Abstract
In this article a comprehensive set of registration methods is utilized to provide image-to-physical space registration for image-guided neurosurgery in a clinical study. Central to all methods is the use of textured point clouds as provided by laser range scanning technology. The objective is to perform a systematic comparison of registration methods that include both extracranial (skin marker point-based registration (PBR), and face-based surface registration) and intracranial methods (feature PBR, cortical vessel-contour registration, a combined geometry∕intensity surface registration method, and a constrained form of that method to improve robustness). The platform facilitates the selection of discrete soft-tissue landmarks that appear on the patient’s intraoperative cortical surface and the preoperative gadolinium-enhanced magnetic resonance (MR) image volume, i.e., true corresponding novel targets. In an 11 patient study, data were taken to allow statistical comparison among registration methods within the context of registration error. The results indicate that intraoperative face-based surface registration is statistically equivalent to traditional skin marker registration. The four intracranial registration methods were investigated and the results demonstrated a target registration error of 1.6±0.5 mm, 1.7±0.5 mm, 3.9±3.4 mm, and 2.0±0.9 mm, for feature PBR, cortical vessel-contour registration, unconstrained geometric∕intensity registration, and constrained geometric∕intensity registration, respectively. When analyzing the results on a per case basis, the constrained geometric∕intensity registration performed best, followed by feature PBR, and finally cortical vessel-contour registration. Interestingly, the best target registration errors are similar to targeting errors reported using bone-implanted markers within the context of rigid targets. The experience in this study as with others is that brain shift can compromise extracranial registration methods from the earliest stages. Based on the results reported here, organ-based approaches to registration would improve this, especially for shallow lesions.
Keywords: registration, target registration error, skin fiducial, face registration, cortical surface, brain deformation, image-guidance
INTRODUCTION
Image-guided neurosurgery (IGNS) systems facilitate the ability to probe visible features of the patient’s physical neuroanatomy while simultaneously establishing the spatial correspondence within the wealth of imaging data obtained preoperatively. The process requires three-dimensional (3D) digitization of landmarks on the patient’s head, usually accomplished by an optically tracked 3D stylus and the designation of corresponding features within the preoperative neuroanatomical image volume (usually magnetic resonance (MR) images). The most common approach is adhering MR-visible synthetic skin markers to the patient’s head prior to imaging that are left in place until after patient fixation within the operating room (OR). The method of registration in this case is a point-based registration (PBR), which minimizes the distance between corresponding surface landmarks in physical and image space through a Procrustes’ least-squares fitting technique.1, 2 As an alternative to skin markers, bone-implanted markers with PBR are also used. This method is currently the “gold standard” for image-to-physical rigid registration.3, 4, 5 A more unusual alternative is the use of cortical surface landmarks whereby vessel bifurcations identified on the patient’s brain and within MR images are used to determine correspondence.6 While PBR is standard in most of today’s ORs, other techniques have also been pursued that use surface contour data to achieve a MR-surface to patient-surface registration.7, 8, 9 These techniques are not as fast but have the advantage of not requiring strict correspondence.
While the field of registration is quite mature, very few studies have attempted to quantify image-to-patient registration accuracy by using “true targets” within an OR setting, i.e., identifiable novel corresponding points on the brain in image space and its intraoperative counterpart in physical space. The most considerable contribution toward this was recently performed by Mascott et al.3, 4 In this work, the authors were able to test several standard registration techniques used most commonly in today’s ORs (surface-to-surface with digitized contours and PBR with synthetic skin fiducials) versus the gold standard of bone-implanted markers. These investigators used cranial holes and implanted 1 mm titanium hemoclips that could be detected on postoperative CT image sets for true targets. The overall findings were that the bone-implanted markers were statistically the best and that little difference existed among the other registration methods. Implanted bone markers results in target accuracy of 1.7±0.7 mm, and other skin-based registration methods amounted to a target accuracy of approximately 4.0±1.9 mm. Despite these results, IGNS has not yet gravitated toward bone-implanted markers as a standard of care. The often-cited reason is the degree of invasiveness that bone markers require, i.e., an additional surgery prior to imaging to implant the markers, which interestingly, is similar to use of standard stereotactic frames. While this is an important consideration, it should also be noted that the value of bone marker registration accuracy degrades significantly in the presence of brain deformations. While brain shift during the course of surgery is well documented10, 11 and ranges from 1–2 cm on average, only recently has it been appreciated that deformation can be present at the earliest instances, e.g., opening of the dura.12, 13, 14 Given the scale of these deformations, IGNS becomes more qualitative than quantitative with respect to guidance. The results presented here represent a first step forward in correction that could be implemented in today’s ORs.
In our previous studies, the clinical deployment of a laser range scanner (LRS) capable of generating textured surfaces (colored Cartesian point clouds) of the surgical field of view has been demonstrated.15, 16, 17 These studies provided a comprehensive experience using LRS technology within the operating room. In Ref. 15, an initial realization of an organ-based registration method was reported whereby textured LRS data of the cortical surface were directly registered to its grayscale-encoded counterpart from the MR images. In recognition of the immediacy of brain shift, this strategy may help to correct some error by aligning the cortical surfaces between physical and image space directly. In Ref. 17, this strategy was deployed in an eight-patient preliminary study to understand whether cortical surface registration could be routinely used, and to perform some qualitative comparisons of three different forms of cortical surface registration (point-based, feature-based, and our novel geometric∕intensity registration method). In addition to rigid registration studies, a preliminary strategy to track intraoperative brain deformations using serial LRS data and nonrigid image registration was also explored in Ref. 16. This study also reported the accuracy of tracking the LRS scanner at approximately 1.0±0.5 mm, which is important for the assessment of tracked LRS as a digitization technology. The report concluded by pointing toward the application of this measurement technology for use in a volumetric brain shift correction strategy (first reported in Ref. 18).
Similar to Mascott et al., this article studies the performance of extracranial registration methods within the context of true targets (not possible in Ref. 17). The registration methods investigated are traditional PBR with synthetic landmarks, and an approach whereby a dense LRS point cloud of the frontal, orbital, and upper nasal region of the face is acquired and registered to the MR counterpart using a surface registration technique (a technique also investigated in Ref. 7, although not within the context of true targets). One difference with Mascott et al. is that the true targets for this study are not on the rigid anatomy, but rather are soft-tissue targets. More specifically, vessel bifurcations and gyri features are identified within MR gadolinium-enhanced image volumes and on the physical patient in the OR as rendered by tracked LRS technology. One of the advantages of using LRS data is that high-resolution textured point clouds are obtained, allowing for detailed study of the cortical surface, thus facilitating corresponding point determination on the MR image volume. The work here confirms the findings of Mascott et al., but also extends them to include cortical surface registration results. Important findings are made when comparing traditional PBR and the face-based registration technique. In addition, the extension toward intracranial organ-based registration demonstrates an impressive correction of registration error due to extracranial methods. Unfortunately, not every method of registration could be achieved with every patient (which in a realistic OR setting is often the case). However, a sufficient sampling of each comparison was possible to suggest important findings regarding our registration platform.
METHODS
Laser range scanning
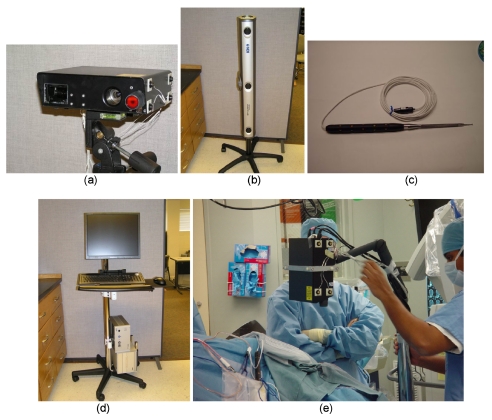
The commercially available LRS system (RealScan3D USB, 3D Digital Corp, Bethel, CT) shown in Fig. 1a is used in this article for intraoperative data acquisition. Briefly stated, each range data acquisition reports five-dimensional (5D) data. The data consist of geometry as measured by a triangulated laserline sweep acquired by CCD camera, and the intensity pattern of the FOV as acquired by a second CCD camera. The first three dimensions are the (x,y,z) Cartesian coordinates of the locations of surface points in LRS space, and the remaining two dimensions (u,v) are the texture-map coordinates dedicated for mapping intensity information of the digital image to the range data. The correspondence between FOV color information and geometric point cloud is established using standard computer graphic techniques. The LRS device is capable of generating point clouds with a resolution of 0.175 mm at a distance of 30 cm and 0.375 mm at the distance of 65 cm. The scanning field consists of 500 horizontal by 1000 vertical lines per scan (this can be adjusted to facilitate faster scanning time). Currently, even the highest resolution scans take less than 10 s. A Cannon Optix 400 color camera is used for capturing texture information with a maximum resolution of 5 megapixels. These specifications generate textured point clouds whereby the digital image of the FOV, i.e., texture, is an order of magnitude more resolved than the spatial resolution of the point cloud (standard interpolation techniques are used to generate Cartesian coordinates for unique texture points). It should also be noted that the point-cloud accuracy and interpoint resolution degrade as the scanner is positioned farther away from the surface of interest or is positioned at increasingly oblique angles to the surface of interest. In this work, the LRS system was routinely positioned normal-to, and between 25 to 40 cm from the face and cortical surface. More detailed descriptions of the type of data produced by the LRS device and its scanning characteristics can be found in Refs. 19, 16, 17, 20. The remaining components of the intraoperative system include the optical tracking system, the digitization stylus, and the surgical platform, as shown in Figs. 1b, 1c, 1d, respectively.
Figure 1.
Components of LRS tracking system. (a) LRS with 12 IRED distribution on the surfaces, (b) optical tracking cameras, (c) pen probe with 20 IRED around the surface, (d) customized cart holding the host computer and control box of the Optotrak Certus system, and (e) arm stand and data collection in the OR.
In order to obtain acceptable data among varied patient head orientations, two types of stands were used to mount the LRS scanner. For the intraoperative clinical face scan, the scanner is mounted on a vibration-damped monopod. For the clinical cortical surface scan, the scanner is mounted on a mobile arm [Fig. 1e] that can be rolled into and out of the surgical FOV easily while allowing for greater extension. Although this setup is somewhat cumbersome at this stage, this scanner was not designed specifically for OR use and all indications are such that approaches with better utility should be possible.
Data acquisition and patient information
The preoperative tomogram used for navigation in all 11 patients was the MR image volume acquired preoperatively taken one day prior to surgery. (Due to patient movement in the MR unit, one patient had the diagnostic scan used for guidance, which was taken two weeks prior to surgery). The MR images were acquired as 1.5 T, T1-weighted, 3D-SPGR, 1×1×1.2 mm voxel, gadolinium-enhanced and nonenhanced image volumes. Approximately ten synthetic skin markers were attached to each patient’s head prior to the MR scan. In this study, the surgical platform shown in Fig. 1e was utilized in parallel with the Stealth Station (Medtronic, Minneapolis, MN) surgical guidance system; therefore, other than some additional time for data acquisition, surgical therapy was not compromised for this study.
The enrolled patients in this study (five men, six women; mean age of 54.7 years) were afflicted with either primary or metastatic brain tumors (see Table 1). All patients were enrolled after obtaining written informed consent, which was approved by the Institutional Review Board of the Vanderbilt University School of Medicine. After anesthetic induction, Mayfield three-pin fixation (Ohio Medical, Cincinnati, OH) was used for each patient with varying head orientations.
Table 1.
Patient Information. Tumor Types: Gr—Grade, Olig.—Oligodendroglioma, Mening.—Meningioma, Astro.—Astrocytoma, GBM—Glioblastoma Multiforme, Met.—Metastatic Tumor. Location: L: left, R: right, F: frontal, T: temporal, P: parietal. Standard of care also involved a gram dose per kg of patient weight of mannitol for all patients, and general anesthesia.
| Patient No. | Age (yr)∕Sex | Tumor type | Craniotomy (diameter, cm) | Location | Lesion size (cm) |
|---|---|---|---|---|---|
| 1 | 22∕Female | Gr(II) Olig. | 7.7 | L, F | 5.2×6.2×6.0 |
| 2 | 52∕Male | Astro. | 8.3 | L, F | 4.9×5.6×5.0 |
| 3 | 58∕Male | Met. | 4.7 | L, P | 3.7×3.5×4.1 |
| 4 | 60∕Female | Mening. | 5.5 | R, F∕T | 4.5×6.4×4.3 |
| 5 | 77∕Male | Gr(IV) GBM | 5.0 | L, T | 3.4×3.6×2.0 |
| 6 | 57∕Female | Gr(II) Astro. | 3.5 | L, F | 1.0×1.4×2.0 |
| 7 | 56∕Female | Met. | 4.5 | L, F | 4.7×3.2×4.0 |
| 8 | 75∕Female | Gr(II) GBM | 6.1 | L, T | 5.0×5.0×5.0 |
| 9 | 23∕Female | Gr(II) Astro. | 6.4 | R, F | 4.0×3.0×3.0 |
| 10 | 46∕Female | Gr(IV) GBM | 4.3 | R, T | 3.0×3.0×3.0 |
| 11 | 26∕Male | Infrastrating | 9.0 | L, T | 6.9×4.0×4.0 |
Upon fixation, reference emitters for the Steath and experimental platform were attached and skin-fiducials were digitized using the Stealth’s stylus, followed by the optical stylus from the experimental registration platform. Once skin-marker localization was complete, the tracked LRS unit was brought into the field and a LRS textured point cloud of the patient’s frontal∕orbital∕nasal facial regions was taken. It should be noted that even though all patients participated in the comparison between face-based registration and traditional skin-marker PBR, the fidelity of LRS face scans (three incomplete face scans) and number of skin markers (five skin fiducials for one patient) was not equivalent. In all cases, if the Stealth’s reference emitter (and fixation attachment) could have been removed, an acceptable face scan could have been achieved. Once draped, the standard of care procedures for opening the scalp, craniotomy, and dura opening were performed uninterrupted. All patients received diuretics and steroids prior to incision. Once dural opening was completed, the LRS unit was brought into the field of view [similar to Fig. 1e] and a cortical surface textured point cloud was acquired. In addition, a second sterilized optically tracked stylus was used to acquire cortical surface features (e.g., blood vessel bifurcations and contours, gyri features, and features on the bone). Finally, a second LRS scan was taken subsequent to the completion of tumor resection. Each of these interrogations of the cortical surface accrued less than 5 min of additional surgical time. The majority of this time was related to preparing for acquisition rather than the data acquisition procedure itself. In each case, the immediate outcome of the patient was neurologic stability or improvement with no added deficits, and no detrimental effects from participation in the study were noted.
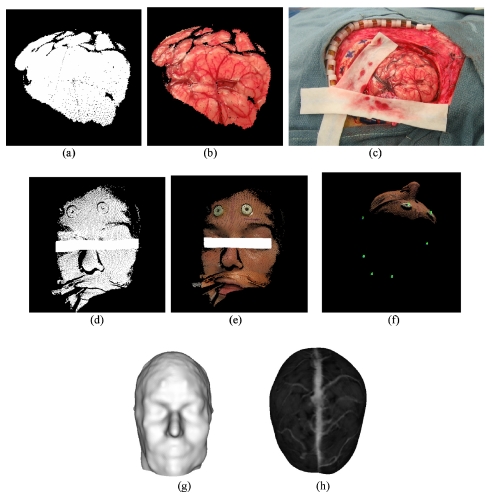
Figure 2 illustrates the stream of data available for intraoperative registration and cortical surface characterization. Figures 2a, 2b, 2c, 2d, 2e, 2f illustrate the textured point-cloud data of the cortical surface [Figs. 2a, 2b, 2c], the textured point cloud of the patient’s face [Figs. 2d, 2e] and the alignment of optically tracked points in reference to tracked textured point clouds in [Fig. 2f]. Figures 2g, 2h illustrate the available data from the preoperative MR that includes a rendering of the patient’s face [Fig. 2g] and brain surface [Fig. 2h].
Figure 2.
Data acquired by LRS intraoperatively (a)–(f) and preMRI (g),(h). (a) 3D point cloud of cortical surface, (b) textured 3D point cloud of the cortical surface, (c) high-resolution 2D image, (d) 3D point cloud of face scan, (e) textured 3D point cloud of face scan, (f) tracked skin markers in patient space, (g) head surface, (h) texture brain surface. The obstructing object in (d), and (e) is to protect patient confidentiality.
One aspect to this type of work (that often goes unappreciated) is the difficulty in acquiring OR data when subjected to this intense, variable, and dynamic environment. As a result, we should indicate some notes regarding nuances within the data that occurred due to environmental, patient, and physician needs. For patient 9, significant care was taken to maintain hair and scalp surgically. As a result, only five fiducials were digitized using our tracked stylus and hence only five fiducials were available for skin-marker registration. This registration was found to be an outlier within PBR results. With respect to face-based LRS data, we were able to obtain eight high fidelity scans. A high fidelity face scan was characterized as a point cloud containing spatial information for the forehead, two orbital areas, two upper infraorbital regions, and the nasal region. Consistency among all 11 patients was compromised due to the presence of two surgical systems, with the Stealth system taking priority over our needs (e.g., reference emitter attachment could obscure face). With respect to patients 3 and 6, high fidelity LRS face scans were not achieved, but partial scans were acquired and used within the results. Patient 7 was in a prone position with a 30° head rotation. This positioning resulted in a very limited LRS face scan. As a result, the remainder of the head was also scanned (shaved scalp was present) and used within the surface registration results. Patient 7 had also incurred movement artifacts in the preoperative MR. As a result, brain features were difficult to identify. For the study of this case, an image-to-image registration was performed with a previously acquired diagnostic scan. This allowed good delineation of the facial and segmented brain surfaces for use in registration and also allowed for the target analysis comparison. With each case, an intramodality distance table among skin fiducial markers and among targets in each separate space was constructed and compared to ensure correspondence and assess the fidelity of the data. For patient 4, the patient needed to be moved from one OR to a second OR after the initial registration and face scans were acquired. Unfortunately, the patient’s skin fiducials were removed before the patient was repositioned in the new OR. As a result, we performed a second face scan and performed a face-to-face LRS surface registration to allow the patient’s skin fiducial marker locations recorded in the first OR reference space to be transformed into the second OR’s patient space. This transformation may have introduced some additional registration error.
Registration methods
Skin marker registration
The synthetic skin markers were localized in physical space by a pen probe that was tracked by the Optotrak Certus system [Figs. 1b, 1c]. The centroid of each skin marker in MR image space was localized using AnalyzeAVW (Mayo Clinic, Rochester, MN). The fiducial localization error has been estimated at approximately 1 mm. Upon completion of localization, the transformation that aligned the tracked marker positions in physical space with corresponding centroids in MR image space was calculated using traditional PBR techniques.2 Generally, not all the skin markers were tracked and localized in the OR since some of the markers were obscured during fixation or had fallen off the patient. Our platform does a combinatorial registration check to generate the proper correspondence among markers when the number of intraoperative and preoperative markers differ. As a confirmation of the accuracy, the fiducial registration error (FRE)21 was reported for each registration with varying numbers of fiducial corresponding points, where FRE is the root-mean-square (RMS) distance between the centroids of the corresponding markers.
LRS face-based registration
The face-based registration approach begins with segmentation of the surface of interest under the guidance of texture information encoded in the 5D point clouds. The facial structures of interest were segmented and processed with a radial basis function (RBF) surface fitting (FarField Technology, Christchurch, New Zealand) to improve the surface quality. The average fitting error associated with these surfaces was 0.2 mm. A second surface using the same RBF fitting techniques was generated from the MR image volume and encompassed the entire segmented head.
After generating the corresponding preoperative and intraoperative datasets, the face-based registration process starts from an initial PBR based on aligning natural landmarks using natural fiducials (e.g., center of orbits, tip of nose, etc.). It should be noted that fiducials are obtained from the LRS and MR point clouds so there is no need for the optically tracked stylus to be used in registration initialization. Following this process, the iterative closest point (ICP) registration algorithm as described by Besl and McKay22 was used to further align the LRS face surface to the MRI head surface counterpart. Although some difficulties in acquiring the face scan were caused by the setup realized in this work, the method is relatively easy to implement and is amenable to the OR environment.
Intraoperative LRS cortical surface registration
Organ deformation is a significant problem for the translation of image-guided surgery techniques. While shift compensation strategies are on the horizon, the ability to maximize the utility of existing surgical platforms is an important concern. In this and previous work, a central hypothesis has been that direct organ-based registration would serve as an improvement over registrations based on the rigid exterior. Based on the recent literature, there is significant quantitative data that suggest the brain surface is often deformed upon opening of the dura. The potential for edema-induced swelling or brain contraction due to diuretics at the earliest stages of surgery would compromise IGNS, even at its initiation. While the potential for predicting these effects and accounting for them has been suggested,18, 23 a cortical surface registration methodology may mitigate some of these effects in the interim, as well as assist in early shift measurements in the future. With respect to the cortical surface registration methods described herein, standard point-based and surface based rigid-body registration methodologies are being explored, as well as a novel intensity based surface method called SurfaceMI15 (described next). In addition, the SurfaceMI method has been extended in this article to incorporate 3D geometric information that has high correspondence confidence as a constraint in the process of the maximization of mutual information.
Registration of the intraoperative cortical LRS data with preoperative MR image data requires the generation of textured brain surfaces from preoperative MR tomograms. It begins with the segmentation of the brain region from the MR image volume using an atlas based segmentation method.24, 25, 26 The average segmentation error associated with this technique is 1 voxel. From this segmented volume, a point-cloud representation of the brain surface geometry is extracted by a marching cubes approach.27 The generated point cloud of the brain surface is fitted by using the RBF surface fitting method for the purpose of smoothing and matching geometry resolution to that of texture resolution. A surface-normal ray-casting algorithm is used to grayscale encode the surface (five voxel intensities were averaged along the ray). At the end of this process, a corresponding MR textured point cloud is generated that contains surface intensity information such as gyri patterns and gadolinium-enhanced vessel patterns.19 These feature patterns are crucial for providing intensity and geometric features for the SurfaceMI algorithm. The generation of color encoded intraoperative LRS cortical surface data begins by segmenting the cortical surface from the LRS 3D point cloud guided by the texture information. The RBF surface fitting is performed and color coding of the surface is reapplied.
After the textured preoperative MR and intraoperative LRS brain surfaces are obtained, a series of rigid-body cortical registration algorithms are used: (1) cortical feature PBR (feature PBR), (2) cortical vessel-contour registration (vessel ICP), (3) SurfaceMI, and (4) constrained SurfaceMI. The latter three registration methods were initialized by face-based registration when available and by PBR of skin-markers when not available. With respect to PBR, except for the localization being performed on the patient’s LRS via a graphical user interface (as opposed to manually demarked with an optical stylus in the OR), it is similar to the method described by Nakajima et al. for organ-based registration.6 This method often produces adequate alignment of the surfaces, but having a sufficient number of cortical landmarks to use for registration is not always assured. In addition, registering the cortical surface using a sparse number of points could be potentially confounded when deformation is present. The second registration method is based on identifying homologous vessel structural patterns on the cortical surface and is then used as the basis for a 3D contour-to-contour registration using the ICP algorithm.22 The third registration method was initially reported in Ref. 15, and refined in Ref. 19 and represents a combined geometric and intensity based registration. This method begins with an initialization performed usually with an initial PBR followed by a second surface-to-surface closest point alignment. Upon completion, a homotopic transformation is applied such that source and target surfaces are on a common spherical surface. The intensity is also mapped to the spherical surface. A constrained registration (three spherical angles) is performed whereby the objective function maximizes the normalized mutual information (NMI) between the LRS and MR textures. The NMI is calculated as reported by Studholmeet al.28 and is written as
| (1) |
with H(I1), H(I2), H(I1,I2) being the two marginal entropies, and joint entropy between the LRS and MR texture on the spherical surface. The registration is multiresolution and multiscale as described in Ref. 19 and is termed SurfaceMI. With respect to the final registration method, its development was a direct result of the study performed by Sinha et al. in Ref. 17. Here it was noted that while SurfaceMI did at times perform well, it could become confounded if intensity configurations were ambiguous. To overcome this, features of confidence have been incorporated to constrain the SurfaceMI registration. More specifically, a simplified version of the approach reported by Hartkens et al.29 has been implemented whereby the objective function is
| (2) |
The second term in expression 2 is
| (3) |
where λ is an empirically determined scaling parameter (0.001 was the value used in the study), and D(T) represents the mean distance between homologous points, p on the LRS and MR 3D textured surfaces after applying the traditional rigid body transformation T. The optimization method employed in this geometry constrained SurfaceMI framework was Powell’s iterative method.30
Target points and analysis methods
Target points
In addition to visual assessment, one aspect that sets this study apart from others is the existence of true soft-tissue targets between image and physical space with respect to the clinical patient. More specifically, the presence of distinct vessel bifurcations and gyri feature points identifiable on both the preoperative MR-encoded grayscale textured surface and the LRS counterpart in physical space provides a measure of true target registration error (TRE). TRE here refers to the RMS distance between the feature points found on the tracked LRS surface and the corresponding segmented textured MR brain surface21
| (4) |
where is the transformed position of targets identified on MR brain surface to LRS physical space, is the position of targets identified on the LRS cortical surface in physical space, and N is the total number of targets. There is little doubt that compounding sources of error such as target localization error, segmentation, surface fitting, tracking, and deformation influence this measure. However, these sources of error influence each of the registrations in a similar way. We hypothesize that information regarding the variability of registration performance may be discernable, despite these collective errors. In this study, for each patient case, several corresponding target points on the LRS cortical surface and the preoperative MR brain surface were picked with the guidance of our neurosurgeon. These targets were spatially distributed and widely spread in the surgical region. The target localization error present during selection was estimated in Ref. 16 at 1.0±0.3 mm. It should also be noted that only in the comparison between skin-fiducial and face-based registration can these unique points be considered true targets. Given that the locations of the targets are on the cortical surface, conclusions regarding the cortical surface registration methods, specifically vessel ICP and surfaceMI, must be tempered.
Registration comparison analysis
With regard to the registration process and the focus of this work, the comparison analysis was divided into two studies, specifically, registration performance analysis (1) prior to craniotomy and (2) postcraniotomy. By separating the work in this way, this article demonstrates how standard IGNS, as practiced today, is a somewhat qualitative experience using standard registration methods (extracranial methods). The extension toward organ-based registration methods demonstrates the potential correction of this experience. However, it should be noted that this trend may not translate to deep subsurface targets and further investigation is required.
With the extracranial registration methods, the study focused on a performance evaluation between two registration methods that could be implemented within standard surgical guidance systems: (1) a traditional skin fiducial marker PBR, and (2) a surface registration method that used the patient’s face. While a gold standard registration is unavailable due to patient-care issues, the true targets in both image and physical space allow for quantitative comparisons to be made as to whether these registration methods would be statistically different. The second series of comparisons was performed among cortical surface registration methods. Similarly, the availability of paired multiple target observations allows for a substantive comparison among the methods.
In order to establish confidence in registration comparison, statistical methods were employed. The first method we used is the paired t-test31 to compare face-ICP and skin fiducial PBR. As alluded to earlier, only five synthetic skin fiducials could be recorded for patient 9. In all other cases seven to ten fiducials were recorded. Given this sparse acquisition, the Sprent32 outlier test was used to determine whether this patient should be included in the t-test (see Appendix0 for explanation). In addition, the paired t-test method is based on the assumption that the two paired datasets satisfy Gaussian distributions. A Chi-Square test was used to determine this relationship.
Similar comparisons can be made among the battery of cortical surface registration methods to assess which are statistically similar or different as the case may be. Before conducting these comparisons, it is still necessary to test whether the observed TRE for the four registration methods, i.e., feature PBR, vessel ICP, SurfaceMI, and constrained SurfaceMI, is Gaussian. The same Chi-Square test was used to evaluate this. In case any one of the four datasets violates the assumed Gaussian distribution, the Friedman test would be used to compare the differences among the four registration methods (see Appendix0 for explanation). If there is any significant difference between these four registration methods, Dunn’s test will be performed as a posthoc test to decide which method significantly differs from others.32
RESULTS
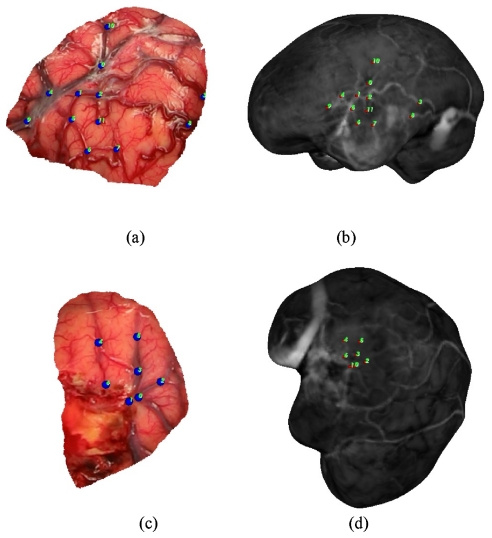
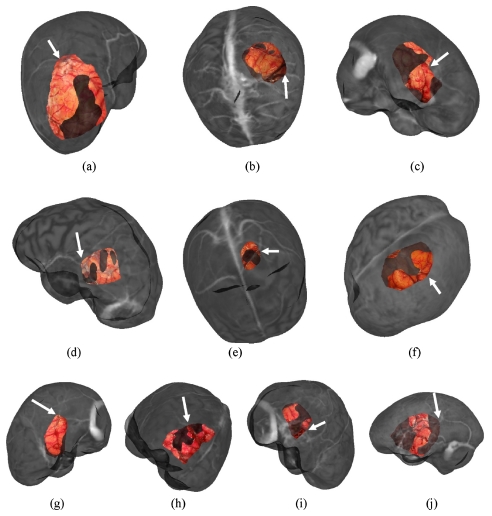
For each patient, six registration methods were utilized and six to 12 target points were identified. These points were primarily located on cortical veins, sulci, and gyri and were identified on intraoperative LRS cortical surface scans and MRI textured brain surfaces. Two representative examples of targets on the LRS cortical surface and MR counterpart are shown in Fig. 3. These target points are truly novel for the extracranial registration methods. In the case of feature PBR, the registration error is a target registration error but was generated using a leave-one out approach whereby one feature is left out of the PBR and used as a target error. The approach cycles through each target being left out, and a RMS TRE can be established. In summary, what are referred to as true targets have high assessment fidelity for extracranial registration methods, a moderate fidelity for vessel ICP∕SurfaceMI methods, and a lower fidelity for feature PBR∕constrained SurfaceMI.
Figure 3.
Target points identified on LRS cortical surface (a),(c) and textured MRI brain surface (b),(d) for Patient 11 and Patient 10, respectively.
Skin fiducial marker and face-based registration
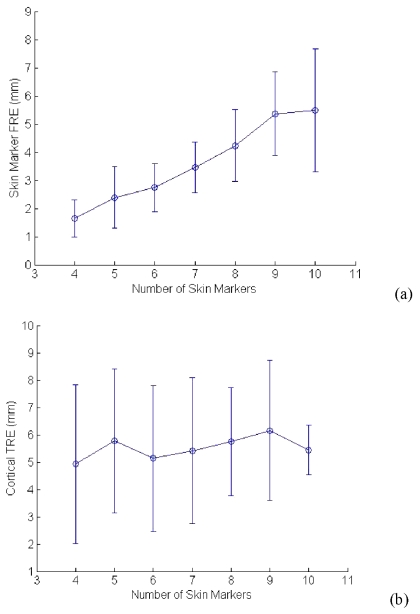
The total number of skin fiducials recorded for each patient case is presented in Table 2. The same skin markers were also localized in MR image space by using Analyze AVW. According to the point-based rigid registration theory, utilization of all fiducials would be the natural choice when performing image-to-physical registration. The third column of Table 2 quantifies the FRE for each case using this method. Figure 4 illustrates the change in average FRE, and TRE with varying numbers of skin fiducials. The soft-tissue RMS TRE for skin fiducial PBR and face-ICP is presented in Table 3 for each case.
Table 2.
Skin marker digitization and registration results using PBR.
| Patient No. | Number of skin markers localized in OR | RMS FRE (mm) |
|---|---|---|
| 1 | 7 | 4.5 |
| 2 | 8 | 4.9 |
| 3 | 9 | 6.1 |
| 4 | 7 | 4.5 |
| 5 | 10 | 7.5 |
| 6 | 8 | 2.5 |
| 7 | 8 | 6.0 |
| 8 | 9 | 7.6 |
| 9 | 5 | 5.5 |
| 10 | 10 | 4.3 |
| 11 | 10 | 5.4 |
| Average | 8 | 5.3±1.5 |
Figure 4.
The relationship between RMS FRE, RMS TRE, and the number of skin fiducial markers. These results show the relationship between standard registration errors and the number of fiducials used for PBR.
Table 3.
Initial registration accuracy for each patient, RMS-TRE in mm.
| Patient No. | Targets | Skin fiducial PBR TRE (mm) | Face-based ICP TRE (mm) |
|---|---|---|---|
| 1 | 6 | 3.7 | 3.5 |
| 2 | 7 | 6.7 | 5.9 |
| 3 | 6 | 3.9 | 3.2 |
| 4 | 7 | 3.7 | 4.8 |
| 5 | 7 | 6.1 | 4.2 |
| 6 | 7 | 2.6 | 3.1 |
| 7 | 9 | 6.9 | 6.8 |
| 8 | 6 | 9.7 | 8.4 |
| 9 | 9 | 6.7 | 8.2 |
| 10 | 7 | 4.9 | 6.6 |
| 11 | 12 | 3.9 | 3.1 |
| Average | 7.5 | 5.3±2.1 | 5.3±2.0 |
Table 4 presents the RMS FRE when the best five skin markers were used with PBR for the 11 patients. The RMS-FRE 5.5 of patient 9 underwent Sprent’s outlier test and a value of 6.55 was obtained, which is larger than the recommended threshold of 5.0. The result indicates that patient 9 is an outlier for the paired t-test of equivalence evaluation between skin marker PBR and face-ICP. Therefore, the nine targets of patient 9 were removed from the 83 paired target registration errors over these 11 patients, as presented in Table 3. For patient 7, even though the brain volume we used was segmented from the MRI image volume taken two weeks prior, the skin fiducials were still localized in the MRI image volume that was taken one day before the surgery. Through coregistration between these two MRI image volumes, the skin fiducials were transformed into the segmented brain image space. According to Sprent’s test on the FRE in Table 4, patient 7 is not an outlier, thus the target points for this patient case were kept in the dataset for comparison analysis. As a result, 74 paired TRE participated in the comparison. Before performing the comparison between these two registrations (Table 3), a test for the Gaussian distribution of the TRE was assessed by using the Chi-square test. The p-values for the Chi-Square distribution are 0.342 and 0.4034, respectively, which is higher than the significance level of p-value 0.10. Thus, an assumption of a normally distributed TRE for the two methods of registration is acceptable.
Table 4.
RMS FRE for skin fiducial PBR when five best skin fiducials are used in PBR.
| Patient No. | RMS FRE (mm) |
|---|---|
| 1 | 2.3 |
| 2 | 2.6 |
| 3 | 3.6 |
| 4 | 2.7 |
| 5 | 2.3 |
| 6 | 1.0 |
| 7 | 2.9 |
| 8 | 2.0 |
| 9 | 5.5 |
| 10 | 1.3 |
| 11 | 1.8 |
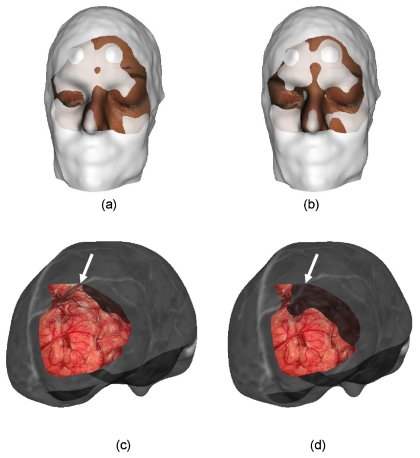
Under the assumption of a normally distributed TRE, the TRE observations underwent the paired t-test. The p-value for the two side test was 0.1167, which is higher than the significant value 0.10, indicating that the hypothesis that face-ICP is equivalent to skin fiducial PBR is accepted. One example visualization of a skin fiducial PBR and face-ICP result for patient 1 is illustrated in Fig. 5. The top pictures illustrate the result for overlaying the patient’s orbital facial region using each method, while the bottom pictures relay the corresponding cortical surface overlays. The visualization qualitatively demonstrates differences but reasonably good alignments for both.
Figure 5.
Visualization of the registration results for both skin fiducial PBR and face-ICP. (a) is the registration result of the face from LRS and MR using skin fiducial PBR. (b) is the registration result of the face from LRS and MR using face-based ICP. (c) uses the same registration transformation as (a) but shows the cortical surface and MR counterpart in the visualization, (d) uses the same registration transformation as (b) but shows the cortical surface and MR counterpart in the visualization. Arrow points to continuation of vessel area between MR and LRS surfaces.
Cortical surface registration results
In the previous subsection, results from extracranial registration methods were presented and compared statistically. The results suggested that a 5 to 6 mm average positional difference exists among cortical surface targets and their preoperative counterparts after opening the dura. Based on the work of others and our own observations, it is likely that a good portion of this positional difference is due to brain deformations. One of the underlying assertions put forth in our work is that it may be advantageous to align the brain using cortical surface information, i.e., the organ itself. Within our registration paradigm, face-ICP or skin fiducial PBR can serve as an initial guess followed by a refining cortical surface registration serving to improve accuracy of the IGNS system. Table 5 shows the RMS-TRE for the four cortical surface registration methods for the 11 patients. The target points are the same as those used in the previous subsection. Once again, it should be noted that labeling these points as targets is somewhat inappropriate given their location relative to the geometric points, contours, and surfaces being utilized for registration, but for consistency with the numbers reported in the previous section, the language was kept here. The numbers reported for TRE feature PBR were generated using a systematic leave-one-out approach whereby a series of PBRs were calculated with one feature left out to represent a target. All features were systematically treated as targets and the average is reported as the target registration error.
Table 5.
Cortical surface registration error results for each patient. The (✓) indicates the best performing cortical surface registration among the four methods based on the value of TRE.
| FRE (mm) | TRE (mm) | ||||
|---|---|---|---|---|---|
| Patient No. | Feature PBR | Feature PBR | Vessel ICP | SMI | Constrained SMI |
| 1 | 1.2 | 2.4 | 2.1 | 1.9 | 1.7✓ |
| 2 | 1.1 | 2.1 | 2.0 | 4.3 | 1.3✓ |
| 3 | 1.0 | 2.0 | 2.0 | 2.4 | 1.4✓ |
| 4 | 0.8 | 1.5 | 1.5 | 1.4 | 1.2✓ |
| 5 | 1.0 | 1.5 | 1.1✓ | 6.2 | 2.7 |
| 6 | 0.7 | 1.0✓ | 1.3 | 1.6 | 1.3 |
| 7 | 0.7 | 1.0 | 0.8✓ | 4.8 | 3.8 |
| 8 | 0.7 | 1.3✓ | 2.1 | 12.8 | 3.5 |
| 9 | 1.1 | 1.7✓ | 2.0 | 3.3 | 1.8 |
| 10 | 0.7 | 1.1 | 1.9 | 1.2 | 1.1✓ |
| 11 | 1.4 | 2.2 | 2.3 | 2.7 | 1.8✓ |
| Average | 0.9±0.2 | 1.6±0.5 | 1.7±0.5 | 3.9±3.4 | 2.0±0.9 |
When using the Chi-Square test method to test for a normal distribution of 83 TRE for each method, the TREs for SurfaceMI were found to be non-Gaussian. Thus, the nonparametric Friedman testing method for multiobservations was applied. As the four methods were ranked through all 83 targets, a corresponding p-value less than 0.001 was obtained that indicated that the four methods are not equivalent and at least one method is significantly different. Following the Friedman test, Dunn’s test was performed to compare the equivalence between any two within the four registration methods, and SurfaceMI was found significantly different from the other three methods, which is somewhat expected given the poor performance especially in patient 5 and patient 8. Dunn’s test also shows that vessel ICP is equivalent to constrained SurfaceMI, and feature PBR. The sequence for TRE average from the high accuracy to the low accuracy is feature PBR, vessel ICP, constrained SurfaceMI, and SurfaceMI. Interestingly, when observing case by case, constrained SurfaceMI performed best in 55% of the cases, with feature PBR performing best in 27%, followed by vessel ICP for the remaining 18%. The original SurfaceMI outperformed some individual registrations, but overall did not perform well when compared to the other methods. This is consistent with preliminary results in Ref. 17. SurfaceMI has the highest registration error for patient 8 and is likely due to the excessive swelling in that case (estimated to be approximately 8 to 9 mm) as well as a lack of texture feature. The addition of features with high confidence in correspondence did improve SurfaceMI to the degree whereby on a case-by-case basis, it performed the best. For completeness, the FRE feature PBR (where all features are used in the registration) was also reported. In some sense, this represents the best possible target error achievable. The results reported have improved over results reported by Refs. 6, 17. While not statistically significant, it should be noted that we have improved the tracking ability of our LRS system by compensating more extensively for lens distortion and by incorporating higher resolution texture, which allows for the identification of more features on the cortical surface. The last aspect to consider is that SurfaceMI and vessel ICP do present more distributed registration methods as opposed to the sparse numbers of points from feature PBR. It is unclear whether feature PBR results translate for the entire surface.
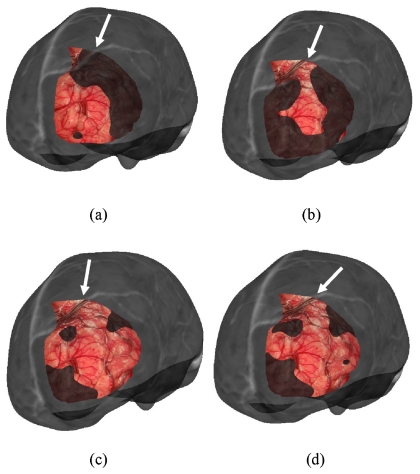
Visualization for the LRS cortical surface overlay on the MRI brain surface is provided in Figs. 67, respectively. For brevity, the results for the four cortical surface registration methods were visualized for patient 1 only, as shown in Fig. 6. Only one registration result for each of the four methods for the remaining cases is illustrated in Fig. 7.
Figure 6.
Visualization of the registration results for patient 1 using all four methods of cortical surface registration. Each image shows overlay between the cortical brain surface as acquired by LRS and the corresponding grayscale encoded MRI brain surface. The transformations are: (a) feature PBR, (b) vessel ICP, (c) SurfaceMl, and (d) constrained surfaceMI. The arrows point to the continuation of vessel area between MR and LRS surfaces.
Figure 7.
Visualization of the registration result for each patient using the same overlay as in Fig. 6. The transformations used for each patient are: (a) patient 2, vessel ICP, (b) patient 3, constrained SurfaceMI, (c) patient 4, constrained SurfaceMI, (d) patient 5, vessel ICP, (e) patient 6, feature PBR, (f) patient 7, feature PBR, (g) patient 8, feature PBR, (h) patient 9, feature PBR, (i) patient 10, SurfaceMI, and (j) patient 11, constrained SurfaceMI. The arrows point to the continuation of vessel area between MR and LRS surfaces. Patient 1 is shown in Fig. 6.
DISCUSSION
The registration platform being developed and the results obtained in this study demonstrate that the tracked laser range scanner does not impede the normal course of surgery. Compared with our previous LRS system, the current system has improved point-cloud resolution and higher resolution texture images. The latter improvement has proved to be very important when identifying feature points on the cortical surface. The current OR system is compact, and relatively easy to setup and move into the OR. However, the difficulty in having two intraoperative guidance systems within the OR does make conducting this research challenging. The newly developed surgical arm to hold the scanner has dramatically improved the quality of the data by allowing the positioning of the scanner normal to the surface. Only with this complete surgical registration platform could we compare extracranial and intracranial registration methods within the context of soft-tissue true TRE. While the evaluation of a gold standard rigid transformation cannot be achieved without bone-implanted markers, even if possible in this study, the reality of brain shift is that any extracranial registration methodology will become confounded. Despite this, the work presented here does represent the first registration study, demonstrating that skin fiducial PBR is equivalent to face-ICP within the context of true target error. It is also the first study to compare extracranial and intracranial registration methods in the presence of true image-to-physical targets. Furthermore, each method is subject to the same instrumentation, segmentation, and tracking errors, which improves the veracity of our conclusions. While more cases are needed to improve the statistical power, the results here do represent a population of a significant size. The most closely related study to the previous findings were that by Mascott et al.3 Their results were achieved under similar OR conditions, however, they were not encumbered with two guidance systems in the OR (which explains our smaller numbers to a degree). While these investigators did not perform LRS faced-based registration specifically or any cortical surface methods, the results are remarkably consistent with our clinical experience, especially when considering our targets were not on the bone but rather on the soft tissue.
Another important aspect of this work is the implication of the reported target error in light of extracranial registration. Figure 4a indicates that with sparse numbers of fiducial points, FRE increases with increasing numbers of fiducials, which correlates with known behavior of FRE (trend is asymptotically constrained as the number of fiducials increases).21, 33, 34, 35 However, Fig. 4b is somewhat more perplexing in that varying the number of fiducials did not seem to influence the TRE. There are several possibilities that may cause this: (1) possible instrumentation error, (2) cortical surface deformation, that would confound all extracranial registrations, and (3) correlated skin marker movement, given the systematic process associated with head placement. At this time, the degree to which each of these factors contributes to the final result is unclear. However, as others have also found, we observe visible shifts associated with swelling and sagging upon opening of the dura, and while qualitative in this article, brain shift is likely for patients afflicted with space occupying lesions (all cases herein). Similarly, the relative ease of skin marker motion when the skin is in retraction provides the impetus to suspect this also. A central tenet in rigid registration work is that fiducial localization error is random and Gaussian. However, with respect to skin fiducials, where error is likely due to skin marker motion (i.e., deformation of the scalp), it is likely that the assumption of Gaussian random noise needs to be challenged. Some initial results by Ref. 36 exist and demonstrate that a simple traction force on a skinlike membrane can increase FRE and TRE for traditional skin markers considerably, as opposed to the more stable bone anchored markers. It is likely that skin markers deform with the scalp and to varying degrees, which would lead to correlated and non-Gaussian error behavior. Understanding this within the context of detailed registration analysis needs to be pursued in future work.
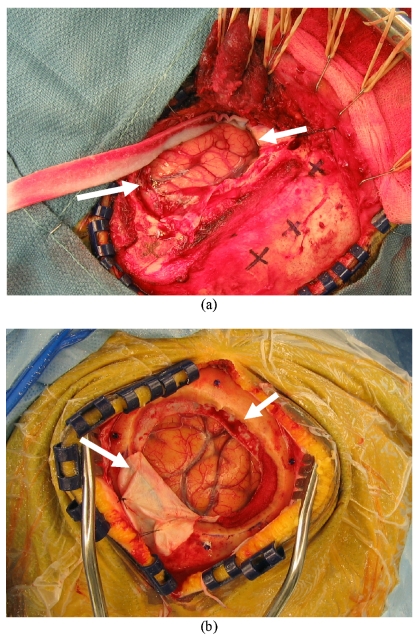
While somewhat more subtle, another important aspect to Table 3 is the qualitative nature of image guidance with respect to brain tumor surgery. The results suggest that soft-tissue deformation compensation strategies may be needed to move beyond qualitative guidance. In our experience, the quantitative use of image guidance is primarily relied on for initial positioning and initial brain entry. The procedural feedback provided during a resection is performed less frequently whereby the surgeon is predominantly interrogating the field using visual and haptic cues. Upon the completion of the resection, the surgeon tends to begin to use the guidance system as an assessment tool. At this stage, in the presence of no shift correction or intraoperative imaging, the assessment relies to a great extent on the experience of the surgeon. Table 3 demonstrates that error can be present even at the point where the most quantitative assessment is performed, and Table 5 indicates that cortical surface registration methods may be quite effective at guidance correction, especially for shallow lesions. Full volumetric compensation is still necessary for deep-seated lesions and strategies to achieve this are in progress.18, 23 As a means to qualitatively demonstrate intraoperative brain surface deformation, the high-resolution texture images of the cases with maximum swelling (patient 8) and sagging (patient 7) are illustrated in Fig. 8. The white arrows in Fig. 8a highlight the brain being nearly level with the extracranial surface, indicating a significant amount of swelling. In comparison, Fig. 8b shows the brain surface well below the extracranial surface and in fact below the intracranial surface. In addition, it is easy to observe the distinct difference in dura flap outcome as result of these very different presentations. The most important aspect with this analysis is that brain shift may often be immediate and that without correction, the reliance on image guidance must be tempered, especially in cases with space occupying lesions with unknown intracranial pressure∕edema tissue deforming effects. Again, it is unclear at this time the level of system error that contributes to these quantified values, but the behavior is consistent with the findings of other investigators.13
Figure 8.
Texture images of the intraoperative cortical surface for patient 8 (a) with swelling and patient 7 (b) with sagging of intraoperative brain surface deformation. These were acquired post dural opening and demonstrate the presence of shift already present at the initiation of surgery. Even the most accurate extracranial registration technique (bone-implanted markers) would still be confounded by shift.
Other observations made during this study demonstrated that the tip of the nose can deform under gravity and can affect registration. It was found that relying on the upper nasal region was required. Similarly, including the most inferior regions of the infraorbital region was also subject to deformation. Another interesting finding is that the RMS TRE averages reflected in Table 3 for both skin fiducial PBR and face-ICP were consistent. While these may seem somewhat large in certain cases, it must be noted that each of these cases involved a substantial tumor (except for patient 6). Furthermore, similar to the experience of others,13 the presence of brain deformation upon opening the dura is somewhat commonplace. These shifts have been relatively easy to witness in the OR and the data here reflect this behavior (e.g., Fig. 8). This is not to say that there are not other sources of possible error that affect these measurements (e.g., segmentation, localization, tracking, etc.) but rather to suggest in cases where large misregistrations occur, undoubtedly portions of this error are induced by intraoperative deformation. One last contribution with respect to the extracranial registration methods concerns the initialization method for cortical surface techniques. In previous work,15, 17 all cortical surface registrations were initialized with a brain feature PBR similar to Nakajima et al.6 In this work, initialization was achieved by face-based ICP, which completes the LRS-based registration platform. While the current setup cannot acquire the face for all surgical orientations due to the constrained design, given the compact nature of the scanner and with the proper integration into the OR design, LRS scanning units could potentially alleviate the need for skin fiducials.
Table 5 presents the extent that refinement using the cortical surface can achieve. Nakajima et al.6 did suggest this also, but they also indicated the difficulty in identifying landmarks. In this work, the tracked textured point cloud combined with the prealignment provided by face-ICP allows for easier feature recognition when compared to the textured MR counterpart. Each of the methods in Table 5 took full advantage of the high-resolution LRS system as well as the textured preMRI brain surface information, which facilitated the registration greatly. In addition, whereas in Nakajima’s work they relied primarily on vessels, we can use all information, including sulcal and gyri information (both in geometry and intensity).
Finally, there are several sources of error within this analysis. These include (1) the tracking of the stylus and LRS unit, (2) the segmentation of the face and brain MR surfaces, (3) the fitting of surfaces, (4) the fiducial∕target localization error, and (5) the LRS range acquisition and texture mapping process. It would be very difficult to isolate these sources of error and understand their compounding effects. However, many of these errors are present in all IGNS systems and the linking between system error and therapy delivery remains to be very challenging. The registration platform we have developed (Fig. 1) allows the collection of many data sources, which when compared under similar conditions is beginning to provide new information regarding image-to-physical space alignment.
CONCLUSION
In this article, results from a new image-to-physical space registration platform are presented for 11 in vivo OR patient cases. Textured point clouds from preoperative MR image volumes and from a laser range scanning device are used to identify novel soft-tissue targets on the cortical surface, thus providing a means to compare methods of image-to-physical alignment. Statistical analysis demonstrates equivalence between face-based surface registration and traditional skin marker PBR, implying that skin markers may no longer be needed if face scans can be routinely acquired. The results suggest that brain shift may confound IGNS from its earliest stages, i.e., post opening of the dura. With respect to addressing the immediate needs within the neurosurgical community, laser range scanning technology coupled with cortical surface registration methods offer a relatively simple way to correct for misregistration for superficial brain tumors. In this article, the TRE for cortical targets ranged from 1.6±0.5–2.0±0.9 mm, which is comparable to the bone-implanted TRE results of 1.7±0.7 mm reported in Ref. 3. This realization has a caveat since the extent of subsurface registration error is not measurable in this study. Nevertheless, the cortical surface methods would improve registration for shallow lesions on the brain surface significantly if implemented within today’s IGNS systems.
Methodologically, this article suggests that organ-based registration would improve IGNS for shallow lesions. It may improve results for subsurface targets too but this remains to be studied. The article confirms the work by Mascott et al. in Ref. 3 in that IGNS using traditional registration methods is somewhat qualitative. While movement toward bone-implanted markers would appear important, the presence of brain shift is significant enough that the added invasiveness is not warranted. As a result, one could argue that bone-implanted markers for use within image-guided tumor surgery will never be warranted until soft-tissue deformation compensation techniques are realized. The interesting aspect to this statement is that the inverse statement may also be true in that, with the realization of compensation strategies, the need for bone-implanted markers may be critical for maximally taking advantage of accuracy gains.
ACKNOWLEDGMENTS
This project is funded by NIH-National Institute for Neurological Disorders and Stroke Grant No. RO1 NS049251-01. The authors acknowledge the Vanderbilt University Medical Center, Department of Neurosurgery, and the operating room staff for their help in data collection.
APPENDIX
The Sprent test32 is a simple and reasonably robust test to classify any observation xi as an outlier if |xi−M|∕MAD, where xi represents the RMS FRE of patient 9 for the skin-marker PBR using five fiducials, M is the median of the RMS FRE of the ten remaining patients using skin-marker PBR with five fiducials, and MAD is the median absolute deviation.
The Friedman test37 is used to compare several related treatments, in this case registration methods, and is an excellent nonparametric two-way analysis of the variance method with observations, in this case target errors above 5 in size. The null hypothesis of the test is that the treatments have identical effects. The alternative hypothesis is that at least one of the treatments tends to differ from the rest. For this study, the test begins by ranking/ordering the target registration errors, {xij}n×k, of the four registration methods for the same target points where n is the number of targets, and k is the method of registration. The ranks for each method are summed and a mean is taken for each method, i.e., . The mean of all target errors over all methods is also calculated, i.e., . The squared difference of the means is taken and multiplied by the total number of targets, . A Chi-Square comparison metric is then determined, Q=(SS)∕[k(k+1)], where k is the number of registration methods and equals 4 in this study. As the four methods were ranked through all 83 targets, a corresponding p-value less than 0.001 was obtained, which indicated that the four methods are not equivalent and at least one method is significantly different.
References
- Arun K., Huang T., and Blostein S. D., “Least-squares fitting of two 3D point sets,” IEEE Trans. Pattern Anal. Mach. Intell. 9, 699–700 (1987). [DOI] [PubMed] [Google Scholar]
- Schonemann P. H., “A generalized solution of the orthogonal procrustes problem,” Psychometrika 31, 1–10 (1966). [Google Scholar]
- Mascott C. R., Sol J. C., Bousquet P., Lagarrigue J., Lazorthes Y., and Lauwers-Cances V., “Quantification of true in vivo (application) accuracy in cranial image-guided surgery: Influence of mode of patient registration,” Neurosurgery 59, 146–155 (2006). [DOI] [PubMed] [Google Scholar]
- Mascott C. R., “In vivo accuracy of image guidance performed using optical tracking and optimized registration,” J. Neurosurg. 105, 561–567 (2006). [DOI] [PubMed] [Google Scholar]
- Maurer C. R., Fitzpatrick J. M., Wang M. Y., Galloway R. L., Maciunas R. J., and Allen G. S., “Registration of head volume images using implantable fiducial markers,” IEEE Trans. Med. Imaging 10.1109/42.611354 16, 447–462 (1997). [DOI] [PubMed] [Google Scholar]
- Nakajima S., Atsumi H., Kikinis R., Moriarty T. M., Metcalf D. C., Jolesz F. A., and Black P. M., “Use of cortical surface vessel registration for image-guided neurosurgery,” Neurosurgery 40, 1201–1208 (1997). [DOI] [PubMed] [Google Scholar]
- Raabe A., Krishnan R., Wolff R., Hermann E., Zimmermann M., and Seifert V., “Laser surface scanning for patient registration in intracranial image-guided surgery,” Neurosurgery 50, 797–801 (2002). [DOI] [PubMed] [Google Scholar]
- Marmulla R., Luth T., Muhling J., and Hassfeld S., “Automated laser registration in image-guided surgery: Evaluation of the correlation between laser scan resolution and navigation accuracy,” Int. J. Oral Maxillofac Surg. 33, 642–648 (2004). [DOI] [PubMed] [Google Scholar]
- Grimson W. E. L., Kikinis R., Jolesz F. A., and Black P. M., “Image-guided surgery,” Sci. Am. 280, 62–69 (1999). [DOI] [PubMed] [Google Scholar]
- Nabavi A., Black P. M., Gering D. T., Westin C. F., Mehta V., Pergolizzi R. S., Ferrant M., Warfield S. K., Hata N., Schwartz R. B., Wells W. M., Kikinis R., and Jolesz F. A., “Serial intraoperative magnetic resonance imaging of brain shift,” Neurosurgery 48, 787–797 (2001). [DOI] [PubMed] [Google Scholar]
- Nimsky C., Ganslandt O., Cerny S., Hastreiter P., Greiner G., and Fahlbusch R., “Quantification of, visualization of, and compensation for brain shift using intraoperative magnetic resonance imaging,” Neurosurgery 47, 1070–1080 (2000). [DOI] [PubMed] [Google Scholar]
- Nauta H. J., “Error assessment during image guided and imaging interactive stereotactic surgery,” Comput. Med. Imaging Graph. 18, 279–287 (1994). [DOI] [PubMed] [Google Scholar]
- Sun H., Roberts D. W., Farid H., Wu Z., Hartov A., and Paulsen K. D., “Cortical surface, tracking using a stereoscopic operating microscope,” Neurosurgery Suppl. 56, 86–97 (2005). [DOI] [PubMed] [Google Scholar]
- Cao A., Miga M. I., Dumpuri P., Ding S., Dawant B. M., and Thompson R. C., “Target error for image-to-physical space registration: Preliminary clinical results using laser range scanning,” Medical Imaging 2007: Visualization and Image-Guided Procedures: Proc. of SPIE, Vol. 6509, 2007.
- Miga M. I., Sinha T. K., Cash D. M., Galloway R. L., and Weil R. J., “Cortical surface registration for image-guided neurosurgery using laser-range scanning,” IEEE Trans. Med. Imaging 10.1109/TMI.2003.815868 22, 973–985 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha T. K., Dawant B. M., Duay V., Cash D. M., Weil R. J., Thompson R. C., Weaver K. D., and Miga M. I., “A method to track cortical surface deformations using a laser range scanner,” IEEE Trans. Med. Imaging 10.1109/TMI.2005.848373 24, 767–781 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha T. K., Miga M. I., Cash D. M., and Weil R. J., “Intraoperative cortical surface characterization using laser range scanning: Preliminary results,” Neurosurgery 59, 368–376 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumpuri P., Thompson R. C., Dawant B. M., Cao A., and Miga M. I., “An atlas-based method to compensate for brain shift: Preliminary results,” Med. Image Anal. 11, 128–145 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha T. K., “Cortical shift characterization using a laser range scanner for neurosurgery,” in Biomedical Engineering (Vanderbilt University, Nashville, 2004), p. 224. [Google Scholar]
- Sinha T. K., Duay V., Dawant B. M., and Miga M. I., “Cortical shift tracking using a laser range scanner and deformable registration methods,” in Medical Image Computing and Computer-Assisted Intervention (Springer-Verlag, Berlin, 2003), Vol. 2879, pp. 166–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick J. M., Hill D. L. G., and Maurer C. R., “Image registration,” in Handbook of Medical Imaging, edited by Sonka M. and Fitzpatrick J. M. (SPIE, Bellingham, 2000), Vol. 2, pp. 447–513. [Google Scholar]
- Besl P. J. and McKay N. D., “A method for registration of 3D shapes,” IEEE Trans. Pattern Anal. Mach. Intell. 10.1109/34.121791 14, 239–256 (1992). [DOI] [Google Scholar]
- Dumpuri P., Thompson R. C., Sinha T. K., and Miga M. I., “Automated brain shift correction using a pre-computed deformation atlas,” Medical Imaging 2006: Visualization and Image-Guided Procedures: Proc. of SPIE, Vol. 6141, 2006.
- Dawant B. M., Hartmann S. L., Thirion J. P., Maes F., Vandermeulen D., and Demaerel P., “Automatic 3D segmentation of internal structures of the head in MR images using a combination of similarity and free-form transformations: Part I, methodology and validation on normal subjects,” IEEE Trans. Med. Imaging 18, 909–916 (1999). [DOI] [PubMed] [Google Scholar]
- D’Haese P. F., Duay V., Li R., du Boise d’Aische A., Cmelak A., Donnelly E., Niermann K., Merchant T. E., Macq B., and Dawant B. M., “Automatic segmentation of brain structures for radiation therapy planning,” Medical Imaging 2003: Image Processing: Proc. of SPIE, Vol. 5032, 2003.
- Hartmann S. L., Parks M. H., Martin P. R., and Dawant B. M., “Automatic 3D segmentation of internal structures of the head in MR images using a combination of similarity and free-form transformations: Part II, validation on severely atrophied brains,” IEEE Trans. Med. Imaging 18, 917–926 (1999). [DOI] [PubMed] [Google Scholar]
- Schroeder W., Martin K., and Lorensen B., The Visualization Toolkit: An Object-Oriented Approach to 3D Graphics (Prentice Hall, New Jersey, 1996). [Google Scholar]
- Studholme C., Hill D. L. G., and Hawkes D. J., “An overlap invariant entropy measure of 3D medical image alignment,” Pattern Recogn. 10.1016/S0031-3203(98)00091-0 32, 71–86 (1999). [DOI] [Google Scholar]
- Hartkens T., Hill D. L. G., Castellano-Smith A. D., Hawkes D. J., Maurer C. R., Martin A. J., Hall W. A., Liu H., and Truwit C. L., “Using points and surface to improve voxel-based nonrigid registrations,” in Proceedings of Lecture Notes in Computer Science: Medical Image Computing and Computer-Assisted Intervention (Springer Verlag, 2002), Vol. 2488, pp. 565–572.
- Press W. H., Teukolsky S. A., Vetterling W. T., and Flannery B. P., Numerical Recipes in C: The Art of Scientific Computing, 2nd ed. (Cambridge University Press, New York, 1992). [Google Scholar]
- Hines W. H. and Montgomery D. C., Probability and Statistics in Engineering and Management Science, 3rd ed. (Wiley, New York, 1990). [Google Scholar]
- Sheskin D. J., Handbook of Parametric and Nonparametric Statistical Procedures, 2nd ed. (Chapman and Hall/CRC, Boca Raton, 2000). [Google Scholar]
- Fitzpatrick J. M. and R. L.Galloway, Jr., “Fiducial-based 3D image- and patient-space matching,” Automedica 2 (2001). [Google Scholar]
- Fitzpatrick J. M., West J. B., and Maurer C. R., “Predicting error in rigid-body point-based registration,” IEEE Trans. Med. Imaging 10.1109/42.736021 17, 694–702 (1998). [DOI] [PubMed] [Google Scholar]
- Sibson R., “Studies in the robustness of multidimensional scaling: Perturbational analysis of classical scaling,” J. R. Stat. Soc. Ser. B (Methodol.) 41, 217–229 (1979). [Google Scholar]
- Rost J., Harris S., Stefansic J., Sillay K., and R. L.Galloway, Jr., “Comparison between skin mounted fiducials and bone implanted fiducials for image-guided neurosurgery,” Medical Imaging 2004: Visualization, Image-Guided Procedures, and Display: Proc. of SPIE, Vol. 5367, 2004.
- Friedman M., “A comparison of alternative tests of significance for the problem of m rankings,” Ann. Math. Stat. 11, 86–92 (1940). [Google Scholar]