Abstract
The type IIA rat brain sodium channel is composed of three subunits: a large pore-forming α subunit and two smaller auxiliary subunits, β1 and β2. The β subunits are single membrane-spanning glycoproteins with one Ig-like motif in their extracellular domains. The Ig motif of the β2 subunit has close structural similarity to one of the six Ig motifs in the extracellular domain of the cell adhesion molecule contactin (also called F3 or F11), which binds to the extracellular matrix molecules tenascin-C and tenascin-R. We investigated the binding of the purified sodium channel and the extracellular domain of the β2 subunit to tenascin-C and tenascin-R in vitro. Incubation of purified sodium channels on microtiter plates coated with tenascin-C revealed saturable and specific binding with an apparent Kd of ≈15 nM. Glutathione S-transferase-tagged fusion proteins containing various segments of tenascin-C and tenascin-R were purified, digested with thrombin to remove the epitope tag, immobilized on microtiter dishes, and tested for their ability to bind purified sodium channel or the epitope-tagged extracellular domain of β2 subunits. Both purified sodium channels and the extracellular domain of the β2 subunit bound specifically to fibronectin type III repeats 1–2, A, B, and 6–8 of tenascin-C and fibronectin type III repeats 1–2 and 6–8 of tenascin-R but not to the epidermal growth factor-like domain or the fibrinogen-like domain of these molecules. The binding of neuronal sodium channels to extracellular matrix molecules such as tenascin-C and tenascin-R may play a crucial role in localizing sodium channels in high density at axon initial segments and nodes of Ranvier or in regulating the activity of immobilized sodium channels in these locations.
Voltage-gated sodium channels mediate the influx of sodium ions that is responsible for the rising phase of the action potential in neurons and other excitable cells. Action potentials are initiated at the axon initial segment (1, 2), where sodium channels are localized in high density (3, 4). In myelinated nerves, rapid conduction of the action potential depends on the high density of voltage-gated sodium channels at nodes of Ranvier (5, 6). The mechanisms by which sodium channels are sorted to the cell surface after synthesis, transported to the membrane, and concentrated in specialized regions of high density are unknown. Interaction with glial cells or extracellular matrix proteins may play a role in this process. In shiverer mice, which lack myelin basic protein and have hypomyelinated axons (7–9), the density of type II sodium channels in affected axons is greatly increased (10, 11). In pure neuronal cell cultures, sodium channels are distributed diffusely along neurites. Addition of Schwann cells, oligodendrocytes, or oligodendrocyte culture supernatants to the neuronal cultures results in the clustering of channels, even in the absence of myelination (12, 13). Identification of proteins that interact with sodium channels during this process would be an important step toward understanding its molecular basis.
The rat brain sodium channel consists of a large pore-forming α subunit of 260 kDa, a noncovalently associated β1 subunit of 36 kDa, and a disulfide-linked β2 subunit of 33 kDa (14). Both β1 and β2 subunits modulate the gating of the α subunit and increase its cell surface expression (15, 16). During biosynthesis of sodium channels in cultured fetal brain neurons, excess free α subunits without disulfide-linked β2 subunits are synthesized and maintained in an intracellular pool (17, 18). Sodium channels that reach the cell surface contain disulfide-linked β2 subunits, consistent with the hypothesis that disulfide linkage with β2 subunits is correlated with cell surface expression in neurons (18, 19). The β2 subunit is found only in neuronal sodium channels (14, 16). It is expressed at high levels during the first 2–3 weeks postnatally in rat brain and retinal ganglion cells (16, 20, 21), corresponding to the period of the largest increase of functional sodium channels on the cell surface and to the period of most active myelination and synapse formation.
The β1 and β2 subunits consist of a single transmembrane segment with a large amino-terminal extracellular domain and a small carboxyl-terminal intracellular domain (15, 16). The amino acid sequences of the extracellular domains of β1 and β2 are predicted to form a single disulfide-linked V-type Ig-like fold similar to cell adhesion molecules, and the amino acid sequence of the β2 subunit is strikingly similar to two separate segments of the neural cell adhesion molecule contactin/F3/F11 (hereinafter called contactin) (16, 22). Ig-like domains of cell adhesion molecules typically bind to extracellular proteins (23–25), and contactin (26) specifically binds the extracellular matrix molecules tenascin-C and tenascin-R via the external loops in its Ig folds (27–31). Tenascin-C, which is secreted by astrocytes and other cell types, is implicated in neuronal migration, axonal outgrowth, and tissue boundary formation (refs. 29 and 32; for review see refs. 33 and 34). Tenascin-R is secreted by oligodendrocytes during myelination and by some astrocytes and neurons. Like tenascin-C, it has complex effects on neuronal cell function (refs. 32 and 35–38; for review see refs. 33 and 34).
It is likely that interactions of sodium channels with extracellular matrix proteins and intracellular cytoskeletal proteins are involved in their localization, immobilization, and function. Although the interaction between sodium channels and the intracellular cytoskeletal protein ankyrin has been established (39), interactions with extracellular matrix proteins have not previously been described. In the experiments presented here, we examined the interaction of tenascin-C and tenascin-R with sodium channels in vitro.
EXPERIMENTAL PROCEDURES
Materials.
Heterotrimeric sodium channels were purified from rat brain through the wheat germ agglutinin chromatography step as described (40). cDNA clones encoding the domains of tenascin-C and tenascin-R were constructed as glutathione S-transferase (GST)-tagged fusion proteins in the pGEX-2T vector (Pharmacia) as described (30, 41). Laminin was obtained from Boehringer Mannheim. Tenascin-C was obtained from Boehringer Mannheim, and tenascin-C and tenascin-R were also isolated from mouse brain as described (42–44).
Protein Expression and Purification.
cDNAs encoding specific regions of tenascin-C or tenascin-R in a modified pGEX bacterial expression vector (30, 41) were used to synthesize fusion proteins of these regions linked to GST by expression in Escherichia coli. The cDNAs were grown in E. coli BL236, a protease-deficient cell line (Pharmacia), in 50-ml cultures overnight at 37°C. The culture was diluted into a 500-ml flask and grown for 2 h at 37°C. Expression of the fusion protein was then induced with 0.1 mM isopropyl thiogalactoside, and the cells were grown for an additional 3 h. Purification steps were carried out on ice or at 4°C. The cells were harvested and washed, and the pellet was resuspended in Tris-buffered saline (TBS) at pH 7.4 with protease inhibitors. Sarkosyl was added to 1.5% and the suspension was sonicated by using a microtip probe at an output level of 4 and a 40% duty cycle. The mixture was sedimented at 12,000 × g for 20 min. The supernatant was collected and Triton X-100 added to a final concentration of 2%. After incubation on ice for 30 min, the lysate was incubated with glutathione-Sepharose 4B beads (Pharmacia) and mixed by rotation at 4°C for 1 h. The beads were washed, and the tenascin moiety was eluted by proteolytic cleavage from the GST epitope tag with thrombin. Pefabloc SC (a serine protease inhibitor from Boehringer Mannheim) was added to the eluate to inhibit any residual thrombin, and the protein concentration was measured by using a Coomassie blue-based assay (Bio-Rad).
The cDNA encoding the extracellular domain of β2 was cloned into the pGEX vector to make a GST–β2 fusion protein. The cells were grown and induced as above. Because of the insolubility of this protein, the cells were resuspended in TBS, pH 7.4, with protease inhibitors, sonicated, and sedimented. The supernatant was discarded, and the pellet resuspended in an 8 M urea solution. After incubation for 1 h at 4°C, the solution was diluted stepwise in TBS with protease inhibitors to a final urea concentration of 1 M. Glutathione-Sepharose 4B beads were added and the mixture was incubated for 4–6 h at 4°C. The GST–β2 protein was eluted with 15 mM glutathione. The protein concentration was determined as for the tenascin fusion proteins.
ELISAs.
Solid-phase binding assays were carried out on Nunc maxisorb plates coated with 100 μl of tenascin-C or tenascin-R, laminin, or purified tenascin domains at the indicated concentrations in TBS, pH 8.0, 1 mM MgCl2, 1 mM CaCl2 for 2 h at 37°C. Nonspecific binding was blocked with 1% gelatin, and the plates were washed with TBS. Sodium channels containing α, β1, and β2 subunits were purified from rat brain and quantified by measurement of [3H]saxitoxin binding activity (40). They were then added at the indicated concentrations in 100 μl of TBS, 1 mM MgCl2, 1 mM CaCl2, 0.1% Triton X-100, and 0.025% phosphatidylcholine for 2 h at 37°C. After washing, the plates were incubated with affinity-purified anti-SP1 antibody, which recognizes the α subunit of the sodium channel (45), in TBS, 5% (wt/vol) nonfat dry milk overnight at 4°C or for 2 h at 37°C. The microtiter plates were washed, and alkaline phosphatase-conjugated anti-rabbit IgG was added in TBS, 5% (wt/vol) nonfat dry milk for 2 h at 37°C. After extensive washing, 100 μl of a developing solution containing 1 mg/ml nitrophenylphosphate was added and incubated for 20 min at room temperature, away from light. The reaction was stopped with the addition of 50 μl of 2.5 M NaOH. Absorbance was measured at 410 nm. All values were obtained in triplicate and specific absorbance calculated by subtracting nonspecific absorbance (in the absence of sodium channel) from total absorbance. Similar experiments were carried out with GST–β2 in place of sodium channels. The bound β2 extracellular domain was quantitated with the anti-β2–2 antibody (16) in an ELISA as described above for intact sodium channels.
RESULTS
Binding of Purified Sodium Channels to Tenascin-C.
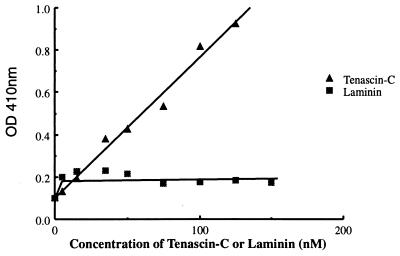
To examine the interaction of the purified sodium channel with tenascin-C, the wells in microtiter plates were coated by treatment with tenascin-C solutions of increasing concentration, and purified sodium channel preparations were incubated in the wells. Unbound proteins were rapidly washed away, and bound sodium channel was quantitated by labeling with a specific antibody and an ELISA as described under Experimental Procedures. Increasing the concentration of tenascin-C adsorbed to the plate caused a progressive, approximately linear increase in binding of sodium channels (Fig. 1). In contrast, increasing concentrations of laminin do not cause an increase in sodium channel binding (Fig. 1). Binding of sodium channels saturated in the range of concentration of 125–150 μM tenascin-C (data not shown). These results demonstrate specific binding of sodium channels to tenascin-C in preference to laminin.
Figure 1.
Specific binding of sodium channels to tenascin-C. Microtiter plates were coated with increasing concentrations of tenascin-C or laminin as indicated on the abscissa. After washing, purified sodium channels were added at a concentration of 40 nM and incubated as described under Experimental Procedures. The bound sodium channels were washed and quantitated by labeling with anti-SP1 antibody in an ELISA as described under Experimental Procedures. The results of a single experiment representative of eight similar experiments are shown.
With a fixed concentration of tenascin-C on the surface of the microtiter plate, binding of purified sodium channels was saturable (Fig. 2). Half-maximal binding was observed at ≈15 nM sodium channel, consistent with a high affinity interaction between the purified channel and tenascin-C. Much lower levels of binding to laminin were observed under the same conditions (Fig. 2).
Figure 2.
High affinity binding of sodium channels to tenascin-C. Microtiter plates were coated with a fixed 100 nM concentration of tenascin-C or laminin. After washing, the indicated concentrations of sodium channels were added, incubated, washed, and quantitated by ELISA as described under Experimental Procedures. Half maximal binding of sodium channels was observed at 15 nM.
Binding of Sodium Channels to Domains of Tenascin-C and Tenascin-R.
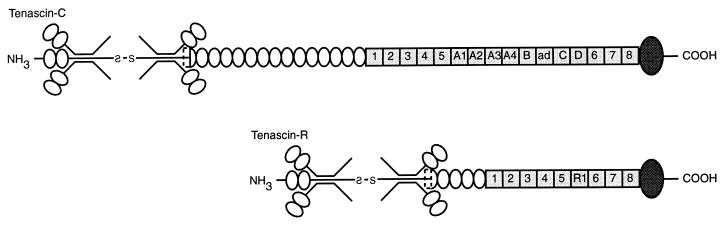
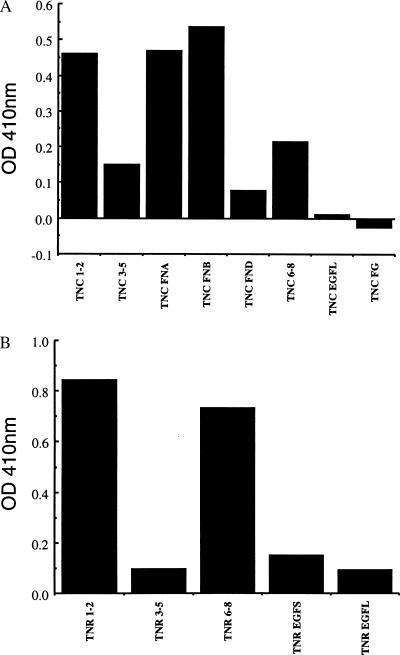
Tenascin-C is composed of a series of epidermal growth factor (EGF)-like repeats, a series of fibronectin type III (FN)-like regions, and a fibrinogen-like region (FG) (Fig. 3). The FN regions may be alternatively spliced, yielding tenascin-C molecules of varying sizes. Tenascin-R is smaller than tenascin-C but has a similar overall organization and structure. The different domains of the tenascins are implicated in different aspects of cell-to-cell signaling in neurons and glia (refs. 30, 35, 41, and 42; for review see refs. 33 and 34). To examine which regions of the tenascins can interact directly with sodium channels, we expressed clusters of the tenascin domains as fusion proteins with GST in E. coli, purified them by affinity chromatography, and cleaved them from the GST tag. Fig. 4A illustrates the binding of a fixed concentration of purified sodium channels to the different regions of tenascin-C in the ELISA assay. Significant binding is observed to fibronectin repeat regions FN1–2, FN-A, FN-B, and FN6–8. We did not observe significant binding to the EGF or fibrogenin domains of tenascin-C under the conditions of these experiments. We conducted a similar survey of sodium channel binding to the domains of tenascin-R, an isoform expressed primarily by glial cells in the nervous system. As for tenascin-C, we observed specific binding to the fibronectin domains FN1–2 and FN6–8, but not to the EGF-like domains (Fig. 4B).
Figure 3.
Domain structure of tenascin-C and tenascin-R. The domain structures of tenascin-C and tenascin-R are illustrated. EGF-like domains, open ovals; fibronectin-like domains, squares; fibrinogen-like domains, solid ovals. The fibronectin domains indicated as A, B, C, and R1 are alternatively spliced. In the text, the fusion protein containing fibronectin domains A1 to A4 is labeled FN-A. Adapted from ref. 49.
Figure 4.
Binding of sodium channels to the domains of tenascin-C and tenascin-R. (A) Microtiter plates were coated with 100 nM solutions of the indicated bacterially expressed domains of tenascin-C. After washing, purified sodium channels were added at 40 nM, incubated, washed and quantitated by ELISA as described under Experimental Procedures. The results of a single experiment representative of five separate experiments are illustrated. Binding to TNC FN1–2, TNC FN-A, TNC FN-B, and TNC FN6–8 were consistently greater than background binding in these experiments. (B) A similar experiment was carried out with the indicated domains of tenascin-R.
Binding of the β2 Subunit of Sodium Channels to Tenascin-C and Tenascin-R.
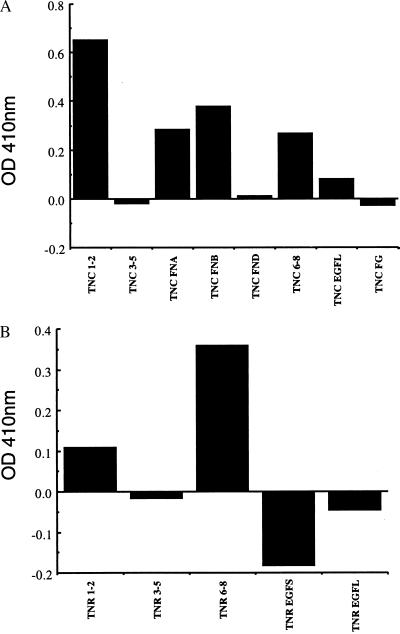
Titration experiments similar to the one in Fig. 1 detected binding of the β2 subunit extracellular domain to tenascin-C (data not shown). To test whether the β2 subunit can bind to the tenascins with the same specificity as the intact sodium channel, we expressed the extracellular domain of the β2 subunit in E. coli as described under Experimental Procedures and examined its binding to immobilized tenascin domains. The expressed extracellular domain of the β2 subunit binds most effectively to FN1–2 of tenascin-C with measurable specific binding also observed for FN-A, FN-B, and FN6–8 (Fig. 5A). Like the intact sodium channel, the β2 subunit binds only to FN1–2 and FN6–8 of tenascin-R, although the binding of purified sodium channel to FN1–2 is greater than observed for β2 (Fig. 5B). There is little or no high affinity binding to FN3–5 or the EGF-like regions. The similarities in binding specificity between the intact sodium channel and the β2 subunit suggest that the interaction of native sodium channel with the tenascins takes place via the extracellular domain of the β2 subunit.
Figure 5.
Binding of GST-β2 to the domains of tenascin-C and tenascin-R. (A) Microtiter plates were coated with 100 nM solutions of the indicated domains of tenascin-C expressed and purified from E. coli. After washing, GST-β2 was added at 40 nM, incubated, washed and quantitated by ELISA as described under Experimental Procedures. The results of a single experiment representative of three separate experiments are illustrated. Binding to TNC FN1–2, TNC FN-A, TNC FN-B, and TNC FN6–8 were consistently greater than background binding in these experiments. (B) A similar experiment was carried out with the indicated domains of tenascin-R.
DISCUSSION
Sodium Channels Bind to Tenascin-C and Tenascin-R.
Our results demonstrate direct binding of neuronal voltage-gated sodium channels to tenascin-C and tenascin-R. In the in vitro binding assays described here, the interaction of purified sodium channels with immobilized tenascin is specific and saturable with an apparent Kd of 15 nM. Because the tenascins are widely expressed and secreted by astrocytes, Schwann cells, and oligodendrocytes in the nervous system, their interaction with sodium channels of neighboring neurons is likely to immobilize them in the cell surface membrane.
Multiple Fibronectin Domains of Tenascin-C and Tenascin-R Participate in the Interaction with Sodium Channels.
The tenascins are modular proteins with many distinct domains that are thought to interact with other molecules. Their EGF, fibronectin, and fibrinogen domains are all candidates for protein–protein interactions. Our results support the idea that the fibronectin domains of tenascin-C and tenascin-R are responsible for the high affinity interaction with sodium channels. The FN1–3 and FN6–8 domains of tenascin-C and the FN1–2 and FN6–8 domains of tenascin-R bind to sodium channels directly. In addition, the FN-A and FN-B domains of tenascin-C are also capable of specific binding to sodium channels. Cerebellar and hippocampal neurons adhere to FN1–2, FN6–8, and the alternative splice variants of tenascin-C (32, 41), and cerebellar neurons adhere to FN1–2 and FN6–8 from tenascin-R (30, 44). Interactions with sodium channels in these neurons may contribute to these cellular interactions.
Expressed β2 Subunits Bind to the Fibronectin Domains of Tenascin-C and Tenascin-R.
The β2 subunits of sodium channels have substantial amino acid sequence similarity with contactin in their Ig-fold motif (16), and contactin binds to the tenascins (27, 28). Therefore, it is likely that the β2 subunits mediate the interaction of the sodium channel with tenascin. Our results provide support for this idea because the specificity of binding of the expressed β2 subunits to the different domains of the tenascins was similar to the specificity of binding of the native sodium channel. The extracellular domain of the β2 subunits bound preferentially to the FN1–2, FN6–8, FN-A, and FN-B domains of tenascin-C and the FN1–2 and FN6–8 domains of tenascin-R. We were not able to compare the affinity of the β2 extracellular domain to that of the native sodium channel because the low level of recovery of correctly folded β2 subunits after expression in E. coli prevented determination of the concentration dependence and stoichiometry of binding to the fibronectin domains of the tenascins. Additional experiments to compare the affinity of the expressed β2 subunits to that of the native sodium channel complex and to test the ability of the β2 subunit to compete with native sodium channels for binding will be required before the relative roles of the α, β1, and β2 subunits in binding of sodium channels to the tenascins can be more critically assessed.
Functional Significance of the Interaction of Sodium Channels with Tenascin-C and Tenascin-R.
High density localizations of sodium channels are found in axon initial segments (3, 4), where they initiate action potentials (1, 2), and in nodes of Ranvier in myelinated axons (5, 6), where they allow discontinuous saltatory conduction. In the mutant mouse shiverer, which lacks myelin basic protein and has hypomyelinated axons (7–9), sodium channels are localized all along the axon instead of clustered at nodes of Ranvier (10, 11). These results implicate interaction with oligodendrocytes in control of sodium channel localization in myelinated central axons. In wild-type mice, sodium channels become clustered in developing myelinated axons in vivo or in cell culture before the myelin sheath is formed (46, 47). Protein factors released by oligodendrocytes stimulate sodium channel clustering in axons developing in cell culture (13). These secreted factors may include the tenascins and related extracellular matrix molecules. Binding of the tenascins to sodium channels may form the initial clusters of sodium channels or may immobilize sodium channels and stabilize sodium channel clusters after they have formed. In addition, binding of the tenascins to sodium channels may initiate local signaling events that regulate cell–cell interactions important for axonal growth and myelination. For example, the EGF-like domains of tenascin-R can modulate adhesion of fibroblast cells expressing sodium channel subunits through a pathway that involves protein kinase C (48). High affinity interaction with the fibronectin domains of sodium channels may allow local signaling through the EGF domains that may interact with sodium channels with an affinity too low to be detected in our in vitro binding experiments or with other local cell surface signaling proteins. Further analysis of the cell–cell interactions and intracellular signaling pathways that depend on interactions of the tenascins with sodium channels may provide new insights into the mechanisms responsible for regulation of sodium channel localization, density, and function in neurons.
Acknowledgments
We thank Dr. U. Dörries and Dr. Z. C. Xiao for cDNAs encoding tenascin-C and tenascin-R fragments. This research was supported by National Multiple Sclerosis Society Postdoctoral Fellowship FA-1220-A-1 (J.S.) and National Institutes of Health Research Grant NS25704 (W.A.C.).
ABBREVIATIONS
- EGF
epidermal growth factor
- FG
fibrinogen
- FN
fibronectin type III
- GST
glutathione S-transferase
- TBS
Tris-buffered saline
References
- 1.Eccles J C. Prog Brain Res. 1964;12:1–31. doi: 10.1016/s0079-6123(08)60614-7. [DOI] [PubMed] [Google Scholar]
- 2.Stuart G, Sakmann B. Neuron. 1995;15:1065–1076. doi: 10.1016/0896-6273(95)90095-0. [DOI] [PubMed] [Google Scholar]
- 3.Catterall W A. J Neurosci. 1981;1:777–783. doi: 10.1523/JNEUROSCI.01-07-00777.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wollner D A, Catterall W A. Proc Natl Acad Sci USA. 1986;83:8424–8428. doi: 10.1073/pnas.83.21.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ritchie J M, Rogart R B. Rev Physiol Biochem Pharmacol. 1977;79:1–49. doi: 10.1007/BFb0037088. [DOI] [PubMed] [Google Scholar]
- 6.Ellisman M H, Levinson S R. Proc Natl Acad Sci USA. 1982;79:6707–6711. doi: 10.1073/pnas.79.21.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bird T D, Farrell D F, Sumi S M. J Neurochem. 1978;31:387–391. doi: 10.1111/j.1471-4159.1978.tb12479.x. [DOI] [PubMed] [Google Scholar]
- 8.Roach A, Takahashi N, Pravtcheva D, Ruddle F, Hood L. Cell. 1985;42:149–155. doi: 10.1016/s0092-8674(85)80110-0. [DOI] [PubMed] [Google Scholar]
- 9.Readhead C, Popko B, Takahashi N, Shine H D, Saarvedra R A, Sidman R L, Hood L. Cell. 1987;48:703–712. doi: 10.1016/0092-8674(87)90248-0. [DOI] [PubMed] [Google Scholar]
- 10.Noebels J L, Marcom P K, Jalilian-Tehrani M H. Nature (London) 1991;352:431–434. doi: 10.1038/352431a0. [DOI] [PubMed] [Google Scholar]
- 11.Westenbroek R E, Noebels J L, Catterall W A. J Neurosci. 1992;12:2259–2267. doi: 10.1523/JNEUROSCI.12-06-02259.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joe E-H, Angelides K. Nature (London) 1992;356:333–335. doi: 10.1038/356333a0. [DOI] [PubMed] [Google Scholar]
- 13.Kaplan M R, Meyer-Franke A, Lamber S, Bennett V, Duncan I D, Levinson S R, Barres B A. Nature (London) 1997;386:724–728. doi: 10.1038/386724a0. [DOI] [PubMed] [Google Scholar]
- 14.Catterall W A. Physiol Rev. 1992;72:S15–S48. doi: 10.1152/physrev.1992.72.suppl_4.S15. [DOI] [PubMed] [Google Scholar]
- 15.Isom L L, De Jongh K S, Patton D E, Reber B F X, Offord J, Charbonneau H, Walsh K, Goldin A L, Catterall W A. Science. 1992;256:839–842. doi: 10.1126/science.1375395. [DOI] [PubMed] [Google Scholar]
- 16.Isom L L, Ragsdale D S, De Jongh K S, Westenbroek R E, Reber B F X, Scheuer T, Catterall W A. Cell. 1995;83:433–442. doi: 10.1016/0092-8674(95)90121-3. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt J, Rossie S, Catterall W A. Proc Natl Acad Sci USA. 1985;82:4847–4851. doi: 10.1073/pnas.82.14.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmidt J W, Catterall W A. Cell. 1986;46:437–444. doi: 10.1016/0092-8674(86)90664-1. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt J W, Catterall W A. J Biol Chem. 1987;262:13713–13723. [PubMed] [Google Scholar]
- 20.Wollner D A, Scheinman R, Catterall W A. Neuron. 1988;1:727–737. doi: 10.1016/0896-6273(88)90171-7. [DOI] [PubMed] [Google Scholar]
- 21.Scheinman R I, Auld V J, Goldin A L, Davidson N, Dunn R J, Catterall W A. J Biol Chem. 1989;264:10660–10666. [PubMed] [Google Scholar]
- 22.Isom L L, Catterall W A. Nature (London) 1996;383:307–308. doi: 10.1038/383307b0. [DOI] [PubMed] [Google Scholar]
- 23.Williams A F, Barclay A N. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- 24.Vaughn D E, Bjorkman P J. Neuron. 1996;16:261–273. doi: 10.1016/s0896-6273(00)80045-8. [DOI] [PubMed] [Google Scholar]
- 25.Schachner M. Curr Opin Cell Biol. 1997;9:627–634. doi: 10.1016/s0955-0674(97)80115-9. [DOI] [PubMed] [Google Scholar]
- 26.Ranscht B. J Cell Biol. 1988;107:1561–1573. doi: 10.1083/jcb.107.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zinsch A H, D’Allessandri L, Ranscht B, Falchetto R, Winterhalter K H, Vaughn L. J Cell Biol. 1992;119:203–213. doi: 10.1083/jcb.119.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vaughan L, Weber P, D’Alessandri L, Zisch A H, Winterhalter K H. Perspect Dev Neurobiol. 1994;2:43–52. [PubMed] [Google Scholar]
- 29.Pesheva P, Gennarini G, Goridis C, Schachner M. Neuron. 1993;10:69–82. doi: 10.1016/0896-6273(93)90243-k. [DOI] [PubMed] [Google Scholar]
- 30.Xiao Z C, Taylor J, Montag D, Rougon G, Schachner M. Eur J Neurosci. 1996;8:766–782. doi: 10.1111/j.1460-9568.1996.tb01262.x. [DOI] [PubMed] [Google Scholar]
- 31.Xiao Z C, Hillenbrand R, Schachner M, Thermes S, Rougon G, Gomez S. J Neurosci Res. 1997;49:698–709. doi: 10.1002/(SICI)1097-4547(19970915)49:6<698::AID-JNR4>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 32.Jung M, Pesheva P, Schachner M, Trotter J. Glia. 1993;9:163–175. doi: 10.1002/glia.440090302. [DOI] [PubMed] [Google Scholar]
- 33.Schachner M, Taylor J, Bartsch U, Pesheva P. Perspect Dev Neurobiol. 1994;2:33–41. [PubMed] [Google Scholar]
- 34.Faissner A. Cell Tissue Res. 1997;290:331–341. doi: 10.1007/s004410050938. [DOI] [PubMed] [Google Scholar]
- 35.Faissner A, Kruse J. Neuron. 1990;5:627–637. doi: 10.1016/0896-6273(90)90217-4. [DOI] [PubMed] [Google Scholar]
- 36.Lochter A, Vaughan L, Kaplony A, Prochiantz A, Schachner M, Faissner A. J Cell Biol. 1991;113:1159–1171. doi: 10.1083/jcb.113.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lochter A, Schachner M. J Neurosci. 1993;13:3986–4000. doi: 10.1523/JNEUROSCI.13-09-03986.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lochter A, Taylor J, Fuss B, Schachner M. Eur J Neurosci. 1994;6:597–606. doi: 10.1111/j.1460-9568.1994.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 39.Srinivasan Y L, Elmer L, Davis J, Bennett V, Angelides K. Nature (London) 1988;333:177–180. doi: 10.1038/333177a0. [DOI] [PubMed] [Google Scholar]
- 40.Hartshorne R P, Catterall W A. J Biol Chem. 1984;259:1667–1675. [PubMed] [Google Scholar]
- 41.Dorries U, Taylor J, Xiao Z, Lochter A, Montag D, Schachner M. J Neurosci Res. 1996;43:420–438. doi: 10.1002/(SICI)1097-4547(19960215)43:4<420::AID-JNR4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 42.Pesheva P, Spiess E, Schachner M. J Cell Biol. 1989;109:1765–1778. doi: 10.1083/jcb.109.4.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faissner A, Kruse J, Kühn K, Schachner M. J Neurochem. 1990;54:1004–1015. doi: 10.1111/j.1471-4159.1990.tb02350.x. [DOI] [PubMed] [Google Scholar]
- 44.Xiao Z C, Revest J M, Laeng P, Rougon G, Schachner M, Montag D. J Neurosci Res. 1998;52:390–404. doi: 10.1002/(SICI)1097-4547(19980515)52:4<390::AID-JNR3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 45.Gordon D, Merrick D, Auld V, Dunn R, Goldin A L, Davidson N, Catterall W A. Proc Natl Acad Sci USA. 1987;84:8682–8686. doi: 10.1073/pnas.84.23.8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vabnick I, Messing A, Chiu S Y, Levinson S R, Schachner M, Roder J, Li C M, Novakovic S, Shrager P. J Neurosci Res. 1997;50:321–336. doi: 10.1002/(SICI)1097-4547(19971015)50:2<321::AID-JNR20>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 47.Vabnick I, Novakovic SD, Levinson S R, Schachner M, Shrager P. J Neurosci. 1996;16:4914–4922. doi: 10.1523/JNEUROSCI.16-16-04914.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao Z C, Ragsdale D S, Brown P E, Isom L L. Soc Neurosci Abstr. 1998;24:1323. [Google Scholar]
- 49.Erickson H P. Curr Opin Cell Biol. 1993;5:869–876. doi: 10.1016/0955-0674(93)90037-q. [DOI] [PubMed] [Google Scholar]