Abstract
BACKGROUND:
The ability to identify low-level somatic DNA mutations and minority alleles within an excess wild-type sample is becoming essential for characterizing early and posttreatment tumor status in cancer patients. Over the past 2 decades, much research has focused on improving the selectivity of PCR-based technologies for enhancing the detection of minority (mutant) alleles in clinical samples. Routine application in clinical and diagnostic settings requires that these techniques be accurate and cost-effective and require little effort to optimize, perform, and analyze.
CONTENT:
Enrichment methods typically segregate by their ability to enrich for, and detect, either known or unknown mutations. Although there are several robust approaches for detecting known mutations within a high background of wild-type DNA, there are few techniques capable of enriching and detecting low-level unknown mutations. One promising development is COLD-PCR (coamplification at lower denaturation temperature), which enables enrichment of PCR amplicons containing unknown mutations at any position, such that they can be subsequently sequenced to identify the exact nucleotide change.
SUMMARY:
This review summarizes technologies available for detecting minority DNA mutations, placing an emphasis on newer methods that facilitate the enrichment of unknown low-level DNA variants such that the mutation can subsequently be sequenced. The enrichment of minority alleles is imperative in clinical and diagnostic applications, especially in those related to cancer detection, and continued technology development is warranted.
A prominent concern confronting clinical and diagnostic applications is the ability to detect clinically significant low-level mutations and minority alleles. The ability to discern mutations is important in many regards, but especially for (a) early cancer detection from tissue biopsies and bodily fluids such as plasma or serum; (b) assessment of residual disease after surgery or radiochemotherapy; (c) disease staging and molecular profiling for prognosis or tailoring therapy to individual patients; and (d) monitoring of therapy outcome and cancer remission/relapse. Efficient detection of cancer-relevant somatic mutations largely depends on the selectivity of the techniques and methods employed. Detection and identification of oncogene and tumor-suppressor gene mutations primarily require analysis of precancerous or cancerous tissue, sputum, urine, stool, and circulating extracellular DNA released in blood. The sample is typically composed of both wild-type and mutant DNA, and the quantity of wild-type DNA often exceeds the mutant DNA contribution. In many cases, wild-type DNA vastly exceeds mutant DNA, making it difficult to detect and identify minority alleles present at extremely low concentrations.
The use of enrichment methods is often beneficial or necessary to increase the mutant concentration to a level at which accurate and precise analysis is feasible. The selectivity of the enrichment and detection methods used must be carefully considered to maintain accuracy. To detect low-level early mutations in tumors or the emergence of resistance mutations (e.g., 10−3 to 10−6 mutant to wild-type DNA), both high selectivity and enrichment of minority alleles are required for successful detection and identification. Furthermore, for a particular approach to be used as a routine diagnostic tool, it must achieve a balance of high selectivity and enrichment while maintaining accuracy, convenience, and low cost.
With these guidelines in mind, we reviewed a selection of PCR-based methods developed to preferentially enrich known or unknown mutations present in low concentrations among wild-type DNA. Parsons and Heflich (1) and Gocke et al. (2) have previously reviewed enrichment methods (also known as “genotypic selection methods”); however, those reviews focused primarily on early methods specifically designed for the enrichment of known point mutations (mutations at predefined DNA positions). Enrichment methods typically segregate by their ability to enrich either known or unknown mutations. The design and development of mutation enrichment methodologies is a much easier task for known mutations than it is for unknown mutations, as sequence data can be used, specific nucleotides can be targeted, and a wider scope of applications are available. As a result, more methods have been developed and modified to preferentially enrich known mutations than unknown mutations. For many cancer-relevant genes, the occurrence of unknown somatic mutations can be very important, and mutation-selectivity is a strong consideration in choosing the appropriate method for routine testing and identification. Selectivity of a mutation detection method refers to the selection of mutation-containing alleles among an excess of wild-type alleles; enrichment refers to a process that increases mutant allele concentration relative to wild-type alleles, such that subsequent mutation detection is facilitated. For purposes of this review, minority allele enrichment methods will be discussed according to their ability to enhance known vs unknown mutations and their degree of selectivity.
MODERATE- TO HIGH-SELECTIVITY METHODS AND ENRICHMENT OF KNOWN MUTATIONS
Many moderate- to high-selectivity PCR methods for known mutations have been developed over the past 2 decades (Table 1). One of the most widely used approaches relies on the use of 3′ terminal nucleotide manipulation to enhance allele-specific amplification of a particular nucleotide variant (i.e., mutant or minority allele). Methods such as amplification refractory mutation system (ARMS)2 (3), allele-specific amplification (ASPCR) (4), allele-specific amplification (ASA) (5), PCR amplification of specific alleles (PASA) (6), and PCR amplification of multiple specific alleles (PAMSA) (7) have the ability to enrich minority alleles present among wild-type DNA at concentrations as low as 0.1% to 1%. Generally, these approaches are relatively easy to use and tend to produce results with high accuracy, although selectivity typically remains low to moderate. Increasing the selectivity further by the inclusion of additional nucleotide mismatches toward the 3′ end is possible, but requires extensive experimentation and optimization. Derivatives of this approach have been developed and include competitive oligonucleotide priming (COP) (8), mutant enrichment PCR [enriched or mutant-enriched PCR (EPCR or ME-PCR)] (9), mismatch amplification mutation assay (MAMA) (10), and mutant allele–specific amplification (MASA) (11). These methods have exhibited similar or higher selectivity and the ability to enrich a single minority allele present among 102 (COP and ME-PCR) to 105 (MAMA) wild-type alleles.
Table 1.
Minority allele enrichment methods.a
| Method | Selectivity | Reference |
|---|---|---|
| Moderate to high selectivity and enrichment of known mutations | ||
| ARMS | 10−1 to 10−3 | Newton et al. (3) |
| ASPCR | 10−1 to 10−3 | Wu et al. (4) |
| ASA | 10−1 to 10−3 | Okayama et al. (5) |
| PASA and PAMSA | 10−1 to 10−3 | Sommer et al. (6), Dutton and Sommer (7) |
| COP | 10−1 to 10−3 | Gibbs et al. (8) |
| E-PCR | 10−1 to 10−4 | Kahn et al. (9) |
| MAMA | 10−1 to 10−5 | Cha et al. (10) |
| MASA | 10−1 to 10−3 | Takeda et al. (11) |
| PNA-mediated PCR | 10−3 to 10−5 | Nielsen et al. (17), Dabritz et al. (20) |
| LNA-mediated WTB-PCR | 10−1 to 10−5 | Dominguez and Kolodney (18), Oldenburg et al. (19) |
| TaqMan RSM | 5 × 10−4 | Wolff and Gemmell (16) |
| TaqMAMA | 5 × 10−5 | Easterday et al. (12) |
| FLAG-PCR | 10−1 to 10−3 | Amicarelli et al. (21) |
| AIRS-RFLP | 10−3 to 10−4 | Haliassos et al. (15) |
| Very high selectivity and enrichment of known mutations | ||
| RSM-PCR | 10−3 to 10−8 | Parsons and Heflich (1), Jenkins et al. (23) |
| APRIL-ATM | 10−3 to 10−6 | Kaur et al. (24) |
| Digital PCR and RMC-PCR | 10−3 to 10−8 | Vogelstein et al. (27), Bielas and Loeb (28) |
| PAP-ASA and bi-PAP-ASA | 10−4 to 10−9 | Liu and Sommer (25), Shi et al. (26) |
| Enrichment and detection of unknown mutations | ||
| Electrophoresis (HET, SSCP, DGGE, dHPLC, CDCE) | 10−1 to 10−2 | Lichten and Fox (29), Orita et al. (30), Cariello et al. (31), Li-Sucholeiki and Thilly (32), Underhill et al. (33), Emmerson et al. (34) |
| Endo V-ligase PCR | 10−1 to 10−2 | Pincas et al. (37) |
| MutY-LM-PCRb | 10−1 to 10−2 | Zhang et al. (36) |
| sRT-MELT | 10−1 to 10−2 | Li et al. (38) |
| iFLP | 10−3 to 10−5 | Liu et al. (39) |
| COLD-PCR | 10−1 to 10−4 | Li et al. (40) |
Selectivity is presented as a range representing the commonly achieved and maximum selectivity of mutant detection of the approach.
LM, ligation-mediated; sRT-MELT, surveyor-mediated real-time melting.
Various combinations of allele-specific PCR with real-time PCR have also been shown to effectively enrich minority alleles with moderate to high selectivity. For example, TaqMAMA (12) combines the real-time scoring attributes of TaqMan® probes with the selectivity of the MAMA approach to preferentially enrich and simultaneously identify the nature of a known nucleotide variant. TaqMAMA can enrich and detect an alternate allele when present in an excess of approximately 2 × 103 wild-type alleles, although it is questionable if this degree of selectivity can be achieved for all mutation screening targets. Another real-time PCR-based approach, antiprimer quenching-based real-time PCR (aQRT-PCR) (13), uses an allele-specific primer for mutant enrichment, real-time genotyping, and real-time product quantification in a single-step, closed-tube format.
Use of thermostable restriction enzymes that selectively destroy wild-type samples during PCR, thereby enriching the mutation frequency, has also led to methods for genotypic selection [restriction endonuclease–mediated selective PCR (REMS-PCR)] (14). When appropriate restriction endonucleases are not available, artificial introduction of a restriction site (AIRS) RFLP (15) can generate an endonuclease recognition sequence by modifying 1 or more nucleotides within the priming region of the wild-type DNA. The selectivity of these approaches was reported to enable detection of 1 mutated cell among 2.5 × 103 wild-type alleles (15). In real-time format, the combination of TaqMan assays and allele-specific restriction by an endonuclease has detection selectivity for an allele contribution of 1 in 2.0 × 103 wild-type alleles (16).
Alternative moderate- to high-selectivity techniques have used physical molecular modifications to allele specificity. For example, peptide nucleic acids (PNAs) (17) and locked nucleic acids (LNAs) (18) both have increased binding affinities. Through a PCR clamping approach, primers can be replaced by LNA or PNA hybridization probes that are specific for the wild type; both PNAs and LNAs can suppress the amplification of wild-type DNA, allowing for increased amplification of the mutant allele. The use of LNAs in wild-type blocking PCR (WTB-PCR) and of PNAs in K-ras amplification has exhibited the ability to identify mutations among 105 wild-type alleles (19, 20), although routine application of these approaches often results in detecting mutants at frequencies of 10−2 to 10−3. Last, the FLAG assay (fluorescent amplicon generation) (21) combines REMS-PCR with incorporation of an exceptionally thermostable endonuclease (PspGI) and PNA probes and can be performed in real-time, high-throughput, and closed-tube format. The FLAG assay has demonstrated the ability to detect 1 mutant in 103 wild-type DNA. Generally, PNA/LNA-based approaches are attractive, although the time and cost required for optimization may hinder their widespread use.
VERY-HIGH-SELECTIVITY METHODS AND ENRICHMENT OF KNOWN MUTATIONS
Some enrichment methods boast very high selectivity and the ability to preferentially enrich and identify known mutations at extremely low levels. RFLP-PCR–based approaches have proven to be simple and inexpensive in their application as well as highly selective for the enrichment of known mutations. The use of thermostable restriction endonucleases either before PCR amplification or concurrently with PCR has been aggressively applied by performing more than 1 round of enrichment, with the aim to increase digestion and suppression of the wild-type allele, thus preferentially amplifying the mutant type. Several derivatives of this approach have been developed; however, one of the first, the restriction site mutation assay (RSM-PCR) (22), has exhibited the capability to enrich mutants present at 1 mutant per 108 wild-type genes (1, 23). The APRIL-ATM method (amplification via primer ligation, at the mutation) (24) uses an inverse approach, mutant-specific RFLP, to digest mutant PCR products rather than wild-type products. Subsequently, oligonucleotides are ligated to the digested fragments at the site of the mutation and subjected to a second PCR, thus preferentially enriching the mutant DNA. APRIL-ATM has exhibited high selectivity, with the ability to detect a frequency of 1.6 × 10−6 mutant alleles among wild-type DNA. Although these RFLP-PCR–based approaches are often advantageous because they are simple in application and low in cost, in some cases they may yield selectivity only on the order of 10−3 to 10−4 mutants per wild-type DNA (25).
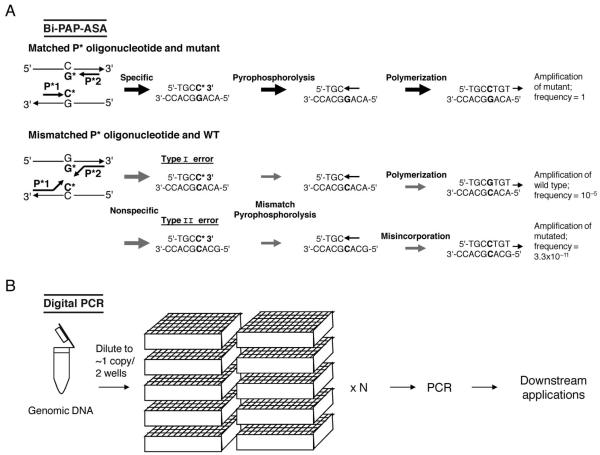
One method that reports very high selectivity and enrichment is based on the combined use of pyrophosphate-activated polymerization (PAP) and ASA (25). PAP-ASA employs an allele-specific oligonucleotide (P*), which is activated by pyrophosphorolysis and DNA polymerization during PCR. A 3′terminal dideoxynucleotide is removed in the presence of pyrophosphate, and the activated P* can then be extended by the DNA polymerase. The authors report that this approach is capable of detecting 1 mutant allele in 106 to 109 wild-type alleles (25). A bidirectional modification of PAP-ASA (bi-PAP-ASA) (26) uses 2 opposing, allele-specific 3′terminal oligonucleotides (P*) to increase selectivity and amplify low-level somatic mutants present among 107 to 109 wild types (Fig. 1A). On the other hand, not all reports achieve this high selectivity using pyrophosphorolysis. One recent study used a PAP-based method to detect low level B-RAF in uveal melanomas. The authors were able to detect low-level mutations that were not detectable by Sanger sequencing; however, their evaluation of the technique exhibited a detection limit of 1 mutant among 104 wild-type alleles.
Fig. 1. Highly selective PCR-based methods for known and unknown mutation enhancement and identification.
(A), Bidirectional pyrophosphorolysis-activated polymerization allele-specific amplification (bi-PAP-ASA) for high selectivity of known mutations (26). P* is a specifically designed oligonucleotide with a 3′-terminal blocker that is activated, but not extended, by pyrophosphorolysis. Downstream and upstream P* contain dideoxy C and G at the 3′ termini that are specific to the mutant but not the wild type. Efficient amplification of the mutant occurs after pyrophosphorolysis (to remove the 3′-terminal ddCMP) and polymerization. Inefficient amplification is denoted by the gray arrows. Nonspecific type I error amplification is rare; type II error is caused by serial mismatch phosphorolysis and misincorporation, which results in the exponential amplification of the mutated product and reduces selectivity. (B), Digital PCR for high selectivity of both known and unknown mutations (27). Genomic DNA is diluted to approximately 1–2 copies per well. The number (N) of required wells varies widely and depends on putative mutant and wild-type concentrations. PCR is performed on each sample well individually. PCR amplicons can be used in many downstream applications such as direct sequencing, pyrosequencing, TaqMan assays, and molecular beacons.
PCR of single DNA molecules may also be considered a form of high mutant enrichment. For example, digital PCR (27) relies on the amplification of individual molecules of DNA (Fig. 1B). In the original report (27), DNA template was diluted to distribute approximately 1 molecule of DNA per reaction, thus allowing detection of approximately 1 mutant in 103 alleles in numerous parallel PCR reactions. In principle, the more reactions that are performed, the higher the selectivity; in practice, the selectivity is limited by the occurrence of PCR errors. Digital PCR is currently difficult to apply in routine applications with conventional thermocyclers, as it requires the analysis of a very large number of samples to detect mutants occurring at very low frequencies relative to the wild-type DNA. However, the latter assessment may eventually change with the onset of nanofluidics. Bielas and Loeb (28) developed [random mutation capture (RMC)], a mutant enrichment method, based on a combination of RSM and digital PCR, that is capable of identifying 1 mutant base among 108 wild-type nucleotides. In this interesting, but complex, approach, the mutant phenotype is enriched through biotin-labeled probes and magnetic bead separation. The collected fraction is then subjected to TaqI cleavage to digest remaining wild-type DNA; the final product is diluted to isolate single molecules, and quantitative PCR is performed to amplify the mutant phenotype.
Overall, there are currently several approaches available that report high selectivity and enrichment of known mutations and that are also applicable for routine application. Many of these assays are simple in their methodology and application (ASA- and RFLP-based approaches, for example); however, their selectivity is often not sufficient for enhancement of extremely low-level known mutations. On the other hand, highly selective approaches for detecting known mutations can be time-consuming and difficult to perform, and therefore may not be appropriate for most routine clinical and diagnostic applications. Accordingly, the selection of a technique depends very much on the intended application.
ENRICHMENT OF UNKNOWN MUTATIONS FOLLOWED BY MUTATION SEQUENCING
Traditionally, the identification of unknown mutations has relied on Sanger sequencing analysis; however, sequencing is reliable only for detecting mutant alleles that exist at concentrations above approximately 20% among wild-type DNA (27). This degree of sensitivity is inappropriate for detecting low-level somatic mutations in several situations, such as in premalignant tissues or during early cancer development, post-treatment tissue, or apoptotic and necrotic circulating DNA molecules or for detecting the emergence of resistance mutations in radiochemotherapy-treated tumors. The development of techniques that can be applied to enrich DNA containing unknown mutations that exist at low concentrations relative to the wild-type DNA, followed by sequencing to identify the exact nucleotide change, is thus of high interest. To this end, although there are techniques that perform mutation scanning with higher selectivity relative to sequencing [e.g., chemical cleavage of mismatches (CCM); cleavage of mismatches using endonuclease V (endo V), endo VII, T4, MutY, thymine DNA glycosylase (TDG), or celery extract I (CEL I) mismatch detection enzymes; high-resolution melting (HRM); and others] the following paragraphs focus specifically on methods that enable nondestructive selection and enrichment of DNA containing unknown mutations, such that they can be followed by sequencing to identify the position and exact nucleotide change.
There are several established approaches to enrich DNA containing unknown mutations via the use of electrophoretic methods of post-PCR products. Among these are heteroduplex analysis (HET) (29), single-strand conformation polymorphism (SSCP) (30), denaturing gradient gel electrophoresis (DGGE) (31), constant denaturing capillary electrophoresis (CDCE) (32), and denaturing HPLC (dHPLC) (33). For example, for the commonly used dHPLC method, mutant and wild-type PCR products can be physically separated via their difference in retention times on polycarbonate columns and subsequently collected on a fraction collector. The mutant DNA can thus be preferentially separated, PCR-amplified, and used in downstream applications. The disadvantages of fraction-mediated dHPLC are the requirement for extra steps in the overall procedure, the limited electro-phoretic separation between mutant and wild-type alleles for certain mutations, and the required equipment expense. When performed accurately, however, this approach has the ability to enrich the mutant fraction 10-fold, from as little as approximately 5% to as much as approximately 50% (34).
Enzymatic approaches using mismatch-detecting enzymes such as immobilized MutS have also been applied to enrich PCR sequences containing unknown mutations (35). In addition, glycosylases MutY or TDG combined with ligation-mediated PCR have also been reported to selectively enrich mutation-containing sequences (36). Unfortunately, the selectivity of MutS is only modest, whereas MutY and TDG-based approaches are restricted to detecting only a fraction of all possible mutations. In another approach, DNA ligation is combined with endo V (37). Endo V detects and cleaves heteroduplex DNA 1 base downstream from the mutation. AK16D DNA ligase is then used to fill in background nicks, increasing assay sensitivity. The second step in this approach uses internally labeled primers to eliminate the endo V cleavage at the 5′ terminus and selectively amplify cleaved fragments, thus allowing for specific mutant detection and amplification. One approach has recently reported the use of a highly selective enzyme, CEL I (Surveyor), in conjunction with ligation of a primer at the 3′OH end of CEL I–digested fragments to enable enrichmentPCR of mutation-containing DNA followed by sequencing (38). This approach detects all mutations and enables sequencing of unknown mutations at levels of 1–5 × 10−2 mutant-to-wild-type ratio.
The sensitivity of approaches employing mismatch-detecting enzymes is ultimately limited by the selectivity and efficiency of the enzymes used. Compared with restriction endonucleases, the selectivity of any available mismatch detecting enzyme is much inferior. An approach that employs a restriction endonuclease to perform a highly selective mutation scanning is iFLP (inverse PCR-based amplified RFLP) (39). iFLP combines inverse PCR, RFLP, and dHPLC. DNA is circularized and subsequently digested by TaqI restriction enzyme. Circularized DNA that does not normally contain TaqI recognition sequences is targeted in this approach. Any sequence that has acquired TaqI sites anywhere on the sequence due to a mutation is recognized by the enzyme and converted into double-stranded linear DNA fragments, which can be ligated to TaqI-specific adaptors and PCR-amplified. This method can detect 1 unknown mutant in 105 wild-type sequences; however, the technique is time-intensive and can detect only a fraction of all possible mutations.
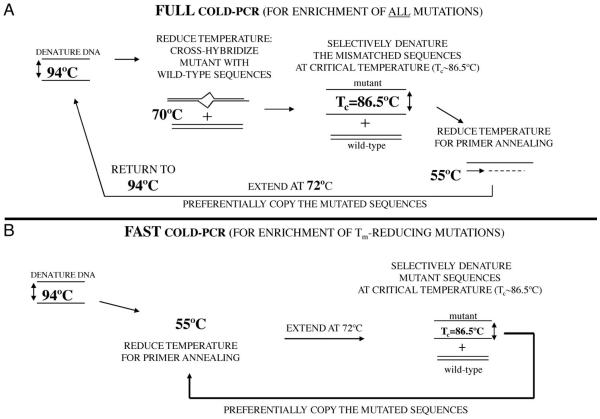
Despite progress provided in the enrichment of unknown minority alleles by methods based on post-PCR capillary electrophoresis or enzymatic recognition followed by second PCR and sequencing, these methods generally require multistep protocols that can be time-consuming to perform. However, a new technique has recently been developed that removes many difficulties associated with the enrichment of unknown mutations. Coamplification at lower denaturation temperature PCR (COLD-PCR) (40) is a single-step method that results in the enhancement of both known and unknown minority alleles during PCR, irrespective of mutation type and position. This approach is based on the observation that, for a given DNA sequence close to a critical denaturation temperature (Tc), the percent denaturation becomes sensitive to the exact DNA sequence, such that even point mutations make a substantial difference. This principle is used for mutation enrichment by inducing the formation of heteroduplexes at positions of mutations, during PCR. Thus by using a lower denaturation temperature during PCR, double-stranded DNA (dsDNA) containing mismatches (heteroduplexes) denature first. True homoduplexes have a higher melting temperature (Tm) and denature less than heteroduplexes at the critical denaturation temperature; thus their amplification is relatively suppressed (Fig. 2). For mutations that tend to lower Tm (such as G:C>T:AorG:C>A:T), which make up approximately 70% of the encountered mutations, COLD-PCR enriches mutant alleles even without formation of heteroduplexes (Fig. 2). As a general rule, a substantial enrichment for most COLD-PCR reactions can be obtained by using a Tc approximately 1 °C lower than the amplicon Tm; for certain sequences, however, fine-tuning of the Tc can be beneficial, and an optimal Tc can vary 0.5 °C to 1.5 °C lower than the Tm. The Tm can be experimentally determined on most real-time thermocyclers by performing a melting curve after PCR.
Fig. 2. COLD-PCR protocol (40). Two forms of COLD-PCR have been developed, Full COLD-PCR (A) and Fast COLD-PCR (B). An example protocol for a 167-bp TP53 amplicon is reviewed here.
(A), Full COLD-PCR has the potential to enrich all possible mutations. Several preliminary rounds of conventional PCR enable an initial buildup of 1 or more target amplicons, then the cycling switches to COLD-PCR. After denaturation at 94 °C, the PCR amplicons are incubated (e.g., 70 °C for 2–8 min) for reannealing and cross-hybridization. Cross-hybridization of mutant and wild-type alleles forms a mismatch-containing structure (heteroduplex) that has a lower melting temperature than a fully-matched structure (homoduplex). The PCR temperature is next raised to the critical denaturation temperature (Tc) (e.g., 86.5 °C) to preferentially denature the heteroduplexed amplicons. The temperature is reduced for primer annealing (e.g., 55 °C) and then raised to 72 °C to extend the amplicon and preferentially amplify the mutation-containing alleles. (B), Fast COLD-PCR is a simpler cycling that can be performed to enrich for mutations that reduce the melting temperature of the wild-type amplicon. Using the mutant Tc, rather than the standard 94 °C denaturation temperature, preferentially denatures the lower-Tm allele. Fast COLD-PCR does not perform the 70 °C incubation step. Fast COLD-PCR amplification and enrichment begins earlier in the cycling than in full COLD-PCR, thus resulting in higher enrichment.
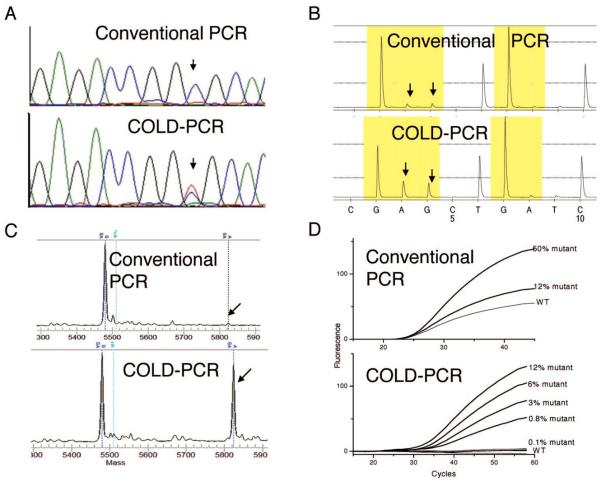
The COLD-PCR principle enables direct PCR from genomic DNA to amplify mutation-containing alleles with a selectivity of up to 100-fold over wild-type alleles. Advantages of COLD-PCR include its simplicity in performance, preferential amplification of mismatch-containing dsDNA containing unknown mutations without the need for lengthy protocols or allele-specific primers, probes, and enzymes, and ability to sequence directly the amplified product. Additionally, COLD-PCR can be used in place of conventional PCR and combined with most existing assays, while it requires essentially no additional cost, time, and labor. COLD-PCR has been used to identify both known and unknown mutations in place of PCR for a variety of techniques such as Sanger sequencing (e.g., Fig. 3A), pyrosequencing (Fig. 3B), MALDI-TOF (Fig. 3C), and TaqMan probe analyses (Fig. 3D) (40). Disadvantages of COLD-PCR include the requirement for precise denaturation temperature control during PCR (to within ±0.3 °C), restriction of analyzing sequences smaller than approximately 200 bp, vulnerability to polymerase errors, and variability of the overall mutation enrichment obtained depending on DNA position and nucleotide substitution. COLD-PCR selectivity for point mutations can increase further if subsequent PCR rounds are performed, as has already been shown for unknown deletions (40). As with deep-sequencing approaches that use single-molecule sequencing (41), COLD-PCR enrichment of mutations is ultimately limited by polymerase-introduced errors. As newer polymerases with very high fidelity are continuously being improved, however, so are the ultimate enrichment abilities of approaches like COLD-PCR. Ultra-deep sequencing following several rounds of COLD-PCR could reveal aspects of cancer biology that are clinically very important (e.g., the origins of resistance to therapy).
Fig. 3. COLD-PCR improves mutation detection in downstream assays (40).
(A), Sanger sequencing detects low-level mutations after COLD-PCR. The HCC2218 cell line (TP53 exon 8; 14516 C>T) was diluted in wild-type DNA. Sanger sequencing was performed on products amplified by both conventional (upper panel) and COLD (lower panel; Tc 86.5 °C) PCR. COLD-PCR sequencing exhibits enrichment of the mutated allele. (B), COLD-PCR improves detection via pyrosequencing. DNA from cell line A549 was diluted 33-fold into wild-type DNA; a 98-bp K-ras exon 2 segment was amplified by both COLD-PCR (lower panel; Tc 80 °C) and conventional PCR (upper panel), followed by pyrosequencing. The G>A mutation of the A549 cell line was visible only when COLD-PCR was applied. (C), COLD-PCR improves the sensitivity of MALDI-TOF genotyping technologies. Fast COLD-PCR (Tc 83.5 °C) was used to amplify an 87-bp fragment in plasma-circulating DNA (hotspot mutation TP53 exon 8, codon 273). Amplicons were genotyped using MALDI-TOF. The G>A mutation was detectable in COLD-PCR amplicons (lower panel); however, it was undetectable in conventional PCR amplicons (upper panel). (D), COLD-PCR improves the sensitivity of TaqMan genotyping technologies. Serial dilutions of the H1975 cell line (containing T790M mutation of EGFR exon 20) in wild-type DNA were screened with conventional and COLD-PCR TaqMan genotyping for T790M mutation. Upper panel: conventional PCR TaqMan genotyping for T790M mutation; lower panel: COLD-PCR TaqMan genotyping for T790M mutation.
In summary, technical developments on many fronts in mutation detection are accumulating at a rapid pace. As knowledge of the biological and clinical impact of low-level mutations in cancer is increasing, the need for further development of methods capable of enhancing low-level minority alleles will continue to grow.
Acknowledgments
Research Funding: NIH grants CA-115439 and CA-111994 and NIH training grant 5 T32 CA09078 (the training grant was to C.A. Milbury and J. Li).
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
Footnotes
Nonstandard abbreviations: ARMS, amplification refractory mutation system; ASPCR, allele-specific enzymatic amplification; ASA, allele-specific amplification; PASA, PCR amplification of specific alleles; PAMSA, PCR amplification of multiple specific alleles; COP, competitive oligonucleotide priming; E-PCR, enriched PCR; ME-PCR, mutant-enriched PCR; MAMA, mismatch amplification mutation assay; MASA, mutant allele–specific amplification; aQRT-PCR, antiprimer quenching-based real-time PCR; REMS-PCR, restriction endonuclease–mediated selective PCR; AIRS, artificial introduction of a restriction site; PNA, peptide nucleic acid; LNA, locked nucleic acid; WTB-PCR, wild-type blocking PCR; FLAG, fluorescent amplicon generation; RSM-PCR, restriction site mutation PCR; APRIL-ATM, amplification via primer ligation, at the mutation; PAP, pyrophosphate-activated polymerization; RMC, random mutation capture; CCM, chemical cleavage of mismatches; endo, endonuclease; TDG, thymine DNA glycosylase; CEL I, celery extract I; HRM, high-resolution melting; HET, heteroduplex analysis; SSCP, single-strand conformation polymorphism; DGGE, denaturing gradient gel electrophoresis; CDCE, constant denaturing capillary electrophoresis; dHPLC, denaturing HPLC; iFLP, inverse PCR-based amplified RFLP; COLD-PCR, coamplification at lower denaturation temperature PCR; Tc, critical denaturation temperature; dsDNA, double-stranded DNA; Tm, melting temperature.
Authors' Disclosures of Potential Conflicts of Interest: Upon manuscript submission, all authors completed the Disclosures of Potential Conflict of Interest form. Potential conflicts of interest:
Employment or Leadership: None declared.
Consultant or Advisory Role: None declared.
Stock Ownership: None declared.
Honoraria: None declared.
Expert Testimony: None declared.
References
- 1.Parsons BL, Heflich RH. Genotypic selection methods for the direct analysis of point mutations. Mutat Res. 1997;387:97–121. doi: 10.1016/s1383-5742(97)00026-4. [DOI] [PubMed] [Google Scholar]
- 2.Gocke CD, Benko FA, Kopreski MS, Evans DB. Enrichment methods for mutation detection. Ann N Y Acad Sci. 2000;9906:31–8. doi: 10.1111/j.1749-6632.2000.tb06587.x. [DOI] [PubMed] [Google Scholar]
- 3.Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, et al. Analysis of any point mutation in DNA: the amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17:2503–16. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu DY, Ugozzoli L, Pal BK, Wallace RB. Allele-specific enzymatic amplification of beta-globin genomic DNA for diagnosis of sickle-cell anemia. Proc Natl Acad Sci U S A. 1989;86:2757–60. doi: 10.1073/pnas.86.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okayama H, Curiel DT, Brantly ML, Holmes MD, Crystal RG. Rapid, nonradioactive detection of mutations in the human genome by allele-specific amplification. J Lab Clin Med. 1989;114:105–13. [PubMed] [Google Scholar]
- 6.Sommer SS, Cassady JD, Sobell JL, Bottema CDK. A novel method for detecting point mutations or polymorphisms and its application to population screening for carriers of phenylketonuria. Mayo Clin Proc. 1989;64:1361–72. doi: 10.1016/s0025-6196(12)65378-6. [DOI] [PubMed] [Google Scholar]
- 7.Dutton C, Sommer SS. Simultaneous detection of multiple single-base alleles at a polymorphic site. Biotechniques. 1991;11:700–2. [PubMed] [Google Scholar]
- 8.Gibbs RA, Nguyen PN, Caskey CT. Detection of single DNA-base differences by competitive oligonucleotide priming. Nucleic Acids Res. 1989;17:2437–48. doi: 10.1093/nar/17.7.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kahn SM, Jiang W, Culbertson TA, Weinstein IB, Williams GM, Tomita N, Ronai Z. Rapid and sensitive nonradioactive detection of mutant K-ras genes via enriched PCR amplification. Oncogene. 1991;6:1079–83. [PubMed] [Google Scholar]
- 10.Cha RS, Zarbl H, Keohavong P, Thilly WG. Mismatch amplification mutation assay (MAMA): application to the c-H-ras gene. PCR Meth Appl. 1992;2:14–20. doi: 10.1101/gr.2.1.14. [DOI] [PubMed] [Google Scholar]
- 11.Takeda S, Ichii S, Nakamura Y. Detection of K-ras mutation in sputum by mutant-allele-specific amplification (MASA) Hum Mutat. 1993;2:112–7. doi: 10.1002/humu.1380020209. [DOI] [PubMed] [Google Scholar]
- 12.Easterday WR, Van Ert MN, Zanecki S, Keim P. Specific detection of Bacillus anthracis using a TaqMan mismatch amplification mutation assay. Biotechniques. 2005;38:731–5. doi: 10.2144/05385ST03. [DOI] [PubMed] [Google Scholar]
- 13.Li J, Wang FF, Mamon H, Kulke MH, Harris L, Maher E, et al. Antiprimer quenching-based real-time PCR and its application to the analysis of clinical cancer samples. Clin Chem. 2006;52:624–33. doi: 10.1373/clinchem.2005.063321. [DOI] [PubMed] [Google Scholar]
- 14.Ward R, Hawkins N, O'Grady R, Sheehan C, O'Connor T, Impey H, et al. Restriction endonuclease-mediated selective polymerase chain reaction: a novel assay for the detection of K-ras mutations in clinical samples. Am J Pathol. 1998;153:373–9. doi: 10.1016/S0002-9440(10)65581-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haliassos A, Chomel JC, Grandjouan S, Kruh J, Kaplan JC, Kitzis A. Detection of minority point mutations by modified PCR technique: a new approach for a sensitive diagnosis of tumor-progression markers. Nucleic Acids Res. 1989;17:8093–9. doi: 10.1093/nar/17.20.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolff JN, Gemmell NJ. Combining allele-specific fluorescent probes and restriction assay in real-time PCR to achieve SNP scoring beyond allele ratios of 1:1000. Biotechniques. 2008;44:193–4, 6, 9. doi: 10.2144/000112719. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted poly-amide. Science (Wash DC) 1991;254:1497–500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 18.Dominguez PL, Kolodney MS. Wild-type blocking polymerase chain reaction for detection of single nucleotide minority mutations from clinical specimens. Oncogene. 2005;24:6830–4. doi: 10.1038/sj.onc.1208832. [DOI] [PubMed] [Google Scholar]
- 19.Oldenburg RP, Liu MS, Kolodney MS. Selective amplification of rare mutations using locked nucleic acid oligonucleotides that competitively inhibit primer binding to wild-type DNA. J Invest Dermatol. 2008;128:398–402. doi: 10.1038/sj.jid.5700920. [DOI] [PubMed] [Google Scholar]
- 20.Dabritz J, Hanfler J, Preston R, Stieler J, Oettle H. Detection of Ki-ras mutations in tissue and plasma samples of patients with pancreatic cancer using PNA-mediated PCR clamping and hybridisation probes. Br J Cancer. 2005;92:405–12. doi: 10.1038/sj.bjc.6602319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amicarelli G, Shehi E, Makrigiorgos GM, Adlerstein D. FLAG assay as a novel method for real-time signal generation during PCR: application to detection and genotyping of KRAS codon 12 mutations. Nucleic Acids Res. 2007;35:e131. doi: 10.1093/nar/gkm809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parry JM, Shamsher M, Skibinski DOF. Restriction site mutation analysis, a proposed methodology for the detection and study of DNA base changes following mutagen exposure. Mutagenesis. 1990;5:209–12. doi: 10.1093/mutage/5.3.209. [DOI] [PubMed] [Google Scholar]
- 23.Jenkins GJS, Hashemzadeh Chaleshtori M, Song H, Parry JM. Mutation analysis using the restriction site mutation (RSM) assay. Mutat Res. 1998;405:209–20. doi: 10.1016/s0027-5107(98)00138-9. [DOI] [PubMed] [Google Scholar]
- 24.Kaur M, Zhang Y, Liu W-H, Tetradis S, Price BD, Makrigiorgos GM. Ligation of a primer at a mutation: a method to detect low level mutations in DNA. Mutagenesis. 2002;17:365–74. doi: 10.1093/mutage/17.5.365. [DOI] [PubMed] [Google Scholar]
- 25.Liu Q, Sommer SS. Pyrophosphorolysis-activated polymerization (PAP): application to allele-specific amplification. Biotechniques. 2000;29:1072–83. doi: 10.2144/00295rr03. [DOI] [PubMed] [Google Scholar]
- 26.Shi J, Liu Q, Sommer SS. Detection of ultrarare somatic mutation in the human TP53 gene by bidirectional pyrophosphorolysis-activated polymerization allele-specific amplification. Hum Mutat. 2007;28:131–6. doi: 10.1002/humu.20423. [DOI] [PubMed] [Google Scholar]
- 27.Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A. 1999;96:9236–41. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bielas JH, Loeb LA. Quantification of random genomic mutations. Nat Methods. 2005;2:285–90. doi: 10.1038/nmeth751. [DOI] [PubMed] [Google Scholar]
- 29.Lichten MJ, Fox MS. Detection of non-homology-containing heteroduplex molecules. Nucleic Acids Res. 1983;11:3959–71. doi: 10.1093/nar/11.12.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci U S A. 1989;86:2766–70. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cariello NF, Scott JK, Kat AG, Thilly WG, Keohavong P. Resolution of a missense mutant in human genomic DNA by denaturing gradient gel electrophoresis and direct sequencing using in vitro DNA amplification: HPRT Munich. Am J Hum Genet. 1988;42:726–34. [PMC free article] [PubMed] [Google Scholar]
- 32.Li-Sucholeiki XC, Thilly WG. A sensitive scanning technology for low frequency nuclear point mutations in human genomic DNA. Nucleic Acids Res. 2000;28:E44. doi: 10.1093/nar/28.9.e44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Underhill PA, Jin L, Zemans R, Oefner PJ, Cavalli-Sforza LL. A pre-Columbian Y chromosome-specific transition and its implications for human evolutionary history. Proc Natl Acad Sci U S A. 1996;93:196–200. doi: 10.1073/pnas.93.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emmerson P, Maynard J, Jones S, Butler R, Sampson JR, Cheadle JP. Characterizing mutations in samples with low-level mosaicism by collection and analysis of DHPLC fractionated heteroduplexes. Hum Mutat. 2003;21:112–5. doi: 10.1002/humu.10159. [DOI] [PubMed] [Google Scholar]
- 35.Wagner R, Debbie P, Radman M. Mutation detection using immobilized mismatch binding protein (MutS) Nucleic Acids Res. 1995;23:3944–8. doi: 10.1093/nar/23.19.3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Kaur M, Price BD, Tetradis S, Makrigiorgos GM. An amplification and ligation-based method to scan for unknown mutations in DNA. Hum Mutat. 2002;20:139–47. doi: 10.1002/humu.10106. [DOI] [PubMed] [Google Scholar]
- 37.Pincas H, Pingle MR, Huang J, Lao K, Paty PB, Friedman AM, Barany F. High sensitivity EndoV mutation scanning through real-time ligase proofreading. Nucleic Acids Res. 2004;32:e148. doi: 10.1093/nar/gnh150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Berbeco R, Distel RJ, Janne PA, Wang LL, Makrigiorgos GM. s-RT-MELT for rapid mutation scanning using enzymatic selection and real time DNA-melting: new potential for multiplex genetic analysis. Nucleic Acids Res. 2007;35:e84. doi: 10.1093/nar/gkm403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu WH, Kaur M, Wang G, Zhu P, Zhang YZ, Makrigiorgos GM. Inverse PCR-based RFLP scanning identifies low-level mutation signatures in colon cells and tumors. Cancer Res. 2004;64:2544–51. doi: 10.1158/0008-5472.can-03-3652. [DOI] [PubMed] [Google Scholar]
- 40.Li J, Wang L, Mamon H, Kulke MH, Berbeco R, Makrigiorgos GM. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat Med. 2008;14:579–84. doi: 10.1038/nm1708. [DOI] [PubMed] [Google Scholar]
- 41.Thomas RK, Nickerson E, Simons JF, Janne PA, Tengs T, Yuza Y, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med. 2006;12:852–5. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]