method ideally suited to the investigation of molecular recognition and to the multiplexed quantification of proteins in complex mixtures. Whereas technical problems concerning protein function microarrays have largely been solved, accurately quantifying proteins in complex mixtures remains a challenge. Antibodies still enjoy their status as the detection reagents of choice, but suffer from cross-reactive binding and therefore require extensive validation.
Overall, the quality of future systems-level investigations will hinge on carefully validated experimental protocols and reagents. Currently, there are no standardized methods for preparing protein-detecting microarrays or widely accepted techniques for analyzing the resulting data. Ensuring stringent standards for protein microarray experiments, and rendering data comparable across different studies, will require a coordinated effort by the scientific community. By far the most pressing need, however, is for a renewable and distributable source of detection reagents. Despite considerable efforts in this area over many years, this remains an unsolved problem. In our opinion, generating this resource will require robust and scalable methods for directed evolution and a well-funded, organized effort by the scientific community. A concerted endeavor of this nature would not only enable microarray-based investigations, but other targeted analyses of proteins in biological contexts as well.
Acknowledgements
This work was supported by awards from the Arnold and Mabel Beckman Foundation, the W.M. Keck Foundation, and the Camille and Henry Dreyfus Foundation and by grants from the NIH (1 R01 GM072872, 1 R33 CA128726, and 1 R21 CA126720).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest published within the period of review have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Kodadek T. Protein microarrays: prospects and problems. Chem Biol. 2001;8:105–115. doi: 10.1016/s1074-5521(00)90067-x. [DOI] [PubMed] [Google Scholar]
- 2.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 3.Espejo A, Cote J, Bednarek A, Richard S, Bedford MT. A protein-domain microarray identifies novel protein-protein interactions. Biochem J. 2002;367:697–702. doi: 10.1042/BJ20020860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- •5.Grigoryan G, Reinke AW, Keating AE. Design of protein-interaction specificity gives selective bZIP-binding peptides. Nature. 2009;458:859–864. doi: 10.1038/nature07885.. This study is a recent example of using protein microarrays to study molecular selectivity within a family of closely related proteins. Microarrays of human basic-region leucine zipper (bZIP) transcription factors were used to characterize the binding selectivity of computationally designed synthetic bZIP ligands.
- 6.Hesselberth JR, Miller JP, Golob A, Stajich JE, Michaud GA, Fields S. Comparative analysis of Saccharomyces cerevisiae WW domains and their interacting proteins. Genome Biol. 2006;7:R30. doi: 10.1186/gb-2006-7-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim CS, Seet BT, Ingham RJ, Gish G, Matskova L, Winberg G, Ernberg I, Pawson T. The K15 protein of Kaposi’s sarcoma-associated herpesvirus recruits the endocytic regulator intersectin 2 through a selective SH3 domain interaction. Biochemistry. 2007;46:9874–9885. doi: 10.1021/bi700357s. [DOI] [PubMed] [Google Scholar]
- 8.Polverini E, Rangaraj G, Libich DS, Boggs JM, Harauz G. Binding of the proline-rich segment of myelin basic protein to SH3 domains: spectroscopic, microarray, and modeling studies of ligand conformation and effects of posttranslational modifications. Biochemistry. 2008;47:267–282. doi: 10.1021/bi701336n. [DOI] [PubMed] [Google Scholar]
- •9.Boutell JM, Hart DJ, Godber BL, Kozlowski RZ, Blackburn JM. Functional protein microarrays for parallel characterisation of p53 mutants. Proteomics. 1958;4:1950–1958. doi: 10.1002/pmic.200300722.. This is one of the first uses of protein microarrays to measure binding affinities. Fifty allelic variants of the p53 tumor suppressor protein were immobilized on a glass support and the resulting microarrays were probed with different concentrations of a radioactively labeled DNA promoter sequence to measure binding curves for each protein in parallel.
- ••10.Jones RB, Gordus A, Krall JA, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177.. This study shows how quantitative protein microarray technology can be used to gain insight into protein domain selectivity. Equilibrium dissociation constants were determined for the binding of peptides representing known sites of tyrosine phosphorylation on the four human ErbB receptors to almost every human SH2 and PTB domain. The resulting data provided insight into systems-level properties of the ErbB receptors.
- ••11.Funeriu DP, Eppinger J, Denizot L, Miyake M, Miyake J. Enzyme family-specific and activity-based screening of chemical libraries using enzyme microarrays. Nat Biotechnol. 2005;23:622–627. doi: 10.1038/nbt1090.. This work provides an example of microarrays of catalytically active enzymes. The authors arrayed six members of the cathepsin family of cysteine proteases and determined rate constants for substrate cleavage. They further demonstrated that a multiplexed screen for enzyme inhibitors could be carried out in a microarray format, and measured dose-response curves for several inhibitors.
- •12.Stiffler MA, Grantcharova VP, Sevecka M, MacBeath G. Uncovering quantitative protein interaction networks for mouse PDZ domains using protein microarrays. J Am Chem Soc. 2006;128:5913–5922. doi: 10.1021/ja060943h.. This study presents a rigorous analysis of the fidelity of protein function microarrays. The authors identified interactions between a set of mouse PDZ domains and a set of mouse protein-derived peptides using both protein microarrays and solution-phase fluorescence polarization. By comparing the two data sets, as well as literature-reported interactions, they established that PDZ domain microarrays identify biophysical interactions with a false positive rate of 19% and a false negative rate of 6%.
- ••13.Stiffler MA, Chen JR, Grantcharova VP, Lei Y, Fuchs D, Allen JE, Zaslavskaia LA, MacBeath G. PDZ domain binding selectivity is optimized across the mouse proteome. Science. 2007;317:364–369. doi: 10.1126/science.1144592.. In this study, the authors used a combination of protein domain microarrays, fluorescence polarization, and computational modeling to generate a high-quality, quantitative dataset of PDZ domain-mediated protein-protein interactions. They further showed that, contrary to the established paradigm, PDZ domains do not fall into discrete functional classes, but instead are evenly distributed across selectivity space.
- 14.Chen JR, Chang BH, Allen JE, Stiffler MA, MacBeath G. Predicting PDZ domain-peptide interactions from primary sequences. Nat Biotechnol. 2008;26:1041–1045. doi: 10.1038/nbt.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andresen H, Bier FF. Peptide microarrays for serum antibody diagnostics. Methods Mol Biol. 2009;509:123–134. doi: 10.1007/978-1-59745-372-1_8. [DOI] [PubMed] [Google Scholar]
- 16.Saxinger WC. Protein tyrosine kinase characterization based on fully automated synthesis of (phospho) peptide arrays in microplates. Methods Mol Biol. 2009;527:257–267. doi: 10.1007/978-1-60327-834-8_19. x. [DOI] [PubMed] [Google Scholar]
- •17.Katz C, Benyamini H, Rotem S, Lebendiker M, Danieli T, Iosub A, Refaely H, Dines M, Bronner V, Bravman T, et al. Molecular basis of the interaction between the antiapoptotic Bcl-2 family proteins and the proapoptotic protein ASPP2. Proc Natl Acad Sci USA. 2008;105:12277–12282. doi: 10.1073/pnas.0711269105.. In this study, peptide microarrays and computational methods were used to uncover the molecular mechanism by which Bcl-2 family proteins interact with ASPP2. These results suggested a model for induction of apoptosis by ASPP2.
- 18.Boisguerin P, Leben R, Ay B, Radziwill G, Moelling K, Dong L, Volkmer-Engert R. An improved method for the synthesis of cellulose membrane-bound peptides with free C termini is useful for PDZ domain binding studies. Chem Biol. 2004;11:449–459. doi: 10.1016/j.chembiol.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 19.Hilhorst R, Houkes L, van den Berg A, Ruijtenbeek R. Peptide microarrays for detailed, high-throughput substrate identification, kinetic characterization, and inhibition studies on protein kinase A. Anal Biochem. 2009;387:150–161. doi: 10.1016/j.ab.2009.01.022. [DOI] [PubMed] [Google Scholar]
- •20.Lowenberg M, Tuynman J, Bilderbeek J, Gaber T, Buttgereit F, van Deventer S, Peppelenbosch M, Hommes D. Rapid immunosuppressive effects of glucocorticoids mediated through Lck and Fyn. Blood. 2005;106:1703–1710. doi: 10.1182/blood-2004-12-4790.. See annotation to [23].
- •21.de Borst MH, Diks SH, Bolbrinker J, Schellings MW, van Dalen MB, Peppelenbosch MP, Kreutz R, Pinto YM, Navis G, van Goor H. Profiling of the renal kinome: a novel tool to identify protein kinases involved in angiotensin II-dependent hypertensive renal damage. Am J Physiol Renal Physiol. 2007;293:F428–F437. doi: 10.1152/ajprenal.00367.2006.. See annotation to [23].
- •22.van Baal JW, Diks SH, Wanders RJ, Rygiel AM, Milano F, Joore J, Bergman JJ, Peppelenbosch MP, Krishnadath KK. Comparison of kinome profiles of Barrett’s esophagus with normal squamous esophagus and normal gastric cardia. Cancer Res. 2006;66:11605–11612. doi: 10.1158/0008-5472.CAN-06-1370.. See annotation to [23].
- •23.Tuynman JB, Vermeulen L, Boon EM, Kemper K, Zwinderman AH, Peppelenbosch MP, Richel DJ. Cyclooxygenase-2 inhibition inhibits c-Met kinase activity and Wnt activity in colon cancer. Cancer Res. 2008;68:1213–1220. doi: 10.1158/0008-5472.CAN-07-5172.. [20–23] are recent reports that use commercially available peptide microarrays to profile the kinomes of biological samples. In each study, the authors identified kinases with altered activities and subsequently investigated their cellular signaling mechanisms to find potential drug targets.
- 24.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Houfek T, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- ••25.Ptacek J, Devgan G, Michaud G, Zhu H, Zhu X, Fasolo J, Guo H, Jona G, Breitkreutz A, Sopko R, et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187.. In this landmark study, Snyder and co-workers used proteome-wide microarrays to identify in vitro substrates for most of the protein kinases in yeast. From the observed 4200 distinct phosphorylation events, the authors assembled a global phosphorylation map, defined consensus sequences for 11 kinases, and identified conserved regulatory modules.
- •26.Popescu SC, Popescu GV, Bachan S, Zhang Z, Gerstein M, Snyder M, Dinesh-Kumar SP. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 2009;23:80–92. doi: 10.1101/gad.1740009.. In this study the authors constructed a protein microarray comprising >2000 Arabidopsis thaliana proteins and probed them with 10 different mitogen-activated protein kinases (MPKs). In doing so, they uncovered both known and new signaling modules encompassing 570 MPK phosphorylation substrates.
- •27.Lin YY, Lu JY, Zhang J, Walter W, Dang W, Wan J, Tao SC, Qian J, Zhao Y, Boeke JD, et al. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell. 2009;136:1073–1084. doi: 10.1016/j.cell.2009.01.033.. This study provides an example of a proteome-wide microarray screen for targets of protein acetylation. Yeast proteome chips were incubated with the NuA4 histone acetyltransferase complex and radioactively labeled acetyl-CoA. Acetylated targets included both chromatin-associated and nonchromatin substrates.
- 28.Hudson ME, Pozdnyakova I, Haines K, Mor G, Snyder M. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc Natl Acad Sci USA. 2007;104:17494–17499. doi: 10.1073/pnas.0708572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson KS, Ramachandran N, Wong J, Raphael JV, Hainsworth E, Demirkan G, Cramer D, Aronzon D, Hodi FS, Harris L, et al. Application of protein microarrays for multiplexed detection of antibodies to tumor antigens in breast cancer. J Proteome Res. 2008;7:1490–1499. doi: 10.1021/pr700804c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen CS, Sullivan S, Anderson T, Tan AC, Alex PJ, Brant SR, Cuffari C, Bayless TM, Talor MV, Burek CL, et al. Identification of novel serological biomarkers for inflammatory bowel disease using E. coli proteome chip. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M800593-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •31.Merbl Y, Kirschner MW. Large-scale detection of ubiquitination substrates using cell extracts and protein microarrays. Proc Natl Acad Sci USA. 2009;106:2543–2548. doi: 10.1073/pnas.0812892106.. This report is the first application of proteome-wide microarrays to investigate the cell cycle. The authors used commercially available human proteome chips to identify differentially ubiquitinated proteins in cells synchronized either at mitotic checkpoints or in anaphase.
- 32.Nielsen UB, Cardone MH, Sinskey AJ, MacBeath G, Sorger PK. Profiling receptor tyrosine kinase activation by using Ab microarrays. Proc Natl Acad Sci USA. 2003;100:9330–9335. doi: 10.1073/pnas.1633513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gembitsky DS, Lawlor K, Jacovina A, Yaneva M, Tempst P. A prototype antibody microarray platform to monitor changes in protein tyrosine phosphorylation. Mol Cell Proteomics. 2004;3:1102–1118. doi: 10.1074/mcp.M400075-MCP200. [DOI] [PubMed] [Google Scholar]
- •34.Chen S, LaRoche T, Hamelinck D, Bergsma D, Brenner D, Simeone D, Brand RE, Haab BB. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nat Methods. 2007;4:437–444. doi: 10.1038/nmeth1035.. This report is a recent example of the use of antibody microarrays to study post-translational modifications. Capture antibodies directed at proteins of interest were printed as microarrays and chemically derivatized to mask their intrinsic glycan modifications. The arrays were then incubated with native protein samples and glycan modifications on the captured proteins were detected using either a glycan-specific antibody or a lectin.
- •35.Knickerbocker T, Chen JR, Thadhani R, MacBeath G. An integrated approach to prognosis using protein microarrays and nonparametric methods. Mol Syst Biol. 2007;3:123. doi: 10.1038/msb4100167.. This work is an example of using antibody microarrays to measure clinically relevant serum biomarkers. The authors use sandwich antibody microarrays to quantify the levels of 14 serum cytokines in 468 patients undergoing kidney dialysis. The resulting cytokine data, together with relevant clinical variables, were used to construct a prognostic model of early mortality.
- 36.Kusnezow W, Banzon V, Schroder C, Schaal R, Hoheisel JD, Ruffer S, Luft P, Duschl A, Syagailo YV. Antibody microarray-based profiling of complex specimens: systematic evaluation of labeling strategies. Proteomics. 2007;7:1786–1799. doi: 10.1002/pmic.200600762. [DOI] [PubMed] [Google Scholar]
- 37.Zhou H, Bouwman K, Schotanus M, Verweij C, Marrero JA, Dillon D, Costa J, Lizardi P, Haab BB. Two-color, rolling-circle amplification on antibody microarrays for sensitive, multiplexed serum-protein measurements. Genome Biol. 2004;5:R28. doi: 10.1186/gb-2004-5-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamelinck D, Zhou H, Li L, Verweij C, Dillon D, Feng Z, Costa J, Haab BB. Optimized normalization for antibody microarrays and application to serum-protein profiling. Mol Cell Proteomics. 2005;4:773–784. doi: 10.1074/mcp.M400180-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez-Carbayo M, Socci ND, Lozano JJ, Haab BB, Cordon-Cardo C. Profiling bladder cancer using targeted antibody arrays. Am J Pathol. 2006;168:93–103. doi: 10.2353/ajpath.2006.050601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghobrial IM, McCormick DJ, Kaufmann SH, Leontovich AA, Loegering DA, Dai NT, Krajnik KL, Stenson MJ, Melhem MF, Novak AJ, et al. Proteomic analysis of mantle-cell lymphoma by protein microarray. Blood. 2005;105:3722–3730. doi: 10.1182/blood-2004-10-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kyo M, Usui-Aoki K, Koga H. Label-free detection of proteins in crude cell lysate with antibody arrays by a surface plasmon resonance imaging technique. Anal Chem. 2005;77:7115–7121. doi: 10.1021/ac050884a. [DOI] [PubMed] [Google Scholar]
- 42.Usui-Aoki K, Shimada K, Nagano M, Kawai M, Koga H. A novel approach to protein expression profiling using antibody microarrays combined with surface plasmon resonance technology. Proteomics. 2005;5:2396–2401. doi: 10.1002/pmic.200401171. [DOI] [PubMed] [Google Scholar]
- 43.Ozkumur E, Needham JW, Bergstein DA, Gonzalez R, Cabodi M, Gershoni JM, Goldberg BB, Unlu MS. Label-free and dynamic detection of biomolecular interactions for high-throughput microarray applications. Proc Natl Acad Sci USA. 2008;105:7988–7992. doi: 10.1073/pnas.0711421105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schweitzer B, Roberts S, Grimwade B, Shao W, Wang M, Fu Q, Shu Q, Laroche I, Zhou Z, Tchernev VT, et al. Multiplexed protein profiling on microarrays by rolling-circle amplification. Nat Biotechnol. 2002;20:359–365. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaudet S, Janes KA, Albeck JG, Pace EA, Lauffenburger DA, Sorger PK. A compendium of signals and responses triggered by prodeath and prosurvival cytokines. Mol Cell Proteomics. 2005;4:1569–1590. doi: 10.1074/mcp.M500158-MCP200. [DOI] [PubMed] [Google Scholar]
- •46.Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–1653. doi: 10.1126/science.1116598.. This study demonstrates how a heterogeneous dataset derived from both antibody microarrays and lower-throughput methods can be used to gain insight into a signal transduction network.
- 47.Paweletz CP, Charboneau L, Bichsel VE, Simone NL, Chen T, Gillespie JW, Emmert-Buck MR, Roth MJ, Petricoin IE, Liotta LA. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. Oncogene. 2001;20:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- 48.Boyd ZS, Wu QJ, O’Brien C, Spoerke J, Savage H, Fielder PJ, Amler L, Yan Y, Lackner MR. Proteomic analysis of breast cancer molecular subtypes and biomarkers of response to targeted kinase inhibitors using reverse-phase protein microarrays. Mol Cancer Ther. 2008;7:3695–3706. doi: 10.1158/1535-7163.MCT-08-0810. [DOI] [PubMed] [Google Scholar]
- 49.Grote T, Siwak DR, Fritsche HA, Joy C, Mills GB, Simeone D, Whitcomb DC, Logsdon CD. Validation of reverse phase protein array for practical screening of potential biomarkers in serum and plasma: accurate detection of CA19-9 levels in pancreatic cancer. Proteomics. 2008;8:3051–3060. doi: 10.1002/pmic.200700951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••50.Chan SM, Ermann J, Su L, Fathman CG, Utz PJ. Protein microarrays for multiplex analysis of signal transduction pathways. Nat Med. 2004;10:1390–1396. doi: 10.1038/nm1139.. This report illustrates the use of lysate microarrays to study signal transduction networks in a time-dependent, multiplexed fashion. The authors profiled the phosphorylation states of 62 signaling proteins in T-cell receptor-activated lymphocytes to delineate downstream signaling events. This work also showed how the low sample consumption of lysate arrays enables the study of rare populations of cells.
- ••51.Sevecka M, MacBeath G. State-based discovery: a multidimensional screen for small-molecule modulators of EGF signaling. Nat Methods. 2006;3:825–831. doi: 10.1038/NMETH931.. This study is the first example of using lysate microarrays for small-molecule screening. By determining the multidimensional phosphorylation profiles elicited by each compound, the authors classified bioactive small molecules according to the ‘states’ they induced in a signaling network. This work also demonstrated that lysate microarrays could be used to perform high-throughput investigations involving large numbers of dilute samples prepared in microtiter plates.
- 52.Ambroz KL, Zhang Y, Schutz-Geschwender A, Olive DM. Blocking and detection chemistries affect antibody performance on reverse phase protein arrays. Proteomics. 2008;8:2379–2383. doi: 10.1002/pmic.200700676. [DOI] [PubMed] [Google Scholar]
- 53.Spurrier B, Washburn FL, Asin S, Ramalingam S, Nishizuka S. Antibody screening database for protein kinetic modeling. Proteomics. 2007;7:3259–3263. doi: 10.1002/pmic.200700117. [DOI] [PubMed] [Google Scholar]