Abstract
Recently, the RNA interference (RNAi) pathway has become the target of small molecule inhibitors and activators. RNAi has been well established as a research tool in the sequence-specific silencing of genes in eukaryotic cells and organisms by using exogenous, small, double-stranded RNA molecules of approximately 20 nucleotides. Moreover, a recently discovered post-transcriptional gene regulatory mechanism employs microRNAs (miRNAs), a class of endogenously expressed small RNA molecules, which are processed via the RNAi pathway. The chemical modulation of RNAi has important therapeutic relevance, because a wide range of miRNAs has been linked to a variety of human diseases, especially cancer. Thus, the activation of tumor-suppressive miRNAs and the inhibition of oncogenic miRNAs by small molecules have the potential to provide a fundamentally new approach for the development of cancer therapeutics.
Key words: cancer, microRNA, RNA, RNA interference, small molecule
INTRODUCTION
RNA interference (RNAi) is a cellular mechanism to regulate eukaryotic gene expression in a sequence-specific fashion by either inducing degradation of a messenger RNA (mRNA) or inhibiting its translation (1–3). RNAi can be triggered by two classes of RNA molecules: (1) small-interfering RNAs (siRNAs) either endogenously encoded (3) or derived from longer double-stranded RNA (dsRNA), which are transcribed from various vectors or are directly introduced into cells by injection or transfection (4–6), and (2) microRNAs (miRNAs) processed from stem-loop precursors that are encoded within the host genome (4,6,7).
Both siRNAs and miRNAs are single-stranded non-coding RNAs of 21–23 nucleotides that negatively regulate gene expression at the post-transcriptional level. While siRNAs are typically exogenous in nature (although endogenous examples have recently been reported), miRNAs represent a new class of endogenous gene regulators that function by binding the 3′ untranslated regions (UTRs) of target mRNAs leading to their downregulation through either suppression of translation or degradation (1,2). However, recent investigations in cell culture showed that the silencing by miRNAs is not confined to 3′ UTRs but can also occur in the 5′ UTR (8). Moreover, miRNAs were recently discovered that actually activate the expression of their target genes (9).
It is estimated that approximately 1,000 miRNAs exist in humans and that they are involved in the regulation of up to 30% of all genes and almost every genetic pathway (10–14). The pivotal role that miRNAs play in the regulation of a wide range of biological processes, including cell cycle progression and proliferation, differentiation, cell survival, and development, has become increasingly evident in recent years (15). Moreover, increasing evidence is accumulating that connects the misregulation of miRNAs to a wide range of human diseases.
The miRNA and siRNA Biogenesis Pathway
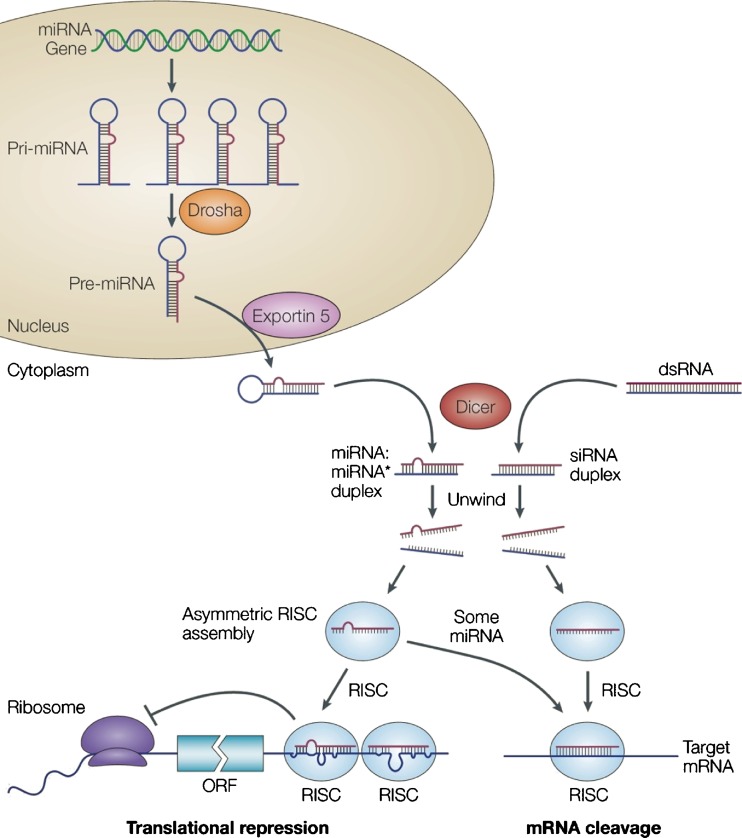
Initially, miRNAs are transcribed by RNA polymerase II as primary transcripts (pri-miRNAs) that require subsequent processing to yield a functionally mature miRNA (Fig. 1) (2,16). The pri-miRNA transcripts contain a 5′ 7-methylguanosine cap and a 3′-polyA tail, range from hundreds to thousands of nucleotides in length, and consist of monocistronic or polycistronic miRNAs. Pri-miRNAs are processed in the nucleus by the RNAse III enzyme Drosha, partnering with DGCR8 (in vertebrates) or Pasha (in invertebrates) (17–19). The Drosha-DGCR8 complex converts pri-miRNAs into shorter, stem-loop-structured dsRNAs of 70–80 nucleotides called precursor miRNAs (pre-miRNAs), with the mature miRNA included in their arms (17–19). Pre-miRNAs contain a 5′-phosphate and a two nucleotide 3′-hydroxy overhang, and are transported from the nucleus to the cytoplasm. Here, they are processed by Dicer, another RNase III enzyme, into mature miRNAs (20–24). Mature 21–23 nucleotide miRNAs enter the effector complex, called the RNA-induced silencing complex (RISC), to target single-stranded complementary mRNAs for translational repression or mRNA degradation, thus downregulating gene expression levels (25–28). In general, incomplete hybridization due to base-pair mismatches between RISC and the target mRNA inhibits mRNA translation rather than inducing mRNA degradation. Several mechanisms of translational silencing have been suggested, including the blocking of translation initiation or elongation and miRNA-mediated mRNA transport to P-bodies, inducing repression of translation and mRNA turnover (8,29–31).
Fig. 1.
The microRNA (left) and small-interfering RNAs (right) pathways. Adapted by permission from Macmillan Publishers Ltd: (2), copyright 2004
The processing and the functional pathway of miRNAs are partly shared by other dsRNAs which can trigger RNAi-induced gene silencing (Fig. 1) (4–7). These typically exogenous dsRNAs are often transfected into cells or expressed as short hairpin RNA (shRNA) from viral expression vectors. They are processed into siRNAs in a similar fashion as pre-miRNAs are processed into miRNAs. In contrast to the often imperfectly paired miRNAs, siRNAs commonly exhibit perfect sequence complementarity to their target. Although both processing pathways share some components in eukaryotic organisms, for example, Dicer processing and loading into RISC, they differ in distinct steps within the nucleus (Fig. 1).
Cellular Regulation of microRNA Biogenesis
The expression levels of mature miRNAs directly affect the level of downregulation of the corresponding miRNA targets. The misregulation of miRNA expression leads to a misregulation of the miRNA target which can have severe implications on cellular homeostasis, with the possibility to turn normal cells into malignant cells. Thus, nature has developed several regulatory mechanisms along the miRNA pathway to control the level of individual miRNA expression. Despite the increasing knowledge in the miRNA field and its direct correlation to a variety of human pathologies, the molecular details of miRNA regulation is one of the least understood aspects of miRNA function. Selected examples are discussed below in order to illustrate the different regulatory mechanisms of miRNA biogenesis and to showcase potential targets for small molecule intervention.
Pre-transcriptional regulation of miRNA expression. The biogenesis of miRNAs at this level is either affected by a change in miRNA gene copy number (frequently found in human cancers), mutations in the miRNA gene, or histone deacetylation and hypermethylation of miRNA promoter regions (32–34). For example, the tumor-suppressive miRNA miR-127 translationally downregulates the human proto-oncogene BCL6. Thus, it is not surprising that miR-127 is either downregulated or completely silenced in a wide range of tumors (35). An analysis of the promoter region specific for miR-127 in the T24 bladder cancer cell line revealed a high level of methylation, effectively inhibiting miR-127 expression. However, when these cells were treated with a DNA-demethylating agent (5-aza-2′-deoxycytidine) and an inhibitor of histone deacetylase (4-phenylbutyric acid, leading to increased histone acetylation), a 49-fold increase in miR-127 expression was observed (35).
Transcriptional regulation of miRNA expression. Recently, a small number of transcription factors that regulate the expression of cancer-related miRNAs have been identified (36). Most of these proteins bind to regulatory motifs upstream of miRNA genes, thus recruiting co-activators and the transcriptional machinery. A prominent example of transcriptional miRNA regulation is that of the oncogenic miR-17-92 cluster by Myc (37,38). The Myc transcription factor is a nuclear protein that is activated in several human malignancies, and elevated levels of Myc lead to the upregulation of the miR-17-92 cluster. Analysis of the DNA upstream of this cluster revealed several putative Myc binding sites, and the direct binding of the transcription factor was confirmed by chromatin immunoprecipitation. Interestingly, Myc also activates expression of the E2F1 gene (encoding another transcription factor regulating tumor suppressor genes), which itself is downregulated by miR-17-92. Thus, the signal transduction between Myc, E2F1, and miR-17-92 provides a complex, tightly controlled regulatory system for cell proliferation and apoptosis.
-
Post-transcriptional regulation of miRNA expression. As shown in Fig. 1, two RNase III endonucleases, Drosha and Dicer, post-transcriptionally process the pri-miRNA transcript to produce mature miRNAs. These enzymes are general factors that non-specifically control miRNA biogenesis, and thus their activity regulates the cellular abundance of all miRNAs. The global analysis of miRNA expression in cancers revealed a widespread downregulation, presumably due to a failure at the Drosha processing step (39). A surprisingly specific post-transcriptional regulation mechanism was found in the processing of pri-miR-21 in human vascular smooth muscle cells. Here, bone morphogenic protein and transforming growth factor β induce an interaction between the SMAD1 protein associated with pri-miR-21 and Drosha through the RNA helicase p68, a subunit of Drosha. This results in an increase in pri-miR-21 processing to mature miR-21, and thus an increased miR-21 level (40).
Another recently discovered post-transcriptional miRNA regulatory mechanism involves the RNA-binding protein KH-type splicing regulatory protein (KSRP), which was found to promote the biogenesis of several miRNAs (41). Transient knockout of KSRP in HeLa cells led to more than 1.5-fold reduction of 14 miRNAs, including let-7a, miR-16, miR-20, miR-21, miR-26b, and miR-106a. KSRP interacts with the terminal loop of the regulated miRNAs and binds preferentially to short G-rich stretches of at least three guanosine residues, although the regulation of miRNAs with other guanosine patterns in the terminal loop was observed as well. Upon binding to the miRNA, KSRP may optimize the positioning and/or recruitment of the miRNA precursor processing complexes through protein–protein interactions (41).
Of the three levels of regulation, both pre- and post-transcriptional regulations are believed to be generally less miRNA-specific, whereas regulation at the transcriptional level offers a higher degree of specificity as transcription factors are presumably involved in the development- and cell-specific regulation of distinct miRNAs (36). All three regulatory mechanisms present potential targets for the activation or deactivation of miRNA function with small molecules.
microRNAs and Human Diseases
Recently, certain miRNAs have been linked to a variety of human diseases, including diabetes, viral infections, as well as neurodegenerative and myocardial diseases. Arguable, the best understood involvement of aberrantly expressed miRNAs is observed in the development and progression of cancer. Here, miRNAs specifically act as tumor suppressors (e.g., let-7, miR-15/16, miR-34a, or miR-143/145) or inhibitors of apoptosis (e.g., miR-21, miR-155, or miR-214) (34,42,43). A list of selected miRNAs with relevance in cancer and cell death is shown in Table I.
Table I.
Selected microRNAs Involved in Cancer
| Cancer | miRNA | Selected targets | Presumed target function |
|---|---|---|---|
| Brain | miR-21↑ | TPM1, PDCD4 | Tumor suppressors |
| miR-221↑ | p27kip1 | Tumor suppressor, cell cycle inhibitor | |
| miR-378↑ | Sufu, Fus-1 | Tumor suppressors | |
| Thyroid | miR-138↓ | hTERT | Oncogenic activity |
| miR-221/222↑ | p27kip1 | Tumor suppressor, cell cycle inhibitor | |
| Breast | miR-10b↑ | HOXD10 | Inhibition of breast cancer metastasis |
| miR-21↑ | TPM1, PDCD4 | Tumor suppressors | |
| miR-27a↑ | ZBTB10, Myt1 | Tumor suppressors | |
| miR-125a/b↓ | HER2, HER3 | Oncogenes, EGF receptor family members | |
| miR-206↓ | ERα | Promoter of cell proliferation | |
| Lung | let-7/miR-98↓ | HMGA2, RAS, c-Myc | Oncogene, related to metastasis, promoter of carcinogenesis |
| miR-29↓ | DNMT3A/B | Oncogenic activity, DNA methyltransferase | |
| miR-34↑ | Bcl2 | Anti-apoptotic factor, promoter of proliferation | |
| miR-221/222↑ | p27kip1 | Cell cycle inhibitor, cell growth inhibitor | |
| Liver | miR-21↑ | PTEN | Tumor suppressor, negative regulation of PI3k |
| miR-122↓ | Cyclin G, Bcl-w | Anti-apoptotic factor | |
| Biliary | miR-21↑ | PTEN | Tumor suppressor, negative regulation of PI3k |
| miR-29↓ | Mcl-1 | Oncogene, anti-apoptotic factor | |
| Pancreas | miR-155↑ | TP53INP1 | Induction of growth arrest, apoptosis |
| Colon/rectum | miR-143↓ | ERK5 | Promoter of cell growth and proliferation |
| miR-145↓ | IRS1 | Oncogene, strong mitogenic activity | |
| Prostate | miR-20a↓ | E2F1, E2F3, E2F3 | Pro-apoptotic factors, regulation of cell cycle |
| miR-125b↓ | Bak1 | Pro-apoptotic factors, induction of apoptosis | |
| miR-126↓ | SLC45A3 | Oncogene | |
| miR-221/222↑ | p27Kip1 | Tumor suppressor, cell cycle inhibitor | |
| Bladder | miR-127↓ | BCL6 | Proto-oncogene, suppression of p53 |
| Testis | miR-372/373↑ | LATS2 | Tumor suppressor |
| Ovarian | let-7/miR-98↓ | HMGA2, RAS, c-Myc | Oncogene, related to metastasis, promoter of carcinogenesis |
| miR-210↓ | E2F3 | Pro-apoptotic factor, regulation of cell cycle | |
| miR-214↑ | PTEN | Tumor suppressor, negative regulation of PI3k | |
| Cervix | miR-218↓ | LAMB3 | Promoter of cell migration and tumorigenicity |
| Lymphoma | miR-143↓ | ERK5 | Promoter of cell proliferation and cell growth |
| Leukemia | miR-15/16↓ | Bcl2 | Anti-apoptotic factor, promoter of cell growth |
| miR-29/181↓ | Tcl1 | Oncogene, co-activator of the Akt oncoprotein | |
| miR-221/222↑ | Kit | Proto-oncogene, tyrosine-protein kinase |
The miRNAs are either increased (↑) or decreased (↓) in the particular cancer, compared to non-cancerous tissue. Adapted by permission from: Cell Cycle 2008, 7, 1529.
Thus, miRNAs not only represent viable markers for the detection and characterization of cancers but also provide potential targets for the development of new cancer therapeutics. Since miRNAs are multi-specific by nature and individual miRNAs are capable of modulating the expression of networks of mRNAs, miRNA-based therapy offers the appeal of targeting entire gene circuits that are controlled by a single, misregulated miRNA.
ASSAYS FOR THE DISCOVERY OF SMALL MOLECULE MODIFIERS OF THE miRNA AND siRNA PATHWAY
Several assays have been developed in order to discover small molecule modifiers of miRNA and siRNA function in either a general or an RNA-specific way.
Enzyme-based Assay for Small Molecule Inhibitors of miRNA Maturation
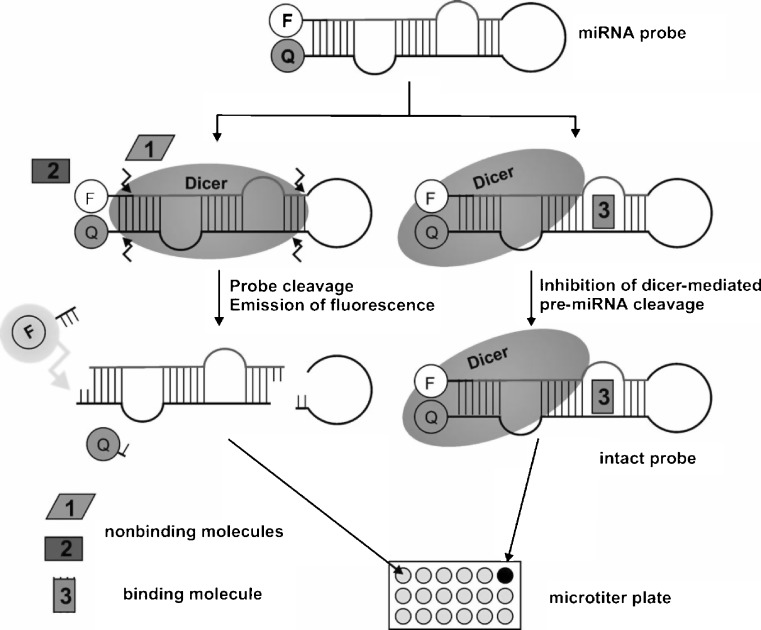
A fluorescence assay was developed to measure the activity of Dicer. Based on the pre-miRNA sequence of let-7, an RNA hairpin was designed which carries a fluorescence emitter (fluorescein, F) at the 5′ terminus and a fluorescence quencher (dabcyl, Q) at the 3′ terminus (Fig. 2) (44). Due to the close proximity of the fluorophore and the quencher, no fluorescence is detected in the case of an intact hairpin. However, upon incubation with Dicer and subsequent processing, an up to tenfold increase in fluorescence was detected. Since the hairpin substrate has no 3′-terminal overhang, it is cleaved several nucleotides further away from the helical end. This results in the cleavage of a total of four phosphodiester bonds, removing the terminal fragments containing the fluorophores from the pre-miRNA and thus prohibiting the quenching of fluorescence (45). The ability to measure the inhibition of pre-miRNA cleavage by Dicer was demonstrated through sequestering the miRNA hairpin by certain peptides, which led to an increase in fluorescence of up to 85% (44). However, so far, no application of this assay towards the screening of small molecule libraries for inhibitors of Dicer has been reported. An advantage of this cell-free assay is its low cost per well, with the disadvantage that miRNA-specific compounds will only be identified if they directly interact with the miRNA itself.
Fig. 2.
microRNA maturation assay. An active Dicer induces cleavage of an RNA hairpin, thus leading to a fluorescence increase (left). Pre-microRNA binding molecules can inhibit Dicer cleavage thus resulting in low fluorescence (right). Adapted by permission from: (44). Copyright Wiley-VCH Verlag GmbH & Co. KGaA
Cell-based Assay for Small Molecule Activators of the RNAi Pathway
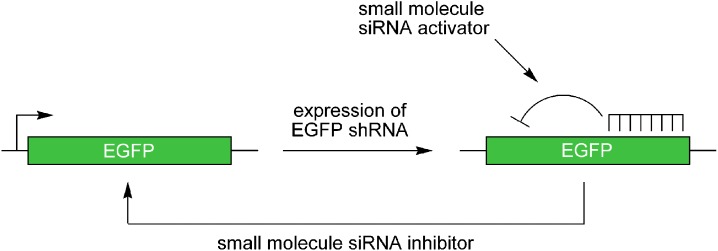
A cellular assay system was developed by generating an HEK293 cell line stably expressing enhanced green fluorescent protein (EGFP) that was subsequently infected with a lentivirus expressing a shRNA which is processed into an siRNA specifically targeting EGFP mRNA (46). The shRNA expression led to reduced levels of EGFP via RNAi. In order to setup a screen for general small molecule activators of the RNAi pathway (Fig. 3), individual cell clones with moderate reductions in GFP expression were selected. The ability of this assay to identify small molecule modifiers of RNAi was validated by transfecting the reporter cells with active and inactive 2′-OMe antisense agents targeting the EGFP siRNA. This assay has been applied in the screening of a small molecule collection for general activators of the RNAi pathway (see below).
Fig. 3.
Assay for activation or inhibition of the small-interfering RNA (siRNA) pathway via control of enhanced green fluorescent protein (EGFP) through lentiviral expression of an EGFP-targeting short hairpin RNA. In the presence of small molecule activators of the siRNA pathway, a decreased level of fluorescence is detected, while siRNA pathway inhibitors will lead to an increase in fluorescence
Cell-based Assay for Small Molecule Inhibitors of Specific miRNAs
Plasmids were constructed based on lentiviral vectors containing the target sequences of mature miRNAs, as well as a control sequence with no detectable recognition by natural miRNAs, downstream of a luciferase reporter gene (Fig. 4) (47). Mature miRNAs bind to their target sequence on the reporter mRNA and either suppress translation or induce the degradation of the mRNAs, thus silencing luciferase. Hence, the luciferase-complementary sequence plasmids can serve as sensors to detect the presence of specific mature miRNAs. Any interference of the miRNA processing pathway, for example, by small molecules, that results in the deficiency of mature miRNA processing can be detected using these reporter systems. This assay has been applied in the screening of small molecule collections for inhibitors of miR-21 function (see below).
Fig. 4.
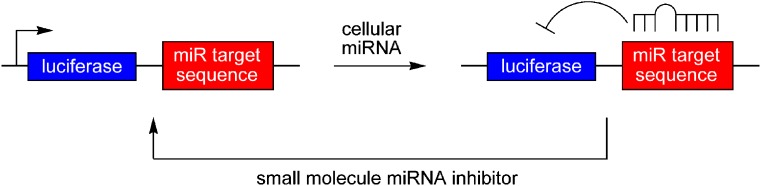
Assay for microRNA (miRNA) function via control of luciferase by a 3′ miRNA target sequence. The presence of the matching miRNA in cells transfected with the reporter construct silences luciferase expression. In the presence of small molecule miRNA inhibitors, the system is reverted back to the expression of the reporter gene which can be detected by an increase in the luciferase signal
Both cell-based assays described here have the advantage that they are not biased towards or against any specific part of the miRNA pathway. Molecules can be identified that can activate or inhibit any component of the pre-transcriptional, transcriptional, and post-transcriptional miRNA processing machinery, as well as the miRNA itself. This increases the chances of identifying hit compounds, and the isolated modifiers of specific miRNAs can subsequently be used as tools to investigate miRNA biogenesis.
SMALL MOLECULE ACTIVATORS OF THE RNA INTERFERENCE PATHWAY
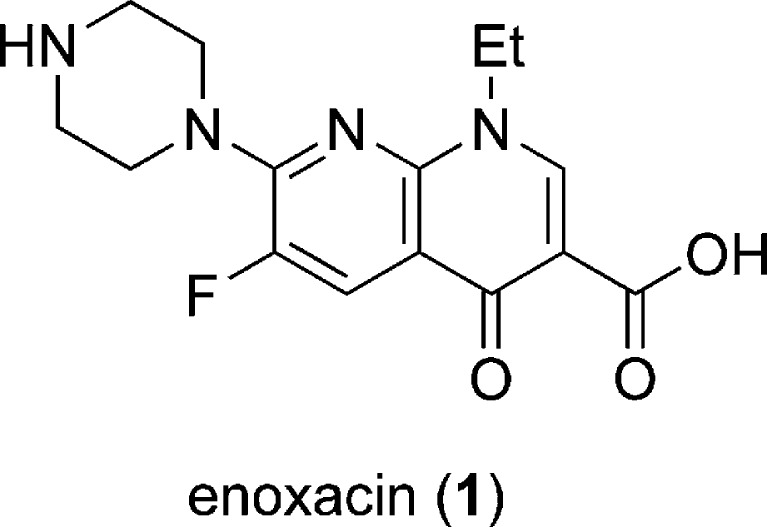
The assay described in Fig. 3 was used to screen a collection of 2,000 small molecules to identify compounds which activate the RNAi pathway and thus induce a further reduction of the GFP signal (46). One molecule, the fluoroquinolone enoxacin (1, see Fig. 5), reduced GFP expression approximately threefold at a 50-μM concentration. The silencing of other endogenous genes by siRNAs either expressed as shRNAs or directly transfected was tested as well. A similar improvement of two- to threefold in the presence of 1 was observed, while unrelated transcripts were not affected, indicating that enoxacin enhances RNAi in a universal fashion. By comparing the gene knockdown efficiency among different concentrations of a transfected siRNA duplex, it was found that enoxacin (1) substantially reduces the siRNA dosage (by a factor of up to 100) required to achieve comparable knockdown efficiency. Fluoroquinolones, like 1, have a broad antimicrobial spectrum and are very successful at treating a variety of bacterial infections (48). The screening of nine additional fluoroquinolones showed no to moderate RNAi-enhancing activity in those molecules. Thus, the RNAi-enhancing activity of 1 does not depend on a general fluoroquinolone activity, but rather on the unique chemical structure of enoxacin and a few related analogs. Importantly, no global effects of 1 on mRNA levels were detected in a microarray-based transcriptional profiling experiment. This is supported by the observation that 1 also does not affect the majority of miRNAs investigated in this study (142/157 miRNAs were unaffected). Further in vitro and in vivo investigations into the mechanism of action of 1 revealed that enoxacin promotes the processing and loading of siRNAs/miRNAs onto RISCs by facilitating the interaction between TAR RNA-binding protein (TRBP) and RNAs. Indeed, it has been shown that the functionality of siRNAs is highly associated with the binding affinity of TRBP (49); therefore, the enhanced interaction between TRBP and RNAs mediated by enoxacin could be the basis of the RNAi-enhancing activity.
Fig. 5.
The activator 1 of the RNAi pathway, discovered from the screening of a small molecule collection
SMALL MOLECULE INHIBITORS OF THE RNA INTERFERENCE PATHWAY
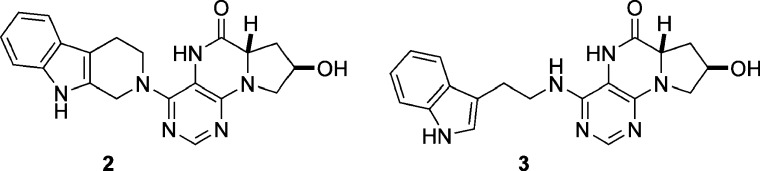
By co-transfecting plasmids expressing RFP and EGFP, together with an siRNA targeting EGFP, in HeLa cells, an assay similar to the activator assay discussed above (Fig. 3) was employed to screen a collection of ATP analogs based on a dihydropteridine scaffold (50). This screen was developed to deliver potential small molecule probes of ATP-dependent events occurring within the RNAi pathway. Two identified compounds, 2 and 3 (Fig. 6), completely inhibited EGFP silencing at 50 μM (and silencing of the endogenous CDK gene at 100 μM). Compound 3 was further investigated for its mode of action, and it was discovered that RISC complexed with single-stranded siRNA or miRNA was not inhibited. However, it appears that 3 specifically affects an early unwinding step in the RNAi pathway, suggesting that its target is an ATP-dependent RNA helicase. Interestingly, no affect of 3 on the miRNA pathway was observed, since let-7 activity was not affected by the inhibitor. The precise molecular target of 3 has not been reported to date.
Fig. 6.
Inhibitors 2 and 3 of the microRNA pathway, discovered from the screening of a small molecule collection of ATP analogs
SMALL MOLECULE INHIBITORS OF A SPECIFIC microRNA, miR-21
miRNA miR-21 was selected as the target for the discovery of specific miRNA inhibitors, because it has been directly linked to several human malignancies. It is upregulated in a large number of primary tumors and cell lines, including glioblastoma (51), breast (43,52), pancreatic (53), head, neck (54), cervical (55), colorectal (56), ovarian (57), and hepatocellular cancer (58). Furthermore, it has been demonstrated that knockdown of miR-21 with antisense agents induced apoptosis and reduced cell growth in glioblastoma (51), breast cancer (43,52), and hepatocellular carcinoma cells (58), presumably by triggering the activation of caspases 3 and 7 (59). This antitumor effect of miR-21 knockdown was also confirmed in xenograft models of glioblastoma (60) and breast cancer (43,52). These reports validated miR-21 as a promising and new target for the development of cancer therapeutics.
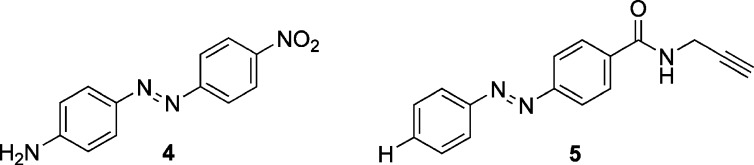
Lentiviral vectors based on the assay shown in Fig. 4 were constructed, containing the complementary sequences of mature miR-21. The Luc-miR-21 reporter, stably introduced into HeLa cells, displayed a 90% decreased luciferase signal in comparison to a control luciferase-linker construct, visualizing a high level of endogenous miR-21 expression in HeLa cells. A primary screen of >1,200 compounds was conducted at a 10 μM compound concentration, and an initial hit compound 4 was discovered (Fig. 7) (47). This diazobenzene led to a 2.5-fold increase of the luciferase signal. Iterations of chemical modification and screening of approximately 100 structurally modified molecules related to the diazobenzene core structure found in 4 delivered the highly active compound 5. This molecule is the most effective small molecule inhibitor of miR-21 to date, inducing an almost fivefold increase in the luciferase reporter signal at a 10 μM concentration, with an EC50 of 2 μM. The diazobenzene 5 is not a general inhibitor of the miRNA pathway but displays specificity for miR-21, as validated by measuring the intracellular miRNA levels via quantitative reverse transcription-polymerase chain reaction. This revealed that levels of the stably expressed, exogenous mature miR-30 and the endogenous mature miR-93 are not reduced by treatment with 5, while miR-21 expression was reduced by 78% in HeLa cells in the presence of 5. In addition, pri-miR-21 levels in cells treated with 5 were reduced by 87%. These results strongly suggest that compound 5 is an inhibitor that targets transcription of the miR-21 gene, but not downstream processes of the common miRNA pathway (47). However, the exact cellular target of 5 is still unknown.
Fig. 7.
microRNA miR-21 inhibitors 4 and 5 discovered from the screening of small molecule collections
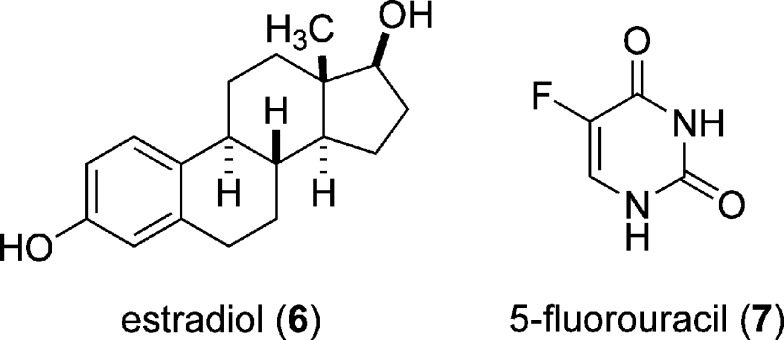
As mentioned above, miR-21 is one of the most significantly upregulated miRNAs in breast cancer, especially estrogen receptor (ER) α positive breast cancer cells (61). However, the mechanisms leading to this upregulation are unknown. Since the development of breast cancer can be stimulated by exposure to estrogens, investigations of the effect of estradiol (6, see Fig. 8) on miR-21 expression have been conducted. Interestingly, two studies came to opposite conclusions, either linking a downregulation of miR-21 (62) or an upregulation of miR-21 after exposure of breast cancer cells to 6 (63). The upregulation of miR-21 levels in the presence of 6 was observed in MCF-7 cells and cell lines derived from MCF-7. An approximately twofold increase in miR-21 expression was observed after a 4 h exposure to 6 (10 nM). Estradiol (6) binds to the ERα, inducing dimerization and translocation into the nucleus, followed by the induction of gene transcription. However, further characterization of the regulation of miR-21 suggests that free ERα represses miR-21 expression (ERα-binding sites were located in the miR-21-regulatory region) and that 6 relieves this repression. It was also found in the same study that 6 induces the expression of Dicer, suggesting that 6 regulates the miRNA pathway on the transcriptional as well as the processing level (63).
Fig. 8.
Chemical structures of estradiol (6) and 5-fluorouracil (7), regulators of miR-21 expression
In contrast, another simultaneously published study on the effect of 6 on miR-21 expression came to the conclusion that 6 leads to a miR-21 downregulation (62). The authors found that exposure of MCF-7 cells to 6 (10 nM, 6 h) reduced miR-21 levels approximately twofold. This estradiol-induced downregulation of miR-21 correlated with an increasing expression of miR-21 target genes, including PDCD4, PTEN, and BCL-2. Moreover, the ER antagonist 4-hydroxytamoxifen inhibited reduction of miR-21 levels by 6 (62). These controversial results reveal the complexity of miRNA regulatory networks and demonstrate the necessity to further investigate miRNA biogenesis and regulation.
Several anti-cancer drugs interfere with nucleic acid function and nucleic acid metabolism, and thus could potentially affect miRNA expression levels. This has been tested for 5-fluorouracil (7, see Fig. 8), an established chemotherapeutic agent that acts as a thymidine biosynthesis inhibitor and is applied in the treatment of colon and pancreatic cancer. Exposure of two colon cancer cell lines, C22.20 and HC.21, to 7 for 6 days at a therapeutic relevant concentration of 10 μM led to a two- to eightfold increase of several miRNAs, including miR-21 (64). The authors of this study speculated that the upregulation of anti-apoptotic miRNAs, like miR-21, could be a cellular response to the cytotoxic effects of the drug.
SMALL MOLECULES THAT DIRECTLY INTERACT WITH miRNAs
Several small molecules have been reported that directly bind to RNAs (65,66), including miRNAs (67,68). Helix-threading peptides were discovered, which consist of an acridine core that acts as a RNA-duplex intercalator, which is flanked by short peptides (e.g., N-Ser-Val-acridine-Arg-C) (67). A selection of RNA binders using the SELEX process followed by RNA footprinting experiments revealed a minimal five base-pair dsRNA sequence for efficient small molecule binding, with a binding constant of 16 μM. The base pairing in that sequence is imperfect and is interrupted with bulges, which is necessary for the peptide-intercalator conjugate to thread through the RNA duplex. The same RNA sequence has been found in pre-miR-39 from Caenorhabditis elegans, and binding of the small molecule might interfere with miR-39 processing. However, no pre-miR-39 binding of the compound has been demonstrated to date.
In another report, an shRNA-targeting luciferase was modified with a G-rich loop and three additional Gs at the 5′ end, in order to enable potential G-quadruplex formation (68). Even though no evidence for actual G-quadruplex formation of this oligomer was provided, a set of classical G-quadruplex small molecule binders were screened for their ability to inhibit processing of the shRNA. Two bisquinolinium compounds and a porphyrazine showed selective inhibition of the processing of the G-modified shRNA compared to a control shRNA. The most selective bisquinolinium displayed complete Dicer inhibition at 7.5 μM and a binding constant of 290 nM to the G-modified shRNA, while no binding could be observed to the control shRNA. However, the same compound did not show any activity in a cell-based luciferase assay. A possible explanation for this observation could be the sequestering of the compound by other intracellular G-quadruplexes (69).
Both approaches described above were developed starting with the small molecules, followed by the identification of suitable miRNA binding sequences. Although these studies provide important information on cellular RNA-small molecule interactions and the design of small molecule binders, both approaches are not as general as selecting a certain miRNA and then developing a small molecule inhibitor of its function by conducting a small molecule screen (as shown in Figs. 3 and 4). Moreover, no miRNA inhibitory activity of the described peptide-intercalator conjugate or the quadruplex binding compounds was reported.
CONCLUSION
The recent development of small molecule activators and inhibitors demonstrates that the miRNA and RNAi pathway represents a viable small molecule target. Several screens have been developed to discover molecules which either activate or inactivate the RNAi pathway in a general fashion or with miRNA sequence specificity. The discovered small molecule modifiers of the miRNA and siRNA pathway represent unique tools to further explore the regulation and biogenesis of RNAs involved in the post-transcriptional silencing of genes and their implication in human pathologies. These small molecules have several advantages over traditional nucleic acid-based tools to manipulate miRNA function, since they are more easily delivered into humans or animals, they are more stable intracellularly, and they are less expensive to manufacture. Based on these promising developments, it can be expected that several new small molecule activators and inhibitors of miRNA and siRNA function will be discovered in the near future.
Acknowledgments
Financial support from the Department of Chemistry at North Carolina State University and the American Chemical Society (TEVA USA Scholars Grant) is acknowledged. The author apologizes to those researchers whose work could not be discussed due to space limitations. AD is a Beckman Young Investigator, a Cottrell Scholar, and a recipient of an NSF CAREER award.
References
- 1.Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16(2):203–8. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 2.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 3.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10(2):94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20(5):515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 5.Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005;122(1):17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 6.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431(7006):343–9. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 7.Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132(21):4645–52. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- 8.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci U S A. 2007;104(23):9667–72. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318(5858):1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 10.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 11.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 12.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294(5543):862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 13.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 14.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94(6):776–80. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Appasani K. MicroRNAs: from basic science to disease biology. Cambridge: Cambridge University Press; 2008. [Google Scholar]
- 16.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16(6):861–5. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432(7014):231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 18.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 19.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 21.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106(1):23–34. doi: 10.1016/S0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 22.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 23.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15(20):2654–9. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16(1):4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404(6775):293–6. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 27.Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297(5589):2056–60. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103(24):9136–41. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan SP, Slack FJ. microRNA-mediated silencing inside P-bodies. RNA Biol. 2006;3(3):97–100. doi: 10.4161/rna.3.3.3499. [DOI] [PubMed] [Google Scholar]
- 30.Kong YW, Cannell IG, de Moor CH, Hill K, Garside PG, Hamilton TL, et al. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci U S A. 2008;105(26):8866–71. doi: 10.1073/pnas.0800650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17(3):118–26. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9(6):435–43. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 36.Shi XB, Tepper CG, deVere White RW. Cancerous miRNAs and their regulation. Cell Cycle. 2008;7(11):1529–38. doi: 10.4161/cc.7.11.5977. [DOI] [PubMed] [Google Scholar]
- 37.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 38.Shah YM, Morimura K, Yang Q, Tanabe T, Takagi M, Gonzalez FJ. Peroxisome proliferator-activated receptor alpha regulates a microRNA-mediated signaling cascade responsible for hepatocellular proliferation. Mol Cell Biol. 2007;27(12):4238–47. doi: 10.1128/MCB.00317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20(16):2202–7. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454(7200):56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, et al. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459(7249):1010–4. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 43.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 44.Davies BP, Arenz C. A homogenous assay for micro RNA maturation. Angew Chem Int Ed Engl. 2006;45(33):5550–2. doi: 10.1002/anie.200601332. [DOI] [PubMed] [Google Scholar]
- 45.Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, et al. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11(5):674–82. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shan G, Li Y, Zhang J, Li W, Szulwach KE, Duan R, et al. A small molecule enhances RNA interference and promotes microRNA processing. Nat Biotechnol. 2008;26(8):933–40. doi: 10.1038/nbt.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A. Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl. 2008;47(39):7482–4. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhanot SK, Singh M, Chatterjee NR. The chemical and biological aspects of fluoroquinolones: reality and dreams. Curr Pharm Des. 2001;7(5):311–35. doi: 10.2174/1381612013398059. [DOI] [PubMed] [Google Scholar]
- 49.Katoh T, Suzuki T. Specific residues at every third position of siRNA shape its efficient RNAi activity. Nucleic Acids Res. 2007;35(4):e27. doi: 10.1093/nar/gkl1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chiu YL, Dinesh CU, Chu CY, Ali A, Brown KM, Cao H, et al. Dissecting RNA-interference pathway with small molecules. Chem Biol. 2005;12(6):643–8. doi: 10.1016/j.chembiol.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 51.Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334(4):1351–8. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 52.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26(19):2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 53.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120(5):1046–54. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tran N, McLean T, Zhang X, Zhao CJ, Thomson JM, O'Brien C, et al. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun. 2007;358(1):12–7. doi: 10.1016/j.bbrc.2007.03.201. [DOI] [PubMed] [Google Scholar]
- 55.Lui WO, Pourmand N, Patterson BK, Fire A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res. 2007;67(13):6031–43. doi: 10.1158/0008-5472.CAN-06-0561. [DOI] [PubMed] [Google Scholar]
- 56.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 57.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67(18):8699–707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 58.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 60.Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67(19):8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 61.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009;37(8):2584–95. doi: 10.1093/nar/gkp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bhat-Nakshatri P, Wang G, Collins NR, Thomson MJ, Geistlinger TR, Carroll JS, et al. Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res. 2009;37(14):4850–61. doi: 10.1093/nar/gkp500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rossi L, Bonmassar E, Faraoni I. Modification of miR gene expression pattern in human colon cancer cells following exposure to 5-fluorouracil in vitro. Pharmacol Res. 2007;56(3):248–53. doi: 10.1016/j.phrs.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 65.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem Rev. 2008;108(4):1171–224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 66.Tor Y. Targeting RNA with small molecules. ChemBioChem. 2003;4(10):998–1007. doi: 10.1002/cbic.200300680. [DOI] [PubMed] [Google Scholar]
- 67.Gooch BD, Beal PA. Recognition of duplex RNA by helix-threading peptides. J Am Chem Soc. 2004;126(34):10603–10. doi: 10.1021/ja047818v. [DOI] [PubMed] [Google Scholar]
- 68.Henn A, Joachimi A, Goncalves DP, Monchaud D, Teulade-Fichou MP, Sanders JK, et al. Inhibition of dicing of guanosine-rich shRNAs by quadruplex-binding compounds. ChemBioChem. 2008;9(16):2722–9. doi: 10.1002/cbic.200800271. [DOI] [PubMed] [Google Scholar]
- 69.Rezler EM, Bearss DJ, Hurley LH. Telomere inhibition and telomere disruption as processes for drug targeting. Annu Rev Pharmacol Toxicol. 2003;43:359–79. doi: 10.1146/annurev.pharmtox.43.100901.135733. [DOI] [PubMed] [Google Scholar]