Abstract
KRAS and BRAF mutations are frequently observed in human colon cancers. These mutations occur in a mutually exclusive manner, and each is associated with distinctive biological features. We previously demonstrated that K-ras can interact with hypoxia to activate multiple signaling pathways. Many hypoxic responses are mediated by hypoxia inducible factor-1α (HIF-1α) and HIF-2α, and we sought to define the roles of mutant KRAS and BRAF in the induction of HIF-1α and HIF-2α in colon cancer cells. Ectopic expression of mutant K-ras in Caco2 cells enhanced the hypoxic induction of only HIF-1α, whereas mutant BRAF enhanced both HIF-1α and HIF-2α. Knockout or knockdown of mutant KRAS in DLD1 and HCT116 cells impaired the hypoxic induction of only HIF-1α. HIF-1α mRNA levels were comparable in cells with and without a KRAS mutation. However, the rate of HIF-1α protein synthesis was higher in cells with a KRAS mutation, and this was suppressed by the PI3K inhibitor LY294002. In contrast, knockdown of mutant BRAF in HT29 cells suppressed both HIF-1α and HIF-2α. Although BRAF regulated mRNA levels of both HIF-1α and HIF-2α, knockdown of BRAF or treatment with the MEK inhibitor PD98059 impaired the translation of only HIF-2α. Our data reveal that oncogenic KRAS and BRAF mutations differentially regulate the hypoxic induction of HIF-1α and HIF-2α in colon cancer, and this may potentially contribute to the phenotypic differences of KRAS and BRAF mutations in colon tumors.
Keywords: KRAS, BRAF, HIF
INTRODUCTION
Activating mutations of KRAS are found in approximately 45% of colorectal cancers (1, 2). Oncogenic activation of KRAS can influence a number of cellular processes that regulate morphology, proliferation and motility, and KRAS mutations are clinically associated with a poor prognosis (3, 4). K-ras interacts with several effector proteins, including Raf kinases and phosphoinositide 3-kinases (PI3K). BRAF, one of the Raf kinases, is also mutated in about 15% of colorectal cancers (5–7). Interestingly, KRAS and BRAF mutations are mutually exclusive (5–7), suggesting they may have similar functions. However, the biological properties of tumors with KRAS or BRAF mutations are quite distinctive. For example, BRAF mutant tumors are more likely to exhibit microsatellite instability, poor histological grade, mucinous histology and a better prognosis (7, 8). However, it remains unclear how these oncogenes that signal through the same pathways can give rise to such distinctive tumor phenotypes.
As tumor cells proliferate, their oxygen and nutrient demands increase, and cells adapt to this hypoxic environment through a switch to anaerobic glycolysis and induction of survival factors and angiogenic growth factors such as vascular endothelial growth factor (VEGF) (9). Hypoxia-inducible factors (HIFs) are thought to play a major role in controlling the transcriptional responses to hypoxia (10, 11). HIF-1 was the first HIF isoform to be recognized, and it is composed of two subunits, HIF-1α and HIF-1α HIF-2α and HIF-3α were identified by homology searches or screens for interaction partners with HIF-1β (12). While HIF-3α is the most distantly related isoform, HIF-1α and HIF-2α are closely related and both activate hypoxia-responsive element (HRE)-dependent gene expression. Although the two isoforms are frequently expressed in human cancers, their functions vary. In colon cancer, we have previously reported that HIF-1α may promote the growth of cancer cells both in vitro and in vivo. However, HIF-2α appears to restrain tumor growth, and loss of expression of HIF-2α but not HIF-1α in human colon cancer tissues is strongly correlated with advanced tumor stage (13). The mechanisms that regulate the expression of HIF-1α vs. HIF-2α in colon cancer are unknown.
Under normoxic conditions, HIF-1α and HIF-2α are hydroxylated by O2-dependent prolyl hydroxylases (PHDs) within the oxygen-dependent degradation domain (ODD) and targeted by the von Hippel-Lindau protein (pVHL) E3 ubiquitin ligase complex, leading to proteasomal degradation. Under hypoxic conditions, PHD activity is inhibited, thereby allowing stabilization and accumulation of HIF-1α and HIF-2α protein (12). In addition to hypoxia, HIF-1α expression can be regulated by certain oncogenic pathways. For instance, insulin, insulin –like growth factor (IGF), epidermal growth factor (EGF) and fibroblast growth factor (FGF) can induce expression of HIF-1α protein in normoxia via mitogen-activated protein kinase (MAPK) and PI3K/Akt pathways (14–16). Transformation by H-ras or mutant K-ras has also been shown to enhance the levels of HIF-1α protein (17, 18). In melanoma cells, mutant BRAF (V600E) can enhance HIF-1α expression in normoxia (19). However, the relevance of these mechanisms in hypoxia as well as the effects on the HIF-2α isoform are unclear. We therefore sought to identify the roles of mutant KRAS and BRAF on the hypoxic induction of HIF-1α and HIF-2α in colon cancer.
MATERIALS AND METHODS
Cell Culture
DLD-1, HCT116, HT29, Caco2, DKs-5, DKO-3, HK2-10 and HKe-3 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Hypoxic conditions were achieved by culturing cell lines in a sealed hypoxia chamber (Billups-Rothenberg, Del Mar, CA) after flushing with a mixture of 1% or 5% O2 and 5% CO2 balanced with N2, as previously described (20).
Plasmid construction and establishment of stable cells
Lentivirus vector pHR-SIN-CSGW GFP-LC3 was kindly provided by Dr. Ramnik Xavier and the GFP-LC3 fragment was excised with BamHI and NotI (21). Human K-ras and BRAF cDNA was amplified by reverse transcription-polymerase chain reaction (RT-PCR) using RNA from Caco2 cells for wild-type K-ras, SW480 cells for mutant K-ras V12, DLD-1 cells for wild-type BRAF, and HT29 cells for mutant BRAF V600E. The PCR primers used were 5’- AAGGAAGGATCCAGGCCTGCTGAAAATGACTG -3’ and 5’- AACCAAGCGGCCGCAAGGCATCATCAACACCCAG -3’ for K-ras and 5’- AAGGAAAGATCTTCTCGGTTATAAGATGGCGG -3’ and 5’- AACCAAGCGGCCGCTCTCCTGAACTCTCTCACTC -3’ for BRAF. The PCR-amplified products were digested with BamHI and NotI for K-as, and BglII and Not I for BRAF and subcloned into the pCSGW vector.
pCSGW K-ras WT, K-ras V12, BRAF WT, BRAF V600E or empty vector were introduced along with packaging plasmid pCMV-dR8.91 and envelope plasmid pMD2.G into HEK293T cells by transfection with the Fugene 6 regent (Roche, Indianapolis, IN). The culture media containing lentivirus was harvested and filtered, and polybrene was added at 8µg/ml. Caco2 cells were infected with the virus for 48 hours.
pSicoR shBRAF plasmids were kindly provided by Dr. Kevin M Haigis. pSicoR shBRAF or empty vector were introduced into HEK293T cells and lentivirus was harvested as described above. HT29, DLD-1 and Caco2 cells were infected with the virus and then selected by puromycin resistance.
RNA interference
Cells were transfected with 20 nM siRNA duplex oligos using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA ), according to the manufacturer’s instructions. Nucleotide sequences of siRNAs were as follows: for K-ras #1, 5’-r(GGUGACUUAGGUUCUAGAU)d(TT)-3’; K-ras #2, 5’-r(GGAAGCAAGUAGUAAUUGA)d(TT)-3’; BRAF #1, 5’-r(GGCCCUAUUGGACAAAUUU)d(TT)-3’; BRAF #2, 5’-r(GGAGGUGUGGAAUAUCAAA)d(TT)-3’ and control, 5’-r(GCGCGCUUUGUAGGAUUCG)d(TT)-3’.
Quantitative polymerase chain reaction analysis (qPCR)
Total RNA was isolated from cultured cells using the ISOGEN kit (Wako, Richmond, VA) and reverse transcription with random hexanucleotide primers and SuperScript Reverse Transcriptase III (Invitrogen) was performed. The resulting cDNA was amplified by real-time polymerase chain reaction using the iQ5 Real-Time PCR Detection System (BioRad, Hercules, CA) and Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). Primer sequences were 5’- GGCGCGAACGACAAGAAAAAG -3’ and 5’- CCTTATCAAGATGCGAACTCACA -3’ for HIF-1α, 5’- GGAGGTGTTCTATGAGCTGG -3’ and 5’- GACAGAAAGATCATGTCGCCA -3’ for HIF-2α, 5’-AGGCCAGCACATAGGAGAGA -3’ and 5’-TTTCTTGCGCTTTCGTTTTT -3’ for VEGF, and 5’-CGGCTACCACATCCAAGGAA-3’ and 5’-GCTGGAATTACCGCGGCT-3’ for 18S rRNA. Transcript levels of HIF-1α HIF-2α and VEGF were normalized to 18S rRNA.
Western blotting
Cells were lysed in chilled lysis buffer supplemented with the Complete protease inhibitor cocktail (Roche). 50 μg of protein extracts were resolved on a NuPAGE 3–8% Tris-Acetate or 10% Bis-Tris SDS polyacrylamide gel (Invitrogen) and transferred onto a polyvinylidene difluoride membrane (Millipore, Bedford, MA). The blots were probed with mouse monoclonal anti-HIF-1α (BD Transduction Laboratories, San Jose, CA; 1:500), rabbit polyclonal anti-HIF-2α (Novus, Littleton, CO; 1:500), mouse monoclonal anti-K-ras (Santa Cruz, Santa Cruz, CA; 1:2,000), mouse monoclonal anti-BRAF (Santa Cruz; 1:10,000), rabbit polyclonal anti-phospho p44/42 kinase (Cell Signaling, Danvers, MA; 1:2,000), rabbit polyclonal anti-p44/42 kinase (Cell Signaling; 1:2,000), rabbit polyclonal anti-phospho Akt (Cell Signaling; 1:1,000), rabbit polyclonal anti-Akt (Cell Signaling; 1:1,000), mouse monoclonal anti-pVHL (BD Pharmingen, San Jose, CA; 1:500), and anti-β actin (Sigma, St. Louis, MO; 1:500,000) antibodies. Immunoreactive proteins were visualized using the Western Lighting Chemiluminescence Reagent Plus (PerkinElmer Life Sciences, Waltham, MA).
MG132 and cycloheximide treatment
Cells were treated with the proteasome inhibitor MG132 (Calbiochem, Gibbstown, NJ, 10µmol/L) for the indicated times. HIF-1α and HIF-2α proteins were measured by western blotting. Band densities were quantified using ImageJ image analysis software and HIF protein level was normalized to β-actin at each time point. Translation of HIF was measured by its accumulation during MG132 treatment. Cells were cultured in hypoxia for 6 hours and the medium was then switched to cycloheximide-containing media under normoxia for the indicated times. HIF-1α and HIF-2α proteins were measured by western blotting, and protein stability of HIF was measured by its degradation during cycloheximide treatment
Statistical analysis
Statistical differences were analyzed by the Student's t-test, and P values < 0.05 were considered statistically significant.
RESULTS
Mutant KRAS enhances the hypoxic induction of HIF-1α but mutant BRAF induces both HIF-1α and HIF-2α
To determine whether activating mutations of KRAS or BRAF regulate the hypoxic induction of HIF-1α and HIF-2α, lentiviral vectors expressing wild type K-ras, mutant K-ras (V12), wild type BRAF or mutant BRAF (V600E) were transduced into Caco2 cells that do not carry mutations in either the KRAS or BRAF gene (Figure 1A). Stably transfected cells were cultured in either normoxia or hypoxia for 12 hours, and hypoxic induction of HIF proteins was analyzed by western blotting. The induction of HIF-1α was enhanced by ectopic expression of K-ras (V12) or BRAF (V600E) (2.11-fold and 1.71-fold, respectively, both P<0.05) in hypoxic conditions (Figure 1B). However, overexpression of wild type K-ras or wild type BRAF did not increase levels of either HIF isoform. Interestingly, BRAF (V600E) enhanced HIF-2α expression in hypoxia (1.81-fold, P<0.05) but also induced expression in normoxic conditions. K-ras (V12) did not upregulate HIF-2α in either condition (Figure 1B).
Figure 1.
Overexpression of K-ras or BRAF and hypoxic induction of HIF proteins. Wild type (WT) K-ras, mutant (V12) K-ras, wild type BRAF, mutant (V600E) BRAF or empty vector (Vec) were lentivirally transduced into Caco2 cells. A, K-ras and BRAF expression was confirmed by western blotting. B, Cells were incubated in either normoxia (N) or hypoxia (H) for 12 hours and HIF-1α and HIF-2α protein induction was analyzed by western blotting. Band densities were quantified and HIF protein levels were normalized to β-actin. Numbers below the bottom panel represent the mean fold change relative to vector control in hypoxia for 2 independent experiments. *P<0.05.
We then evaluated whether the hypoxic induction of HIF-1α and HIF-2α was regulated by endogenous KRAS mutations in colon cancer cells. DKO-3 and HKe-3 cells are derived from DLD-1 and HCT116 cells, respectively, in which the endogenous mutant KRAS allele was disrupted by homologous recombination, and control DKs-5 and HK2-10 cells have a mutant KRAS allele (22). These cells were incubated in either normoxia or hypoxia. HIF-1α induction was impaired by genetic disruption of mutant KRAS in both DKO-3 and HKe-3 cells (62% and 74% reduction, respectively, both P<0.05), whereas the induction of HIF-2α was unaffected (Figure 2A). To verify this observation, DLD-1 and HCT116 cells were transfected with 2 independent siRNA oligos targeting K-ras and then cultured in either normoxia or hypoxia. The hypoxic induction of HIF-1α was suppressed by knockdown of K-ras by 47–76% (P<0.05), whereas HIF-2α was unaffected (Figure 2B). Interestingly, knockdown of wild type K-ras only slightly suppressed HIF-1α expression by 15–26% (P=NS) in Caco2 and HT29 cells that do not carry a KRAS mutation (Supplemental Figure S1A). These data suggest that activating mutations in the KRAS gene play an important role in enhancing the hypoxic induction of HIF-1α but not HIF-2α.
Figure 2.
Depletion of mutant KRAS and BRAF oncogenes and hypoxic induction of HIF proteins. Hypoxic induction of HIF-1α and HIF-2α proteins was analyzed by western blotting. A, DKs-5, DKO-3, HK2-10 and HKe-3 cells were incubated in either normoxia (N) or hypoxia (H) for 12 hours. The weak K-ras bands seen in the DKO-3 and Hke-3 cells correspond to the remaining wild-type KRAS allele. B, DLD-1 and HCT116 cells were treated with siRNA targeting K-ras (K-ras #1 and #2) or control siRNA. Thirty-six hours after transfection, cells were subjected to either normoxia (N) or hypoxia (H) for 12 hours. C, shRNA against BRAF (shBRAF #1 and #2) or empty vector was lentivirally transduced into HT29 cells. Stable cells (HT29 pSicoR) were incubated in either normoxia (N) or hypoxia (H). D, HT29 cells were treated with siRNA targeting BRAF (BRAF #1 and #2) or control siRNA, and then cells were subjected to either normoxia (N) or hypoxia (H). Band densities were quantified and HIF protein levels were normalized to β-actin. Numbers below the bottom panel represent the mean fold change in hypoxia relative to control for 2 independent experiments. *P<0.05.
To address the role of endogenous mutant BRAF (V600E) in HT29 cells, we established stable knockdown cells using the pSicoR lentivirus expressing BRAF shRNA. The stable cells (HT29 pSicoR) were incubated in either normoxia or hypoxia. Hypoxic induction of both HIF-1α and HIF-2α was impaired by the stable knockdown of BRAF by 56–88% and 56–70% (both P<0.05), respectively (Figure 2C). This suppression of HIF-1α and HIF-2α was also observed when BRAF was transiently knocked down by siRNA in HT29 cells (52–59% and 47–69% reductions, respectively, both P<0.05) (Figure 2D). Interestingly, stable knockdown of wild type BRAF in DLD-1 and Caco2 cells that do not carry a BRAF mutation minimally suppressed HIF-1α expression by 21% and HIF-2α expression by 20–29% (Supplemental Figure S1B). These data indicate that activating mutations in BRAF play an important role in enhancing the hypoxic induction of both HIF-1α and HIF-2α.
Mutant KRAS does not regulate HIF-1α mRNA expression in either normoxia or hypoxia
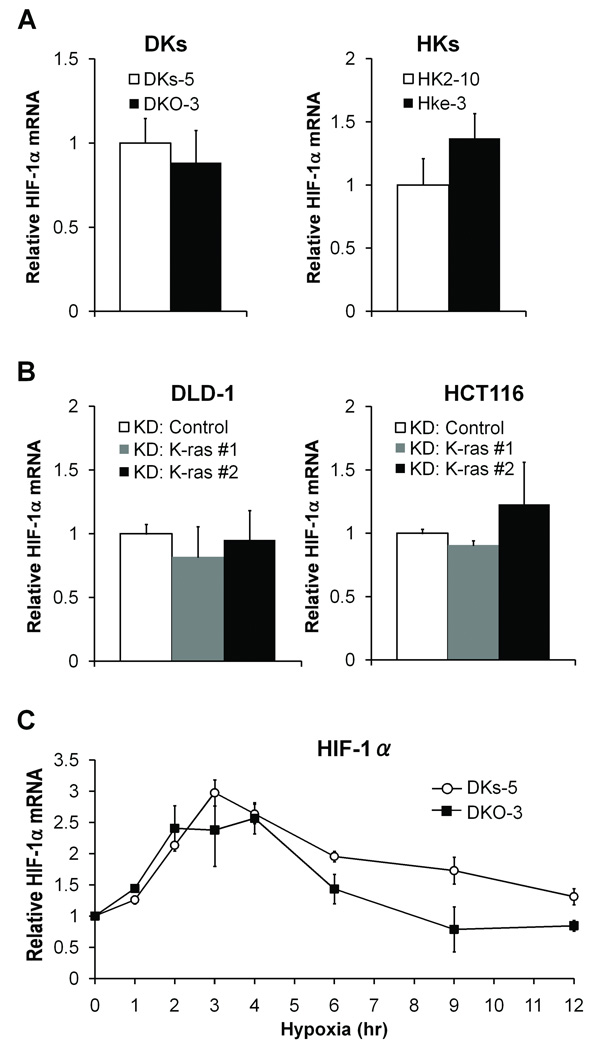
We next addressed the mechanisms by which mutant KRAS enhances the hypoxic induction of HIF-1α. DKs-5, DKO-3, HK2-10 and HKe-3 cells were cultured in normoxia and relative levels of HIF-1α mRNA were measured by qPCR. HIF-1α mRNA levels were comparable between DKs-5 and DKO-3 cells as well as between HK2–10 and HKe-3 cells (Figure 3A). DLD-1 and HCT116 cells were treated with siRNA oligos against K-ras, and HIF-1α mRNA levels were measured. HIF-1α mRNA expression was not affected by knockdown of K-ras in either cell line (Figure 3B). It has previously been shown that HIF-1α mRNA can be induced early in the response to hypoxia (0.5–3 hours) in pulmonary artery smooth muscle cells (23). To investigate whether the hypoxic regulation of HIF-1α mRNA in colon cancer may also be time-dependent and whether this can be affected by mutant KRAS, DKs-5 and DKO-3 cells were cultured in hypoxia for 1–12 hours. HIF-1α mRNA was induced approximately 3-fold at 3 hours after exposure to hypoxia in both DKs-5 and DKO-3 cells, and there were no significant differences in mRNA levels at any of the time points analyzed (P = NS) (Figure 3C). These data suggest that mutant KRAS does not regulate HIF-1α mRNA expression in either normoxia or hypoxia.
Figure 3.
Depletion of mutant K-ras and mRNA expression of HIF-1α Total RNA was extracted from DKs-5 and DKO-3 cells (DKs) or HK2-10 and HKe-3 (HKs) cultured in normoxia (A), or from DLD-1 and HCT116 cells treated with control siRNA or K-ras siRNA (B). Relative levels of HIF-1α mRNA were determined by qRT-PCR and data were normalized to the levels of 18S rRNA. All reactions were performed in triplicate and data represent the mean ± SD for 3 different cDNA samples. C, DKs-5 and DKO-3 cells were incubated in hypoxia for the indicated times. Relative levels of HIF-1α mRNA were determined by qRT-PCR and data were normalized to the levels of 18S rRNA at each time point. Graph shows the mean ± SD relative to time 0 in each cell line for 2 different cDNA samples.
Mutant KRAS regulates the translation of HIF-1α through the PI3K pathway
Although the levels of mRNA were similar, HIF-1α protein was induced by hypoxia to a greater extent in DKs-5 compared to DKO-3 cells (Supplemental Figure S2A). This resulted in higher induction of VEGF mRNA (Supplemental Figure S2B). These data suggest that mutant KRAS may not play a key role in the hypoxic induction of HIF-1α mRNA, but may regulate the accumulation of HIF-1α protein at the translational level. To further address this hypothesis, DKs-5 and DKO-3 cells were treated with MG132 for 0, 10, 20 and 30 min to block proteasomal degradation of HIF-1α protein. The rate of translation of HIF-1α in normoxia was measured by its accumulation during MG132 treatment. The rate of HIF-1α protein synthesis was significantly lower in DKO-3 cells (45% and 39% reductions at 10 and 20 min, respectively, both P<0.05) compared with DKs-5 cells (Figure 4A). DLD-1 cells were then treated with the MEK inhibitor PD98059 or the PI3K inhibitor LY294002 for 9 hours, followed by MG132 treatment. HIF-1α protein synthesis was inhibited by LY294002 (63% and 55% reductions at 20 and 30 min, respectively, both P<0.05) but not PD98059 (Figure 4B). DLD-1 cells were treated with PD98059 or LY294002 and then incubated in hypoxia for 6 hours. Hypoxic induction of HIF-1α protein was reduced by 61% (P<0.05) with LY294002 but not PD98059, despite the activation of ERK in hypoxia (Figure 4C). These data suggest that mutant KRAS may regulate the translation of HIF-1α through the PI3K pathway.
Figure 4.
Knockout of mutant KRAS and translation of HIF-1α. A, DKs-5 and DKO-3 cells were cultured in normoxia and treated with MG132 for the indicated times. Protein levels of HIF-1α were analyzed by western blotting (left panel). Band densities were quantified and HIF-1α protein level was normalized to β-actin at each time point. Protein synthesis of HIF-1α was measured by its accumulation during MG132 treatment and data represent the mean ± SD for 2 independent experiments (right panel). *P<0.05. B, Inhibition of MAPK and PI3K and translation of HIF-1α DLD-1 cells were cultured in normoxia and treated with DMSO, PD98059 or LY294002, and then treated with MG132 for the indicated times. Protein levels of HIF-1α were analyzed by western blotting (upper panel). Protein synthesis of HIF-1α was measured as indicated above and data represent the mean ± SD for 2 independent experiments (lower panel). *P<0.05. C, Inhibition of MAPK and PI3K and hypoxic induction of HIF-1α DLD-1 cells were treated with DMSO, PD98059 or LY294002 for 6 hours, and then cultured in normoxia (N) or hypoxia (H) for 6 hours. Hypoxic induction of HIF-1α and HIF-2α proteins and phosphorylation of p42/p44 (ERK) and Akt were analyzed by western blotting. Band densities were quantified and HIF-1α protein levels were normalized to β-actin. Numbers below the bottom panel represent the mean fold change relative to DMSO control for 2 independent experiments. *P<0.05.
HIF-1α protein is unstable in normoxic conditions, and the half life of HIF-1α protein is estimated at 2–5 min. To examine whether mutant KRAS affects the stability of HIF-1α protein, DKs-5 and DKO-3 cells were incubated in hypoxia for 6 hours and then cultured in the presence of the translational inhibitor cycloheximide. The half life of HIF-1α protein was comparable between DKs-5 and DKO-3 cells (Supplemental Figure S3A). Furthermore, pVHL protein levels were not affected by either knockout of mutant KRAS or knockdown of K-ras in DLD-1 and HCT116 cells (Supplemental Figure S3B). These data indicate that mutant KRAS does not regulate HIF-1α protein stability.
Mutant BRAF enhances mRNA expression of HIF-1α and HIF-2α in normoxia
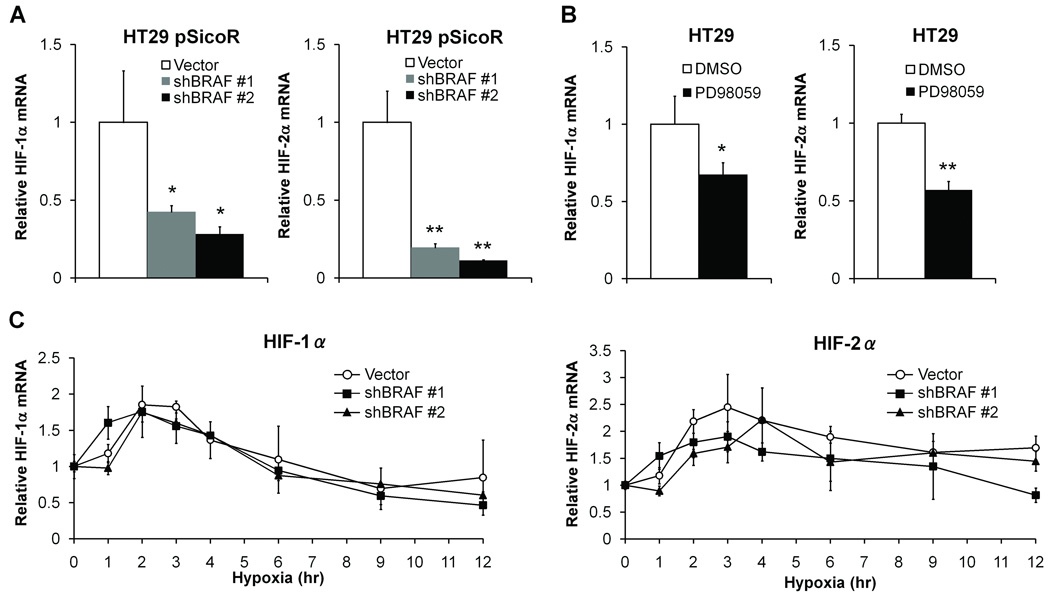
We next addressed the mechanisms by which mutant BRAF enhances the hypoxic induction of both HIF-1α and HIF-2α. HT29 pSicoR cells were cultured in normoxia and HIF mRNA levels were measured. The mRNA levels of both HIF-1α and HIF-2α were significantly reduced in BRAF knockdown cells (57–72% reduction in HIF-1α (P<0.05) and 80–89% reduction in HIF-2α (P<0.005)) compared with control cells (Figure 5A). BRAF signals through MAPK, and we tested the significance of this pathway with the MEK inhibitor PD98059. The mRNA levels of both HIF-1α and HIF-2α in HT29 cells were suppressed by PD98059 (33% reduction in HIF-1α (P<0.05) and 43% reduction in HIF-2α (P<0.005)) (Figure 5B). To investigate whether the early induction of HIF mRNA is enhanced by mutant BRAF, HT29 pSicoR cells were cultured in hypoxia for 1–12 hours. HIF-1α and HIF-2α mRNA were induced 2-fold and 2.5-fold, respectively, at 3 hours after exposure to hypoxia, but these changes were not affected by knockdown of mutant BRAF (Figure 5C). These data suggest that although mutant BRAF can enhance HIF-1α and HIF-2α mRNA expression in basal normoxic conditions, it does not enhance the induction of HIF mRNA in hypoxia.
Figure 5.
Knockdown of BRAF and mRNA expression of HIF-1α and HIF-2α. Total RNA was extracted from HT29 pSicoR (Vector, shBRAF #1 and shBRAF #2) cells cultured in normoxia (A), or from HT29 cells treated with DMSO or PD98059 (B). Relative levels of HIF-1α and HIF-2α mRNA were determined by qRT-PCR and normalized to the levels of 18S rRNA. All reactions were performed in triplicate and data represent the mean ± SD for 3 different cDNA samples. *P<0.05, **P<0.005. C, HT29 pSicoR cells were incubated in hypoxia for the indicated times. Relative levels of HIF-1α and HIF-2α mRNA were determined by qRT-PCR and normalized to the levels of 18S rRNA at each time point. Graph shows the mean ± SD relative to time 0 in each cell line for 2 different cDNA samples.
Mutant BRAF enhances the translation of HIF-2α through the MAPK pathway
Although there were no changes in the hypoxic induction of HIF mRNAs, mutant BRAF did regulate the hypoxic induction of HIF-1α and HIF-2α protein (Supplemental Figure S2C), which was accompanied by an increase in the levels of VEGF mRNA (Supplemental Figure S2D). To examine whether mutant BRAF controlled the translation of HIF protein, HT29 pSicoR cells were treated with MG132 for 0, 10, 20 and 30 min. Interestingly, the rate of HIF-1α protein synthesis was not affected, but the rate of HIF-2α protein synthesis was reduced by the knockdown of BRAF at 20 minutes (Figure 6A, P<0.05). The rate of HIF-2α protein synthesis was also suppressed at 10 and 20 min when HT29 cells were treated with PD98059 (Figure 6B, both P<0.05). Interestingly, PD98059 treatment suppressed the hypoxic induction of HIF-1α protein by 68% and HIF-2α by 58% in HT29 cells (both P<0.05) (Figure 6C). These data suggest that mutant BRAF regulates the translation of HIF-2α through MAPK pathways.
Figure 6.
Knockdown of BRAF and translation of HIF-1α and HIF-2α. A, HT29 pSicoR cells were cultured in normoxia and treated with the proteasome inhibitor MG132 for the indicated times. Protein levels of HIF-1α HIF-2α and β-actin were analyzed by western blotting (upper panel). Band densities were quantified and HIF-1α and HIF-2α protein levels were normalized to β-actin at each time point. Protein synthesis of HIFs was measured by its accumulation during MG132 treatment and data represent the mean ± SD for 2 independent experiments (lower panels). *P<0.05. B, Inhibition of MAPK and translation of HIF-1α and HIF-2α. HT29 cells were treated with DMSO or PD98059 in normoxia, and then treated with MG132 for the indicated times. Protein levels of HIF-1α and HIF-2α were analyzed by western blotting (upper panel). Protein synthesis of HIFs was measured as indicated above and data represent the mean ± SD for 2 independent experiments (lower panels). *P<0.05. C, Inhibition of MAPK and hypoxic induction of HIF-1α and HIF-2α HT29 pSicoR cells were treated with PD98059 for 6 hours and then incubated in normoxia (N) or hypoxia (H) for 6 hours. Hypoxic induction of HIF-1α and HIF-2α proteins and phosphorylation of p42/p44 (ERK) were analyzed by western blotting. Band densities were quantified and HIF protein levels were normalized to β-actin. Numbers below the bottom panel represent the mean fold change relative to DMSO control for 2 independent experiments. *P<0.05.
To examine whether mutant BRAF affects the stability of HIF proteins, HT29 pSicoR cells were incubated in hypoxia for 6 hours and then cultured in the presence of cycloheximide. The half life of HIF-1α or HIF-2α protein was not altered by stable knockdown of BRAF (Supplemental Figure S3C). Furthermore, pVHL expression was not affected by either stable or transient knockdown of BRAF in HT29 cells (Supplemental Figure S3D). These data indicate that mutant BRAF does not affect the stability of either HIF-1α or HIF-2α protein.
DISCUSSION
Although HIF-1α and HIF-2α are both induced by hypoxia in colon cancer cells, they have distinctive biological functions (13). We sought to determine whether the induction of these factors may be influenced by the underlying tumor genotype. In particular, KRAS and BRAF are among the most frequently mutated oncogenes in colon cancer, and tumors with these mutations are associated with distinct clinical phenotypes (1, 2, 5–8). Our previous studies have demonstrated a unique interaction between K-ras and hypoxia (20, 24). We now show that oncogenic KRAS functions primarily to induce HIF-1α at the level of translation. In contrast, oncogenic BRAF can enhance the mRNA expression of both HIF-1α and HIF-2α, but at the translational level, only HIF2α is induced by BRAF.
Transformation with H-ras or mutant K-ras in fibroblasts enhances protein levels of HIF-1α (17, 18), which is consistent with our results in Caco2 cells. However, ectopic overexpression of mutant K-ras can produce misleading results (25, 26). Thus, it is essential to evaluate the oncogenic regulation of HIF-1α at physiologic levels of mutant K-ras. In the present study, we used mutant KRAS knockout cells or transiently knocked down endogenous K-ras in DLD-1 and HCT116 cells. The effects of endogenous mutant KRAS on the hypoxic induction of HIFs were consistent. Importantly, these effects on HIF appear to be specific to mutant KRAS, as no such regulation of HIF was seen with the wild-type KRAS gene.
A previous report suggested that mutant BRAF may regulate HIF-1α expression in melanoma cells, but its role in colon cancer was unknown (19). In our study, stable knockdown of BRAF in HT29 cells suppressed mRNA expression of HIF-1α but did not regulate its translation, which is consistent with the previous report (19). However, HIF-1α protein stability or pVHL protein expression was not affected by knockdown of BRAF in HT29 cells, whereas stable knockdown of BRAF in WM793 melanoma cells increased pVHL protein, resulting in lower stability of HIF-1α protein (19). These findings suggest that the effects of mutant BRAF on HIF-1α protein stability and pVHL protein expression are cell-specific.
In our study, mutant KRAS and BRAF appear to regulate HIF protein primarily at the level of translation. Whereas KRAS primarily regulates the translation of HIF-1α, BRAF may selectively regulate the translation of HIF-2α. Global protein translation is generally suppressed under hypoxic conditions (27), and in our study, p70 S6 kinase, which regulates global translation and can be phosphorylated by mammalian target of rapamycin (mTOR), was inactivated in hypoxia (data not shown). Recent reports have shown that the translation of HIF-1α may also be regulated by mTOR-independent pathways (28,29). In addition, hypoxia increases HIF-2α translation by disrupting the iron-regulatory protein 1 (IRP1)-HIF-2α iron-responsive element (IRE) interaction (30). This may explain how HIF-2α protein can accumulate during hypoxia while global translation is suppressed. However, it is unknown whether oncogenic signaling regulates the binding activity of IRP1.
Although HIF-1α and HIF-2α are similarly induced by hypoxia and both can bind HREs at target gene loci, a number of reports have shown that they have distinct expression patterns and functions (31–41). Our previous study suggested that HIF-1α may promote the growth of colon cancer cells, while HIF-2α may restrain growth. Expression of HIF-2α in human colon cancer tissues was inversely correlated with tumor stage (13). In the present study, we show that mutant BRAF but not KRAS upregulates HIF-2α in hypoxia. These differential effects of oncogenic KRAS and BRAF on the hypoxic induction of HIF-2α may potentially contribute to the more favorable clinical behavior associated with mutant BRAF tumors (7). It is important to recognize that BRAF mutations typically occur in the setting of high levels of MSI, but there are likely to be other genetic alterations associated with MSI as well as additional targets of BRAF that may contribute to the unique clinical phenotype. Of note, our studies of BRAF were performed in HT29 and Caco2 cells, neither of which exhibits MSI, suggesting that the observed relationship between BRAF and HIF isoforms does not depend upon the presence of MSI.
In conclusion, we have demonstrated that oncogenic KRAS and BRAF mutations differentially regulate the hypoxic induction of HIF-1α and HIF-2α in colon cancer. Mutant KRAS enhances the hypoxic induction of HIF-1α by regulating its translation through PI3K pathways. Mutant BRAF enhances HIF-1α and HIF-2α mRNA expression, but more importantly, regulates HIF-2α translation. These differential effects on HIFs highlight the unique interaction between oncogenes and the tumor microenvironment and may potentially contribute to the some of the phenotypic differences in mutant KRAS and BRAF colon tumors.
Supplementary Material
ACKNOWLEDGEMENTS
We thank following individuals for sharing these plasmids; Dr. R. Xavier (pHR-SIN-CSGW GFP-LC3, pCMV-dR8.91 and pMD2.G) and Dr. K. Haigis (pSicoR shBRAF). We thank Dr. M. Mino-Kenudson for helpful discussions. This work was supported in part by NIH CA92594
REFERENCES
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 2.Forrester K, Almoguera C, Han K, Grizzle WE, Perucho M. Detection of high incidence of K-ras oncogenes during human colon tumorigenesis. Nature. 1987;327:298–303. doi: 10.1038/327298a0. [DOI] [PubMed] [Google Scholar]
- 3.Benhattar J, Losi L, Chaubert P, Givel JC, Costa J. Prognostic significance of K-ras mutations in colorectal carcinoma. Gastroenterology. 1993;104:1044–1048. doi: 10.1016/0016-5085(93)90272-e. [DOI] [PubMed] [Google Scholar]
- 4.Pricolo VE, Finkelstein SD, Wu TT, et al. Prognostic value of TP53 and K-ras-2 mutational analysis in stage III carcinoma of the colon. Am J Surg. 1996;171:41–46. doi: 10.1016/S0002-9610(99)80071-3. [DOI] [PubMed] [Google Scholar]
- 5.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 6.Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418(934) doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 7.Yuen ST, Davies H, Chan TL, et al. Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res. 2002;62:6451–6455. [PubMed] [Google Scholar]
- 8.Li WQ, Kawakami K, Ruszkiewicz A, Bennett G, Moore J, Iacopetta B. BRAF mutations are associated with distinctive clinical, pathological and molecular features of colorectal cancer independently of microsatellite instability status. Mol Cancer. 2006;5:2. doi: 10.1186/1476-4598-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shweiki D, Itin A, Soffer D, Keshet E. Vascular endothelial growth factor induced by hypoxia may mediate hypoxia-initiated angiogenesis. Nature. 1992;359:843–845. doi: 10.1038/359843a0. [DOI] [PubMed] [Google Scholar]
- 10.Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 11.Cummins EP, Taylor CT. Hypoxia-responsive transcription factors. Pflugers Arch. 2005;450:363–371. doi: 10.1007/s00424-005-1413-7. [DOI] [PubMed] [Google Scholar]
- 12.Maxwell PH, Ratcliffe PJ. Oxygen sensors and angiogenesis. Semin Cell Dev Biol. 2002;13:29–37. doi: 10.1006/scdb.2001.0287. [DOI] [PubMed] [Google Scholar]
- 13.Imamura T, Kikuchi H, Herraiz MT, et al. HIF-1alpha and HIF-2alpha have divergent roles in colon cancer. Int J Cancer. 2009;124:763–771. doi: 10.1002/ijc.24032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feldser D, Agani F, Iyer NV, Pak B, Ferreira G, Semenza GL. Reciprocal positive regulation of hypoxia-inducible factor 1alpha and insulin-like growth factor 2. Cancer Res. 1999;59:3915–3918. [PubMed] [Google Scholar]
- 15.Fukuda R, Hirota K, Fan F, Jung YD, Ellis LM, Semenza GL. Insulin-like growth factor 1 induces hypoxia-inducible factor 1-mediated vascular endothelial growth factor expression, which is dependent on MAP kinase and phosphatidylinositol 3-kinase signaling in colon cancer cells. J Biol Chem. 2002;277:38205–38211. doi: 10.1074/jbc.M203781200. [DOI] [PubMed] [Google Scholar]
- 16.Jiang BH, Jiang G, Zheng JZ, Lu Z, Hunter T, Vogt PK. Phosphatidylinositol 3-kinase signaling controls levels of hypoxia-inducible factor 1. Cell Growth Differ. 2001;12:363–369. [PubMed] [Google Scholar]
- 17.Chen C, Pore N, Behrooz A, Ismail-Beigi F, Maity A. Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J Biol Chem. 2001;276:9519–9525. doi: 10.1074/jbc.M010144200. [DOI] [PubMed] [Google Scholar]
- 18.Chan DA, Sutphin PD, Denko NC, Giaccia AJ. Role of prolyl hydroxylation in oncogenically stabilized hypoxia-inducible factor-1alpha. J Biol Chem. 2002;277:40112–40117. doi: 10.1074/jbc.M206922200. [DOI] [PubMed] [Google Scholar]
- 19.Kumar SM, Yu H, Edwards R, et al. Mutant V600E BRAF Increases Hypoxia Inducible Factor-1{alpha} Expression in Melanoma. Cancer Res. 2007;67:3177–3184. doi: 10.1158/0008-5472.CAN-06-3312. [DOI] [PubMed] [Google Scholar]
- 20.Mizukami Y, Li J, Zhang X, Zimmer MA, Iliopoulos O, Chung DC. Hypoxia-inducible factor-1-independent regulation of vascular endothelial growth factor by hypoxia in colon cancer. Cancer Res. 2004;64:1765–1772. doi: 10.1158/0008-5472.can-03-3017. [DOI] [PubMed] [Google Scholar]
- 21.Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirasawa S, Furuse M, Yokoyama N, Sasazuki T. Altered growth of human colon cancer cell lines disrupted at activated Ki-ras. Science. 1993;260:85–88. doi: 10.1126/science.8465203. [DOI] [PubMed] [Google Scholar]
- 23.Belaiba RS, Bonello S, Zahringer C, et al. Hypoxia up-regulates hypoxia-inducible factor-1alpha transcription by involving phosphatidylinositol 3-kinase and nuclear factor kappaB in pulmonary artery smooth muscle cells. Mol Biol Cell. 2007;18:4691–4697. doi: 10.1091/mbc.E07-04-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizukami Y, Fujiki K, Duerr EM, et al. Hypoxic regulation of vascular endothelial growth factor through the induction of phosphatidylinositol 3-kinase/Rho/ROCK and c-Myc. J Biol Chem. 2006;281:13957–13963. doi: 10.1074/jbc.M511763200. [DOI] [PubMed] [Google Scholar]
- 25.Hua VY, Wang WK, Duesberg PH. Dominant transformation by mutated human ras genes in vitro requires more than 100 times higher expression than is observed in cancers. Proc Natl Acad Sci U S A. 1997;94:9614–9619. doi: 10.1073/pnas.94.18.9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arena S, Isella C, Martini M, de Marco A, Medico E, Bardelli A. Knock-in of Oncogenic Kras Does Not Transform Mouse Somatic Cells But Triggers a Transcriptional Response that Classifies Human Cancers. Cancer Res. 2007;67:8468–8476. doi: 10.1158/0008-5472.CAN-07-1126. [DOI] [PubMed] [Google Scholar]
- 27.Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8:180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- 28.Shafee N, Kaluz S, Ru N, Stanbridge EJ. PI3K/Akt activity has variable cell-specific effects on expression of HIF target genes, CA9 and VEGF, in human cancer cell lines. Cancer Lett. 2009 doi: 10.1016/j.canlet.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pore N, Jiang Z, Shu HK, Bernhard E, Kao GD, Maity A. Akt1 activation can augment hypoxia-inducible factor-1alpha expression by increasing protein translation through a mammalian target of rapamycin-independent pathway. Mol Cancer Res. 2006;4:471–479. doi: 10.1158/1541-7786.MCR-05-0234. [DOI] [PubMed] [Google Scholar]
- 30.Zimmer M, Ebert BL, Neil C, et al. Small-molecule inhibitors of HIF-2a translation link its 5'UTR iron-responsive element to oxygen sensing. Mol Cell. 2008;32:838–848. doi: 10.1016/j.molcel.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iyer NV, Kotch LE, Agani F, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang V, Davis DA, Haque M, Huang LE, Yarchoan R. Differential gene up-regulation by hypoxia-inducible factor-1alpha and hypoxia-inducible factor-2alpha in HEK293T cells. Cancer Res. 2005;65:3299–3306. doi: 10.1158/0008-5472.CAN-04-4130. [DOI] [PubMed] [Google Scholar]
- 33.Wiesener MS, Jurgensen JS, Rosenberger C, et al. Widespread hypoxia-inducible expression of HIF-2alpha in distinct cell populations of different organs. Faseb J. 2003;17:271–273. doi: 10.1096/fj.02-0445fje. [DOI] [PubMed] [Google Scholar]
- 34.Sowter HM, Raval RR, Moore JW, Ratcliffe PJ, Harris AL. Predominant role of hypoxia-inducible transcription factor (Hif)-1alpha versus Hif-2alpha in regulation of the transcriptional response to hypoxia. Cancer Res. 2003;63:6130–6134. [PubMed] [Google Scholar]
- 35.Flamme I, Frohlich T, von Reutern M, Kappel A, Damert A, Risau W. HRF, a putative basic helix-loop-helix-PAS-domain transcription factor is closely related to hypoxia-inducible factor-1 alpha and developmentally expressed in blood vessels. Mech Dev. 1997;63:51–60. doi: 10.1016/s0925-4773(97)00674-6. [DOI] [PubMed] [Google Scholar]
- 36.Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci U S A. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 38.Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. Embo J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng J, Zhang L, Drysdale L, Fong GH. The transcription factor EPAS-1/hypoxia-inducible factor 2alpha plays an important role in vascular remodeling. Proc Natl Acad Sci U S A. 2000;97:8386–8391. doi: 10.1073/pnas.140087397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian H, Hammer RE, Matsumoto AM, Russell DW, McKnight SL. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 1998;12:3320–3324. doi: 10.1101/gad.12.21.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Compernolle V, Brusselmans K, Acker T, et al. Loss of HIF-2alpha and inhibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nat Med. 2002;8:702–710. doi: 10.1038/nm721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.