Abstract
Whole-body autoradiography ((WBA) or quantitative WBA (QWBA)), microautoradiography (MARG), matrix-assisted laser desorption/ionization mass spectrometric imaging (MALDI-MSI), and secondary ion mass spectrometric imaging (SIMS-MSI) are high-resolution, molecular imaging techniques used to study the tissue distribution of radiolabeled and nonlabeled compounds in ex vivo, in situ biological samples. WBA, which is the imaging of the whole-body of lab animals, and/or their organ systems; and MARG, which provides information on the localization of radioactivity in histological preparations and at the cellular level, are used to support drug discovery and development efforts. These studies enable the conduct of human radiolabeled metabolite studies and have provided pharmaceutical scientists with a high resolution and quantitative method of accessing tissue distribution. MALDI-MSI is a mass spectrometric imaging technique capable of label-free and simultaneous determination of the identity and distribution of xenobiotics and their metabolites as well as endogenous substances in biological samples. This makes it an interesting extension to WBA and MARG, eliminating the need for radiochemistry and providing molecular specific information. SIMS-MSI offers a complementary method to MALDI-MSI for the acquisition of images with higher spatial resolution directly from biological specimens. Although traditionally used for the analysis of surface films and polymers, SIMS has been used successfully for the study of biological tissues and cell types, thus enabling the acquisition of images at submicrometer resolution with a minimum of samples preparation.
Key words: MALDI-MSI, microautoradiography, SIMS-MSI, whole-body autoradiography
INTRODUCTION
Autoradiography (ARG), matrix-assisted laser desorption imaging mass spectrometry (MALDI-MSI), and secondary ion mass spectrometric (SIMS) imaging technologies are used across various disciplines of science to identify and/or quantify molecular entities in various environments, and they have become powerful tools to characterize pharmaceutical entities during drug discovery and development. Specifically, these technologies shed the highest resolving light on the quantitative distribution of parent molecules and their metabolites in biological samples, such as the entire body, and organ systems, at the tissue and cellular level. No other current imaging technologies match the image resolution and/or compound identification and quantitation provided by these technologies, and their combined use and continued improvements will certainly prove to be more valuable in drug discovery and development in the future. This paper will discuss the history, validation, current applications, and limitations of ARG, MALDI-MSI, and SIMS molecular imaging in pharmaceutical discovery and development.
AUTORADIOGRAPHY
ARG is truly the first molecular imaging technique used for the localization of radioactivity in biological specimens. ARG is defined as “self” “radioactivity” “writing,” and it has become a collection of techniques where radioactive particles/energy (beta/gamma) localized within a solid substance is imaged by close apposition to a detection media. The original detection media was X-ray film, or another type of media that contained a photographic emulsion, and the radioactivity was derived from a compound with an attached radioisotope label of relatively low energy, such as a beta particle. Simply stated, ARG is a set of “photographic” methods for recording the spatial distribution and relationships of radioisotopes within or on a (solid) specimen (1). Although ARG has applications in botany, entomology, microbiology, biology, and the material sciences, this review will focus on its use for the discovery and development of large and small molecule pharmaceutical compounds. Over the years, technology has vastly improved the utilization of the autoradiographic process, and today, it can be generally divided into macroautoradiography, which includes whole-body autoradiography (WBA), and microautoradiography (MARG).
WBA entails the spatial imaging of radiolabeled substances in large samples, such as the entire body of lab animals (such as arthropods, fish, rodents, lagomorphs, canines, and nonhuman primates). WBA has the ability to show the distribution of radioactivity in all organs and tissues of an intact animal carcass. The original WBA method required long exposure times (from weeks to months), but the images were of exquisitely high resolution. However, phosphor-imaging technology has all but replaced the photographic techniques and also provides quantitative WBA (QWBA) tissue concentration data, which can be obtained in days (typically 4–7 days; 2). This was an important advancement that has advanced the study of tissue distribution of drugs in lab animals. The former standard method of assessing drug distribution utilized organ homogenates obtained from dissected animals. That technique provides limited and sometimes misleading information because it is an organ homogenate. Unfortunately, outdated organ homogenate techniques are still being used and are misrepresented as “tissue distribution” studies. This assay is also limited in the data it provides because it can only provide information on organs that are collected, and so, a lot of information related to the remaining tissues is lost after the carcass is discarded. A more detailed description of QWBA will be presented later in this review.
MARG is a histological technique that provides an image of the distribution of radiolabeled compounds in tissues and organs at the cellular level. This technique, which relies on direct exposure of the radioactive entity to a photographic emulsion, is prone to artifacts and cannot provide quantitative data. However, its strength lies in its ability to examine the spatial distribution of radioactivity at the tissue and cellular layer, and it serves as an important informational link from the whole-body, organ, and tissue levels to the cellular level. A further description of this technique and it applications and limitations will also be presented later in this manuscript.
Quantitative Whole-Body Autoradiography
A brief description of the QWBA technique (1) begins with the administration of a radioactive test substance (beta particle emitters, such as 14C, 3H, and 125I) to a lab animal followed by euthanasia and snap freezing of the carcass (typically in a dry ice–hexane bath). Frozen carcasses are then freeze embedded in a carboxymethylcellulose block, cryosectioned at 30–50 µm, and sections are collected onto adhesive tape. The sections on tape are dehydrated and then apposed to phosphor-imaging plates, or X-ray film, along with a set of radioactive calibration standards. Exposure times vary from a few days to a few weeks for phosphor-imaging plates, or for weeks to months for X-ray film. Digital images are obtained to enable tissue quantification of radioactivity concentrations by image analysis.
WBA was a crude method in the beginning. In 1867, Niepse de Saint Victor first described the phenomenon of autoradiography, which he described as the “persistent activity due to an unknown chemical radiation” (3). This observation lead London to performing an experiment in which an autoradiographic image of a frog treated with radium was first produced in 1904 (4). To that end, ARG is truly the first molecular imaging technique used for the localization of radioactivity in biological specimens. Fifty years later, Dziewaitkovski used beta radiation to investigate the localization of compounds in biological samples (5) followed by the development of the whole-body autoradiography technique by Ullberg in 1954 (6) who pioneered the technique by administering 35S-penicillin to mice followed by freeze embedding them in water-soaked cotton using dry ice. He then sectioned their entire body bodies using a large microtome in a walk-in freezer. The whole-body sections he produced were then opposed to X-ray film which produced the autoradiographs of tissue distribution. Methods for sectioning the whole bodies of lab animals were also developed and methods included the use of dry ice-cooled microtomes (7); exposing the surface of ground down frozen carcasses (8); abrasion of resin-embedded carcasses (9); thick sectioning using a circular saw (10); and finally, the development of a large microtome held inside a chest freezer (11). Leica Microsystems, Inc. (Nussloch, Germany) began manufacturing commercially available large format cryomicrotomes, and they are the past and current leading provider of large cryomicrotomes used for WBA today.
Quantitation of autoradiographs was the next challenge for pioneering macroautoradiographers and Berlin and Ullberg (12), and Kutzim (13) made the first attempts to quantify tissue concentrations in autoradiographs using early image analysis techniques. Unfortunately, the results achieved were only semiquantitative, but in the following years (1974–1987), several investigators (14–19) researched methods to better determine quantitative data from autoradiographs with limited success owing to the inherent nonlinearity of film. Also during this time, Schweitzer (20) developed an image calibration method using 14C-spiked blood standards at concentrations bracketing the sensitivity of phosphor-imaging plates. This robust technique continues to be used by many investigators today. Luckey revolutionized WBA in 1975 by developing and patenting phosphor-imaging technology or radioluminography (2), which provided images capable of providing digital images from whole-body sections within days. Most importantly, these images enabled the direct determination of quantitative tissue concentrations over four to five orders of magnitude, and validation efforts began. A technical validation of the phosphor-imaging instrument, and QWBA methods, which more completely described the principals, specifications, and limitations of the instrumentation and QWBA, was published in 2000 in a special edition of the Journal of Regulatory Toxicology and Pharmacology (21). In 1994, a group of autoradiographers in the pharmaceutical industry formed the Society for Whole-Body Autoradiography (SWBA) whose mission was to promote the use of QWBA over traditional organ dissection homogenate methods to determine tissue distribution of new drugs. Further work parameterized quantitative aspect to meet stringent bioanalytical expectations (22–27), and in 1990, a Japanese collaboration of >20 companies proposed that QWBA should replace the use of traditional dissection and liquid scintillation counting (LSC) assay to determine true tissue distribution during the drug development (28). Dr. Yasuo Ohno of the National Institute of Health Science (Tokyo, Japan) concluded his presentation at the 1997 meeting of the Society for Whole-Body Autoradiography by stating that the Japanese Ministry of Health and Welfare would accept QWBA data in lieu of traditional organ dissection distribution studies for the approval of new drugs as long as the procedures were appropriately validated. Today, pharmaceutical companies have almost entirely eliminated the use of dissection studies to determine the distribution of new pharmaceuticals, and anecdotal information to the author has suggested that the Food and Drug Administration is now requesting that “QWBA” studies be provided especially to answer certain questions that arise during drug development.
In addition to autoradiography and autoradioluminography, direct nuclear imaging technologies were also developed. These instruments utilize ionization chambers and different imaging technologies (e.g., scintillating sheet, charge-coupled device camera) and were developed by Jeavons in 1983 (29). Today's instrument consists of a parallel plaque avalanche chamber, which is based on the invention by Charpak in 1989 (30). These instruments, which are currently sold by Biospace Lab (Paris, France), also image radioactivity in whole-body and smaller tissue sections, and have the ability to acquire quantitative images in real time. These instruments have proved most useful in drug discovery because they are capable of dual-isotope analysis, and they provide data quickly. One limitation for these systems is that only a limited number of sections can be analyzed at one time, and the instruments require regular and careful maintenance.
The key strength of the QWBA technique is that it shows true tissue distribution of radiolabeled test articles in a relatively unadulterated, in situ sample. Phosphor imaging has also been shown to be a very robust yet sensitive technology for the quantification of radioactivity in whole-body sections. Its wide linear range and sensitivity that can reliably quantify ∼45 dpms distributed over an area of one half square centimeters far exceeds that achieved by LSC for similar sized samples. Phosphor imaging is also able to image the relatively weak energy of 14C and 3H, which fortunately are also long-lived isotopes so that drugs/metabolites with very long half-lives can be tracked in the body of animals over years, which is not possible using the relatively short-lived isotopes used for in vivo positron emission tomography (PET) and single-photon emission computed tomography imaging. In general, QWBA provides a much higher quality data set than that provided by organ dissection and homogenate assay by providing concentration data on all tissues; not just those chosen for typical dissection studies, which eliminates the possibility of missing organs that may have high concentrations. The images can also be reviewed and analyzed at any time so that if an issue arises after the tissue distribution has been completed, then investigators can go back and review and quantify the nonroutine tissue(s) of interest. QWBA also eliminates the variables of cross-contamination of organs and the variable effects of organ exsanguination that inevitably occurs during organ removal. The whole-body-freezing procedure, which takes approximately 5–15 min, which depends on body weight, to halt biological and metabolic processes, also decreases the interanimal variability of the data, and this has been routinely experienced by whole-body autoradiographers. This observation supports the use of a study design where one animal/time point and more time points are used, which in turn provides more reliable pharmacokinetic (PK) parameters for a more accurate description of tissue compartment PK. This approach also satisfies and supports the clinical use of lab animals. The techniques used for QWBA can vary across different laboratories, however, the combined methods have been thoroughly developed, tested, and reviewed, and are very robust (31,32). In short, QWBA provides a higher quality, more detailed, and more useful data, while using less animals, than is required for organ homogenate assay studies.
Of course, QWBA has several limitations that need to be considered. The first, which is common to all radiolabeled studies, is that the technique provides data on the concentration of radioactivity only. That is to say, investigators do not know the actual molecular identity of the radioactivity they are tracking and/or quantifying. Tissue concentrations determined by QWBA (or LSC) can include parent drug, plus its metabolites, and/or degradation products. This is most problematic when using 125I-labeled peptides and proteins, and to a lesser extent, when using 3H, because the in vivo stability of these two radiolabels is often not too good, and the radiolabel can be lost as free 125I or 3H2O. When the radiolabel dissociates from the parent molecule, then investigators are faced with the responsibility of characterizing the extent to which the radiolabel has been lost and then adjusting their tissue concentration appropriately and/or stating the fact that the results are semiquantitative. Additionally, QWBA sections are typically dehydrated to facilitate further processing, and so, volatile metabolites are lost, although this situation probably does not occur that often and may be able to be predicted when the chemistry is well understood. This is where the strengths of MALDI-MSI can be realized, especially because the same sections used for QWBA can be used for analysis by MALDI-MSI as will be presented in the next section of this article. Another limitation of QWBA is that it is difficult to evaluate short-lived isotopes, like those used for radiopharmaceuticals, due to the processing time required, although it is possible to alter the processes to obtain data more quickly, but it requires special attention. For example, when using 90Y, 11C, or 18F as the radiolabel, which have half-lives of 2.67 days, 21 min, and 60 min, respectively, as the radiolabel, which have a half-life of 2.67 days, respectively, the frozen “wet” sections may need to be exposed to imaging plates immediately and while under freezer conditions, otherwise it could take too long to dehydrate the sections, and half of the radioactivity could be lost thus decreasing the sensitivity of the technique. Despite these technical drawbacks, Kaim et al. (33) were able to demonstrate that the uptake of 18F-FET in nonneoplastic inflammatory cells in an experimental soft tissue infection model was lower than that of 18F-FDG, allowing thus to predict a higher specificity for the detection of tumor cells using 18F-FET.
Another limitation is cost. The instrumentation for QWBA is relatively expensive, and highly trained staffs are needed to perform the processing and analysis, which can be a big limitation for some labs. QWBA is also limited in that it cannot adequately provide data at the microscopic level due to the freezing technique. Despite the “snap-freezing” techniques used, the freezing of all tissues is too slow to prevent cellular damage due to ice crystal formation. This makes receptor localization/staining difficult and cellular identification much more difficult, so that radiolabeled test articles cannot be easily colocalized to specific molecular targets. Other limitations, that also affect dissection studies, include the influence of the experimental conditions, such as radiochemical purity of the test compound, nominal and radioactive doses given to the animals, anesthesia, and methods of euthanasia. Investigators need to ensure that they are working with radiolabeled material that has the proper specific activity and radiopurity so that the proper dose of radioactivity can be given and to make sure that the radioactivity being quantified is actually compound related and not a radioimpurity to ensure quantifiable results related to the test compound. The use of anesthesia and method of euthanasia can also affect the tissue distribution of any test article and should be adjusted in cases where the investigator(s) think it may affect distribution of their test article. For example, it is commonly thought that CO2 alters the permeability of the blood–brain barrier (acidosis) and therefore, alters normal brain penetration. Thus, euthanasia by CO2 inhalation may not be a good technique to use if the compound is targeted for the brain. Likewise, euthanasia by exsanguination, which must be used for tissue dissection studies, will undoubtedly have an effect on tissue distribution due to the massive fluid changes that take place in the body during that process. This is another benefit of QWBA over organ dissection because most often animals for QWBA are not exsanguinated and remain intact.
There are also limitations that are related to the instrumentation and methods. Difficulties can arise during cryomicrotomy, where there are limitations on section thickness, sample size, and sample characteristics. The thickness of whole-body sections, which is determined by the cryomicrotome settings, needs to be thick enough to be collected on tape, and also so, there is enough tissue and radioactivity present to produce an image for quantitation. The section thickness and isotope of the calibration standards must also match the test sample to ensure proper imaging calibration and determination of tissue concentrations of drug-derived radioactivity. Additionally, the cryomicrotome has its own functional limitations, such as how thin it can section, vertical blade movement (for thicker samples), blade sharpness, cryochamber size and ability to dehydrate sections, and overall sample size that can fit in the microtome. These are just a few of the several functional limitations that must be considered when using the microtome. The phosphor imager used to obtain the images for analysis also has its limitations, such as image resolution (pixel size), lower limits of detection (typically ∼2,220 dpm/g/0.5 mm2 image region from sections that are ∼40-µm-thick section). Typical exposure times for QWBA range from 3 to 4 days depending on the isotope and radioactive dose administered to the animal. However, despite the current limitations, QWBA still provides the best way to determine tissue concentrations in situ.
Quantitative Whole-Body Autoradiography Applications
QWBA offers many applications for researchers in drug discovery and development and can best answer questions related to the biodistribution of new or old drugs. Schweitzer et al. (34) investigated the distribution of [14C] Diclofenac sodium (Voltaren®), which is a drug that has been marketed for years, after a single oral administration in rats. These investigators showed that Diclofenac preferentially distributed into the inflamed tissues and achieved exposures that were 26- and 53-fold higher in the inflamed neck and inflamed paws of treated animals than in control rats. All other tissues in treated animals and control animals showed similar distribution and exposure. Most often, QWBA is used during the drug development stages and to support regulatory submissions. The data are used to show tissue pharmacokinetics and to predict human radiation dosimetry that might occur during human-radiolabeled absorption, distribution, metabolism, and excretion (ADME) studies. A routine tissue distribution performed using QWBA may routinely evaluate 35–40 tissues over various study periods that often go as long as 35 days postdose, and by having ten or more time points, which provides more reliable pharmacokinetic parameters for all tissues that can be measured. However, QWBA data is also used in drug discovery to support the selection of new drug candidates, based on tissue distribution/uptake/retention, and also to help evaluate and identify potential toxicological issues (35). Foster et al. (36) demonstrated that the immunomodulatory drug FTY720 and its phosphorylation product were specifically localized to the central nervous system white matter, and preferentially along myelin sheaths, following single and multiple oral doses. Weiss et al. (37) showed that the bisphosphonate zoledronic acid was initially rapidly eliminated from plasma and noncalcified tissue but only slowly from bone, whereas the terminal half-lives of elimination from these tissues were similar, which suggest redistribution of drug from the bone rather than prolonged retention in the latter. Potchioba and Nocerini (38) have described how tritiated compounds can be used to determine tissue distribution characteristics early on during the drug discovery phase to identify lead compounds. The key observation and rational for the use of 3H was that the tritium-labeled material could be obtained much quicker than the more stably labeled 14C compound (2 weeks versus 2–3 months), however, they did not discuss how they assured in vivo radiostability of the 3H, which is known to be exchanged with water in vivo, and which can result in a loss of the 3H, thus affecting tissue quantitation. This is something that needs to be addressed whenever 3H will be used for quantitative tissue distribution by QWBA. The paper also failed to mention that the cost of using tritiated compounds for QWBA can be very expensive because the phosphor-imaging plates used for 3H can be used only once. Furthermore, these imaging plates are relatively small and can fit only about six whole-body sections on them, and so, two or more plates are required for each rat. These authors also reported that a 17-day exposure time was used, which is much longer than that used for 14C (typically approximately a 1–4-day exposure). The use of tritiated compounds to support drug discovery is not new though and was being routinely used by pharmaceutical companies (E. Solon, presented at the European Society for Autoradiography meeting in Heidelberg, 1998).
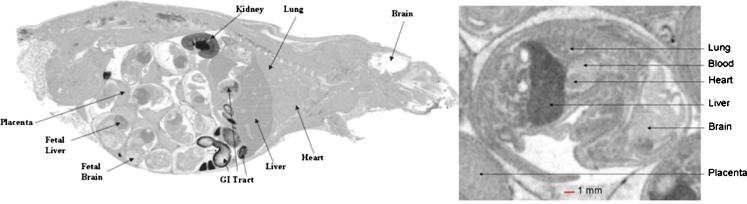
QWBA is a very good technique to study placental and milk transfer of xenobiotics in rodents, owing to the high resolution offered so that small tissues of the fetus can be positively identified and quantified (see Fig. 1). Bruin et al. (39) showed that following oral dosing of 14C-labeled deferasinox (Exjade, ICL670) to rats, that placental transfer was limited, but that approximately 3% of the dose was transferred into the breast milk. Phosphor imaging can also be used on sections of individual organs, such as kidney, brain, and eyes, where high resolving power may be needed to screen drug penetration into the finer structures of organs. Figure 2 presents an example of an in vitro competitive binding assay in brain sections that was imaged and quantified using phosphor imaging of brain sections incubated in different solution of test drug and known receptor antagonists. Figure 3a shows the utility of using cryosectioning and phosphor imaging of dog eyes that have been exposed to various test articles to evaluate ocular distribution. This level of detail can also be obtained in rat eyes (Fig. 3b), and it can be very important when the data will be used to determine human dosimetry exposure and there is an association of the test article with melanin. QWBA is the probably the best technique currently available to enable the evaluation of test article association to melanin. This is particularly important because while tissue dissection may not detect concentrations of drug-derived radioactivity in the pigmented uveal tracts of lab rats (due to the small sample size and relative low sensitivity), QWBA can image these small pigmented tissues and can provide the concentration data necessary to enable a more precise prediction of a possible health risk to human volunteers participating in a human radiolabeled ADME study.
Fig. 1.
An example of an image obtained during a placental transfer study conducted using QWBA. Whole-body autoradioluminographs of a pregnant rat (day 17; left) and a 17-day-old rat fetus (right) showing differential distribution of 14C-AZT-derived radioactivity in liver and brain
Fig. 2.

An example of the type of results that can be obtained after an in vitro rat brain section competitive binding assay screen using quantitative phosphor imaging. Control rat brain sections (∼40 µm thick) can be incubated in solutions with a radiolabeled test compound known to bind brain receptors in the presence or absence of competing ligands to screen new compounds in drug discovery
Fig. 3.
Dog and rat eyes shown by autoradioluminography. a Dog eye individually cryosectioned at 40 µm thick and imaged using phosphor storage technology. Adjustments to the gray scale appearance facilitate tissue identification for quantification. b Melanin association. A pigmented and albino rat eye exposed to the same test article. In situ samples as imaged by phosphor storage technology from whole-body cryosections. Differential distribution of the test article shows a high concentration in the pigmented uveal tract, while a lower concentration of radioactivity is seen in the uveal tract of an albino rat
125I-labeled proteins and peptides are being used more often these days in QWBA analyses to study tissue distribution and pharmacokinetics in the Biotech arena. Although these studies are not often required for regulatory submission, Biotech companies are finding that the data gleaned from these studies have helped them to the better understand and select their compounds in development. Although the in vivo stability of 125I on most large molecule xenobiotics is not 100%, the image data can still be useful, especially when target tissue/organ distribution data are needed, and semiquantitative data can be obtained with confidence when the in vivo stability and amount of free 125I circulating in the body is estimated. However, it is important for investigators using 125I to consider that the following tissues/organs either contain iodine symporters, and/or organify free iodine, thus these tissue concentrations must be interpreted with caution: thyroid gland, stomach, kidneys, mammary gland, salivary gland, thymus, epidermis, and choroid plexus (40).
QWBA studies are also routinely used to evaluate tissue distribution of xenobiotics in large lab animals such as rabbits, dogs, and monkeys (see Fig. 4). In these cases, the animal's carcass must be able to fit into a freezing frame block of approximately 40 × 15 × 15 cm for sectioning. However, if the frozen carcass is larger than those dimensions, then it can be subdivided so that all parts of the carcass can be sectioned and analyzed. In this way, even larger samples, such as the distribution into pigs, can be analyzed (41).
Fig. 4.
Monkey QWBA. The image on the left shows a monkey carcass being cryosectioned for QWBA, and a sample phosphor image of a monkey is on the right
QWBA has also been used to answer very specific questions related to issues that can arise during drug discovery. For example, target organ penetration (tumor, brain), tracking the distribution of oligonucleotides and nanoparticles, and drug–drug and drug–food interactions (40).
QWBA is a versatile tool which can provide pharmaceutical scientists with quantifiable high-resolution images of the distribution of xenobiotics in practically any biological sample, large or small, and when the proper study design is applied, a wealth of knowledge can be obtained from a single study. The benefits of QWBA and updated study designs over the outdated tissue dissection technique are numerous and substantial. To that end, regulatory authorities should encourage drug developers use QWBA instead of organ dissection and homogenization to conduct tissue distribution studies whenever possible. This is because QWBA provides the highest quality quantitative data and a more complete analysis of true tissue distribution, which is especially important when trying to predict human exposure to radiation during human-radiolabeled studies.
Microautoradiography
MARG provides pharmaceutical scientists with a high-resolution tool to investigate spatial localization of radiolabeled drugs at a tissue and cellular level. MARG is especially good at providing insight regarding in vivo receptor binding in various cell types and has predictive value for specific drug targeting. In this respect, it has been used widely in academic settings where it can provide important information on cellular mechanisms. MARG has applications in all areas of science, but this report will discuss examples in drug metabolism, pharmacology, toxicology, and molecular biology.
The methods used by the author which are described below are based on the methods of Appleton (42) and Stumpf (43), but it is important to realize that there are many variations of the method presented in the literature. To begin, an animal is dosed with a radiolabeled substance (typically 3H, 14C, 35S, or 125I), the animal is exsanguinated, and tissues are dissected and snap frozen in isopentane that is chilled in liquid nitrogen. The tissue is then cryosectioned at −20°C (or the optimal cutting temperature for a given tissue/organ) to obtain 4−5-µm-thick sections. Then, under darkroom conditions, sections are thaw mounted onto dry glass microscope slides that have been precoated with nuclear photographic emulsion. The slides are placed into a light-tight box with desiccant and allowed to expose for an appropriate amount of time. The collection of sections onto dry slides is a key step developed by Appleton (42), and it eliminates the possibility of diffusion of soluble compounds, which can happen during slide and section dipping into an aqueous emulsion. In contrast, the original Stumpf and Roth method (44) involved collection of the section into vials for freeze drying, which required very careful section handling and was very time consuming and prone to sample destruction. Following exposure, the slides are developed in a manner similar to developing photographic film before being stained using conventional histological staining protocols. This may include immunostaining techniques that can provide positive colocalization of drug-derived radioactivity to known cell types, receptors, and/or other structures/markers for which antibody staining protocols exist (43).
The first microautoradiographic data were produced by Lacassagne in 1924 (45), which lead to further work by Bélanger and Leblond (46), who poured liquid photographic emulsion onto histological sections to reveal the location of radioactive substances in the tissues. Joftes and Warren in 1955 (47) revised that technique and dipped slides into photographic emulsion, which gained wide use due to its ease of manipulation. This technique has survived the years and is sometimes used today. However, if diffusible radiolabeled compound are imaged, the results can be useless owing to the relocation of the radiolabeled substance, which produces telltale artifacts that can invalidate experiments and discourage investigators. During the 1940s, methods utilizing strips of dried emulsion were developed where the histological sample that contained a radioactive substance was placed in direct contact with the dried emulsion strip and allowed to expose it over time. This method proved cumbersome though because the technician would need to maintain the precise position of the strip on the samples during exposure, development, and staining, and often the alignment could not be maintained and the entire process would end in failure. However, in 1964, Appleton (48) first developed the technique of collecting cryosections onto slides covered with strips of dried emulsion using a thaw-mounting technique. This required sectioning and collection of sections in a darkroom and under safelight conditions, which requires a dedicated staff who can master and maintain their skills. The use of cryopreservation and cryosectioning remains critical to the study of diffusible substances because it maintains the spatial locale of the radiolabeled substance in the matrix whereas liquid tissue fixation steps most often solubilize and relocate the diffusible test article. However, when substances are tightly bound to cellular structures (e.g., receptor proteins), positive results may still be obtained from samples processed using conventional histology techniques. MARG procedures are also still used reliably by molecular biologists to detect RNA molecules in situ and to study the localization of genes and DNA sequences in histological preparations owing to their relatively stable positions in cells. Further refinement of MARG techniques occurred during the 1960s by Caro (49), and shortly after that, Stumpf and Roth (44) made additional improvements to establish receptor autoradiography as a more reliable technique. This established the basis for the current MARG techniques. Numerous elaborations on the techniques have been presented since then by different investigators (50), but the basic principals have remained unchanged for >30 years. Today, as in the past, the MARG technique is very difficult to master, which continues to hamper its use. Researchers must use caution when reviewing the literature and relying on articles that used the emulsion-dipping technique and claim quantitative data. The conclusions may be questionable and may be disputed in some cases. Validation of the technique is lacking in most labs, and results can be very subjective. Claims of truly quantifiable results have never been clearly shown by any investigator, and they are at best semiquantitative. This is due to the lack of thickness uniformity of both the tissue sections and the emulsion detection media used. Additionally, there are few, if any, instances where calibration and/or quality control standards have been coexposed within the samples, which is the only way to clearly prove quantitation of samples.
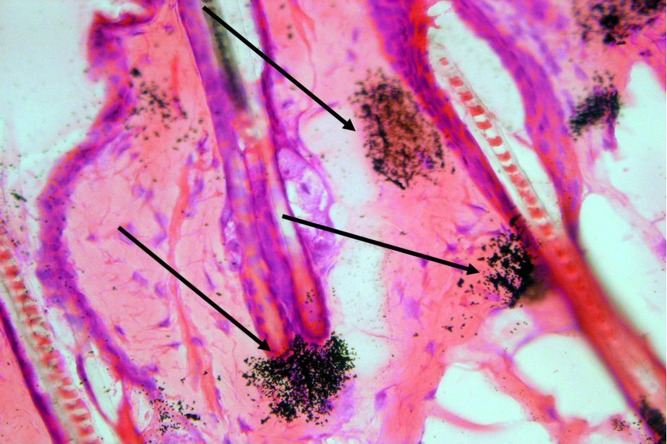
Nevertheless, MARG has made important contributions to drug discovery and development over the years and has provided insight into the localization of pharmaceuticals to support proof of concept studies, mechanisms of toxicity, efficacy, physiology of hormone action, and cell regulation. For example, MARG is useful to study skin penetration of various compounds and is routinely used in the cosmetic industry, physiology research, pharmacology, and safety studies. Unilever is a company that makes consumer skin care products and as such is responsible to prove safety of substances that are applied to skin. They have developed MARG techniques to examine skin penetration in different in vitro test models in rats, pigs, and human skin samples. They have also coupled MARG with confocal microscopy and other techniques to extend the usefulness of their studies (H. Minter presented at the 2007 Meeting of the Society for Whole-Body Autoradiography Meeting). Linoleic acid (LA) is commonly used in cosmetics, but its in vivo human skin penetration characteristics were not very well demonstrated. However, in 2006, Rauvast and Mavon (51) used a unique, in situ, “virtual” microautoradiographic slide to examine transfollicular delivery of LA in human scalp. They combined their MARG data with an in vitro permeation experiment and compartmental analysis to show that most of the LA was localized to the hair sheath, but that none was present in the dermal compartment and that 10% of the total LA recovered was found in the stratum corneum and dermis after 6 h. This supported their notion that the diffusion of LA occurred by a transfollicular route. It also demonstrated the value of high-resolution MARG for providing detailed cellular localization of the molecule. Skin receptor MARG techniques have also been used to study the absorption, penetration, and cellular localization of 3H-Maxacalcitol, which is a vitamin D analog used for the treatment of psoriasis. Hayakawa et al. treated the dorsal skin of rats with a 3H-Maxacalcitol ointment and examined skin exposed for periods of 0.5, 2, 8, 24, 48, or 168 h (52). They discovered two routes of skin penetration; one via epidermal cell layers and the other via hair follicles. They were also able to distinguish very fine regions of cellular localization, which supported theories on the mechanism of efficacy. Figure 5 shows an example of a 14C-labeled test article localized to the sebaceous gland in the skin of a rat after a dermal application (unpublished data from author). This example demonstrates the fine detail of localization that can be obtained with MARG.
Fig. 5.
MARG showing localization of a 14C-labeled test article in the sebacious glands of rat skin after a topical application. Arrows point to localizations of radioactivity
Both renal function and localization of various substances in the kidney have been studied by MARG. Young et al. (53) used 14C-iodoantipyrine as a tracer to study intrarenal blood flow in nephrectomized rats, and they used their microautoradiographs and standards to determine blood flow rates. They noted how MARG was helpful in defining the morphological location of blood flow, and they made several conclusions regarding changes in regional renal blood flow; “the interaction between vasoactive mediators and the autonomic nervous system”; and “that medullary blood flow was dependent on local prostaglandin production and is also influenced by sympathetic nervous supply.”
Another laboratory used in vitro MARG to show localization and density of atrial natriuretic peptide (ANP) in nephrectomy biopsy samples obtained from patients with renal disease (54). These investigators used [125I]-alpha-human (1–28) ANP, and they found localization in the glomerulus and tubular regions in the human biopsy specimens. They also observed that density of ANP binding generally decreased in patients with renal dysfunction and hypertension. Overall though, their study established an in vitro MARG method to assess ANP binding in human biopsy specimens.
More recent work by Yamamoto et al. (55) used MARG in combination with immunohistochemistry, macroautoradiography, and positron emission topography to examine intestinal ulceration and healing in the rat. They used 18F-FDG to examine ulcerations in the small intestine of rats, which were induced using indomethacin. MARG combined with immunohistochemistry showed an accumulation of 18F-FDG in inflammatory cells and in granular tissue-forming cells, forming granulation tissue, and around ulcers. 18F-FDG was also found to be present in proliferating intestinal crypt cells and in intact intestinal tissue taken from indomethacin-treated and control animals. They concluded ulceration could be visualized early by the prominent uptake of 18F-FDG by inflammatory cells, and by the formation of granulation tissue by cells in, and around ulcers. This work also demonstrated the value of combining both in vivo and ex vivo imaging techniques, which provided robust data sets for analysis.
Several limitations have impeded the progress and wider use of MARG in drug discovery and development. These include: the processing time required to obtain results; the inability of the technique to provide quantitative results, which includes the inability to assure and prove uniformity of tissue and emulsion thicknesses, and lack of internal calibration and/or quality control standards; the high rate and ease of artifact production; and difficulties in collection tissue sections under darkroom conditions. The processing time to obtain results by MARG is a difficult thing to gauge as each tissue must be treated and evaluated differently depending on how much radioactivity is present. The exposure time can take anywhere from days to weeks and even months to obtain optimal results. This often discourages the drug discovery scientist who often works under much shorter timelines, and it becomes an overwhelming amount of work for the scientists in the development area, who need strong study designs, which require many more samples be included in the evaluation, and who may be challenged by regulators to assure high quality results, through the use of validated procedures, and quality control and calibration standards for each sample. Technology may help to solve some of these problems if the detection media (e.g., emulsions) can be more uniformly produced and made to have inherently linear quantitation. Technology may also help to develop easier methods of collecting uniformly thick tissue sections that can be automatically mounted onto slides for processing. Although this would be quite a challenge due to the varying matrices to be sectioned (e.g., hard bone, adipose, and eyes). Dependable microsized calibration and quality control standards that can be coexposed with every section would also need to be developed to assure reproducibility of quantitation. Finally, the new methods would need to enable a significant reduction in the amount and types of artifacts that are produced. Currently, the following types of artifacts (43) must be controlled: (1) effects on emulsion by slight variations in light, humidity, temperature, tissue characteristics, fixation, freezing, chemicals, pH, developer, fixer, and miscellaneous debris in developer solutions; (2) tissue condition (e.g., freezing technique, fixation, autolysis, sectioning temperature, improper section mounting); (3) light leaks; (4) latent image fading; (5) reticulation of emulsion; (6) positive chemography; (7) negative chemography; (8) deviations of pH in processing fluids; (9) pressure artifacts; (10) ice crystals on knife; (11) crystalline deposits from developing process. Some of these are more easily controlled than others, but together they require a high level of skill by the analyst to overcome. Until methods and/or technologies can be developed that can better control tissue section and emulsion uniformity and also reduce the sources of and occurrence of artifacts, the current technique will remain strictly qualitative and will prove to be too daunting for routine use in pharmaceutical discovery and development. The lack of new developments in MARG methods has continued to make MARG an underutilized technique in drug discovery and development, but when performed correctly, the results can be of utmost value in promoting a drug candidate and in answering some pivotal questions for pharmaceutical investigators.
MATRIX-ASSISTED LASER DESORPTION/IONIZATION MASS SPECTROMETRIC IMAGING: LABEL-FREE MEASUREMENT OF COMPOUND AND METABOLITE DISTRIBUTIONS
WBA is an established technology for assessing pharmacokinetic and distribution properties of drug candidates in small animals and is therefore widely applied in pharmaceutical research. Resulting images reflect radioactivity distribution at high spatial resolution with a wide adjustable dynamic range (56). Despite the method's many benefits, one intrinsic limitation of this technology is the inability to distinguish the parent compound distribution from potential metabolite distributions. For this reason, WBA data are often combined with identification data obtained from tissue extracts. These extracts are typically analyzed by liquid chromatography mass spectrometry (LCMS), which allows the identification and quantification of the compound and its metabolites. Using LCMS, identification is done either based on the exact mass of the analyte, or more often by fragmenting the ions in the MS instrument and measuring the specific fragment ion pattern (also termed MS/MS). In contrast to WBA, this technology delivers high specificity, but a spatial resolution which is limited to the size of the tissue sample being extracted. For this reason, the combination of the two techniques is typically favored to gain specific compound and metabolite distribution information.
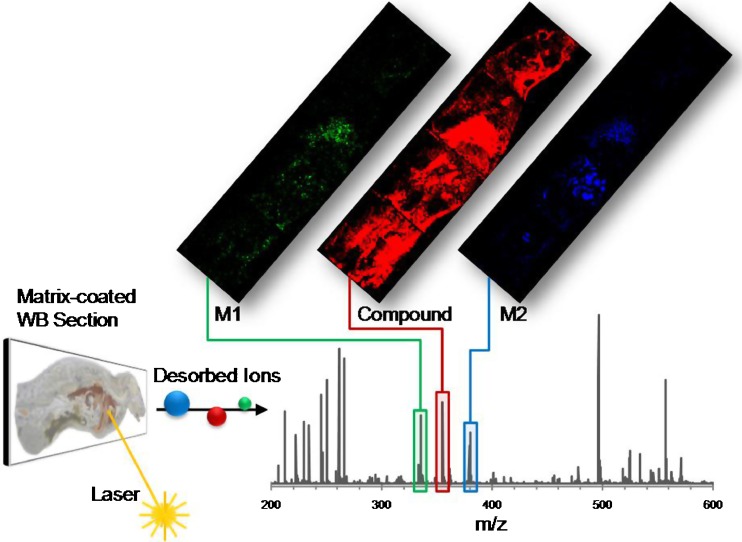
Based on this background, a new technology termed MSI (57) finds its way in a growing number of analytical laboratories doing ADME and pharmacokinetic compound assessments (58–60). The technology combines the advantages of two techniques: the spatial resolution of WBA and the specificity of mass spectrometry. In brief, sections of dosed animals are placed in a mass spectrometer, where thousands of mass spectra are acquired from a raster of positions over the sample. These spectra are assembled into a data set, from which one can extract multiple specific distributions of a particular mass (Fig. 6). With this technology come two advantages over existing methods: The mass and the fragmentation pattern are intrinsic properties of each molecule, and by using mass spectrometry as a detection system, the need for compound labeling is eliminated. And second, a mass spectrometer can acquire full spectra (or multiple MS/MS transitions) from the same sample, allowing parallel analysis of hundreds of analytes. It is for these advantages that the technology was specifically developed for the application in drug distribution studies, where the label-free simultaneous detection of compound and its metabolites poses an interesting alternative to well-established technologies.
Fig. 6.
MALDI-MSI process. Analytes are simultaneously desorbed from the animal section in a defined raster. Out of the acquired spectra-specific molecular images are calculated
The major steps leading to the success of MSI were the development of suitable MS instrumentation allowing fast data acquisition, the availability of software for data evaluation, and the optimization of sample preparation protocols for interfacing the tissue sections with the mass spectrometer. First, the sections must be dehydrated before the introduction in the vacuum chamber of the mass spectrometer, and second, the analytes must be transferred to the gas phase and ionized to allow a measurement. From the selection of available surface desorption mechanisms, MALDI (61) is currently most widespread. This technology offers unmatched sensitivity and allows ionization of a wide substance range, and in addition, a broad selection of commercial instruments is available. Alternative desorption technologies suitable for MSI, including surface sampling (62) and desorption electrospray (63), are still in early development stages and are not discussed in this article. Secondary ion mass spectrometry (64) operates in a different spatial resolution regime and is described in the following section.
Experimental
Whole-body autoradiography and mass spectrometric imaging both start with whole-body tissue sections from dosed animals (11). For a direct comparison of the techniques, the animals are dosed with radioactive compound, and adjacent sections from the same animals can be utilized for both methods. MALDI-MSI alone does not require radioactive compound, which is a significant advantage over WBA. The sections are mounted on tape and are immediately lyophilized for WBA, while for MALDI-MSI analysis, they are first frozen and then later lyophilized just before further processing. In MALDI, a matrix is required in order to desorb and ionize the analytes from the surface upon impact of laser pulses. From the wide variety of different matrices and matrix application protocols, the manual application of α-cyano-4-hydroxycinnamic acid solution using a conventional thin-layer chromatography sprayer represents a robust and reproducible method. If properly applied, this matrix leads to excellent sensitivity for a large number of compounds desorbed from tissue sections. The matrix-coated sections are then rastered over with a laser in the mass spectrometer, acquiring either spectra over the mass range of interest or multiple MS/MS spectra of the analytes of interest. The latter mode is especially suited for simultaneous imaging of a compound and its metabolites since both are often known in advance, and in addition, MS/MS images offer a high confidence in the specificity of the result. Such an image acquisition may take from 10 mins up to several hours, depending on the imaging resolution. A typical experiment on a rat section with a raster of 0.5 mm takes about 1 h.
The dataset is visualized and analyzed using dedicated software written by the author, which is available for free download at http://www.ms-imaging.org. For each of the masses of interest, a specific MS image is calculated and displayed, similar to WBA images. The advantage of MSI is its high specificity, which allows the assignment of exact distributions for any analyte of choice. Images of multiple analytes may also be combined in a single red–green–blue image to allow an information-rich display of the distribution data (Fig. 7).
Fig. 7.
Comparison of MSI, WBA, and MALDI-MSI results obtained from the same compound-dosed animal. MALDI-MSI allows distinguishing between compound (red) and its metabolites (green, colocalized with compound (yellow)), label-free and simultaneous
Advanced Image Processing
MS images obtained by the process described above do not consider analyte-specific ion suppression effects by the biological matrix and therefore give only qualitative information on the analyte distribution. Since these suppression effects can by quite dramatic (up to 95% signal loss were observed), additional experiments are required to evaluate and compensate for these effects. One method (59) is to measure a control section homogeneously coated with compound. The resulting image displays the analyte suppression at every single location. This information, together with an internal standard spiked on the section of the dosed animals, allows a quantitative assessment of the analyte concentration. This method compensates suppression at the tissue level and not on single pixels, since all the pixels in the same tissue are normalized by the same correction factor obtained in the control experiment. A more accurate quantification can be achieved by coating sections with a stable isotope-labeled analyte before processing and then calculating the peak ratio for each pixel.
Discussion
The beauty of MALDI-MSI results from the ability to simultaneously measure multiple label-free analytes and thereby assess distributions of administered compounds and their metabolites in the same experiment. Obviously, this approach is only applicable to molecules that are ionizable by the MALDI process, either through protonation or deprotonation. In the author's experience with small molecule drugs undergoing development as candidate therapeutics in multiple indications, about 80% of the studies resulted in information-rich MS images. A careful interpretation of the data is necessary especially if no control spiking experiments are available for compensation of suppression effects. Quantitative WBA suffers much less from these effects, and the author considers it an excellent choice if quantification of the total compound and metabolite pool is required. Also, in terms of sensitivity, MALDI-MSI falls short compared to quantitative WBA. In the latter, sensitivity may be increased to the required level by prolonging the exposure time to the detector system. Unfortunately, this expedient cannot be applied to MALDI-MSI, where every discrete spot is usually exhausted after 200 laser shots (depending on the laser energy).
Using the MALDI-MSI process, images are generated in a rastered mode compared to imaging mode in WBA. The achievable spatial resolution depends on the diameter of the focused laser spot, which is typically in the range of 100 to 20 μm for commercial MALDI-MS instruments. Acquiring images from a full rat section at this resolution would require days of acquisition time and is therefore not practical. In practice, a raster of 500 μm to 1 mm results in an acceptable scanning time of less than 1 h. This resolution matches with the obtainable resolution limited by the matrix deposition method, during which some delocalization of the analytes is observed. Higher resolution images may be achieved using special matrices (65,66), or by switching to SIMS as described in the following section.
While the application described in this article focuses on administered compounds and metabolites, MS images contain much more information. Hundreds of endogenous substances are measured in each full scan imaging experiment, but this information is currently not being utilized. For example, MSI could be used to localize endogenous metabolites previously detected in body fluids by a metabolomics approach. Or statistical tools could be applied to image data sets of multiple animals to correlate metabolite distribution patterns with disease status and compound efficacy.
MSI of compounds and metabolites is a new application and is currently applied to provide additional ADME information in combination with more established methods. Based on experience collected during application in drug discovery, it is expected to find its way into more laboratories and to be broadly applied in pharmaceutical industry and academic research.
SECONDARY ION MASS SPECTROMETRIC IMAGING
Introduction
Secondary ion mass spectrometric imaging (SIMS-MSI) offers a complementary or alternative method to MALDI-MSI for the acquisition of higher spatial resolution images directly from biological tissue. Although traditionally used for the analysis of surface films and polymers, in recent times, SIMS has been successfully applied to the analysis of many biological tissue and cell types enabling the acquisition of images of submicrometer spatial resolution with a minimum amount of sample preparation (67).
In the SIMS ionization process, a beam of ions are directed at the tissue surface resulting in the desorption of ionized atoms, clusters, and molecules which are traditionally measured by quadrupole or time of flight mass spectrometry. As the mode of operation is by analysis of the material removed by sputtering, it is a locally destructive technique. The sputtered products include atoms, molecules, and molecular fragments that are characteristic of the surface composition within each volume element sputtered by the ion beam, a small fraction of which are ionized as positive or negative ions. Secondary ion sputtering yields (and thus the sensitivity of the analysis) depend upon the nature, energy, and incident angle of the primary ion beam as well as the properties of the target tissue and the application of any yield enhancement steps such as a metal or organic matrix coating (68). Images can be acquired at resolution of a few hundred nanometers (far superior to MALDI-MSI) due to the use of primary ion sources which can be focused to spot sizes of ≤50 nm.
Key stages required for the acquisition of a high quality SIMS image include:
preparation of the sample (i.e., sectioning of tissues, freeze fracturing of cells to provide samples stable under high vacuum conditions which are required for SIMS analysis)
ionization of the sample by the primary ion beam
mass separation (predominantly by time of flight (TOF))
ion detection in positive or negative polarity modes
data handling and image processing
In contrast to MALDI, SIMS is a hard ionization method which results in heavy fragmentation of desorbed ions making the desorption of large intact molecules very difficult. The first SIMS instruments were developed in the 1960s utilizing an argon primary ion beam (69). Subsequent developments included the use of time of flight mass separation (70), pulsed ion beams, and liquid metal ion sources to increase the sensitivity, spatial, and mass resolution of the analysis (71).
The recent development of ion sources producing cluster ions as their primary beam has lead to an increase in the range of both samples and analytes which can be studied. Commercially produced polyatomic ion sources such as Au+n, Bi+n, SF+5, and C+60 are easy to operate and have long operational lifetimes (72). They have the great advantage of producing a larger secondary ion yield and reducing sample damage and subsequent molecule fragmentation in comparison to the traditionally used liquid metal ion sources such as gallium and indium, which produce fewer peaks above m/z 100 (73). This increase in mass range means SIMS-MSI is ideally applicable for drug, metabolite, and lipid analysis.
There are two modes of SIMS available for imaging analysis: static SIMS and dynamic SIMS. Static SIMS utilizes a low intensity primary ion beam which only removes a few monolayers of the surface but generates larger mass fragments. Dynamic SIMS utilizes a higher intensity primary ion beam and results in an increased secondary ion production, but with increased fragmentation of molecular species. As the method is more surface destructive and results in the sputtering of species from deeper within the tissue, it can be used for depth-profiling analysis, whereby the data is acquired as a function of its depth through the tissue (74). Problems have been encountered in recent studies applying SIMS-MSI for the depth profiling of biological tissue (75). While good signals were imaged for lipids in rat brain sections in the lipid-dense first 300 nm tissue depth, deeper profiling below 1 µm was not possible due to low lipid concentrations present and accumulative damage induced by the sputter process.
Submicrometer Cellular and Tissue Imaging
Previous molecular imaging analysis of individual cells required fluorescent labeling of the molecule of interest which is problematic due to limited available antilipid antibodies and alterations in the lipid chemical properties due to insertion of fluorescent probes (76). Using this method, analysis is completely restricted to the labeled molecule. SIMS has been applied to the imaging of elements in cells at high spatial resolution (77,78). However, analyzing elements in isolation only provides limited information to the biochemist who is usually more concerned with larger biomolecules and/or compounds.
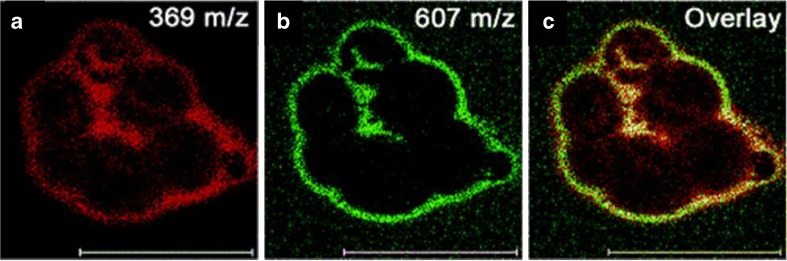
Due to the advancements in primary ion source technology, it is also possible to acquire high spatial resolution microprobe images of important biological molecules in individual cells, and both static and dynamic SIMS have been used for this purpose. Lipids and sterols are the predominantly analyzed species, examples of which include cholesterol (79), sphingomyelin (80), and phosphatidylcholine (81). Recent studies utilizing these capabilities include the analysis of cellular membrane components to understand the behavior and function of the major lipid constituents, and separate research groups have imaged individual glial (82) and neuroblastoma (83) cells (as shown in Fig. 8). Relative semiquantitative SIMS imaging analysis of cholesterol has been reported in the membranes of macrophage cells treated to contain elevated levels of cholesterol with respect to control cells (79). By using the signal of C5H+9 (at m/z 69) as an internal standard, the authors observed a statistically significant signal increase of over 100% in images from the treated cells.
Fig. 8.
SIMS images of individual neuroblastoma cells after deposition of 1 nm gold. a Cholesterol [M-OH]. b Diacyglcerol. c Overlay showing the colocalization of the two species. Scale bar 100 µm. (Adapted with permission from Ref. 18)
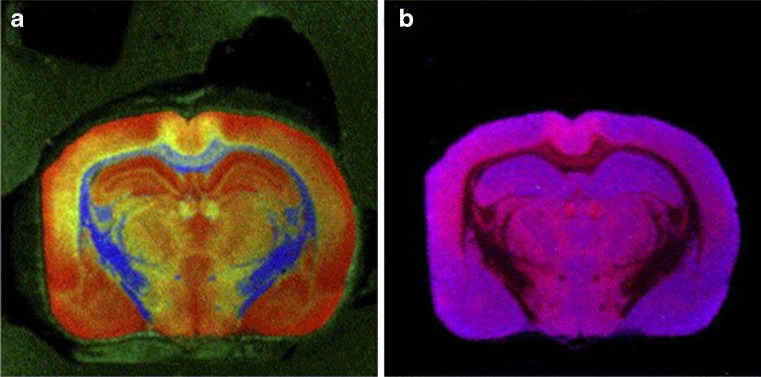
Lower resolution imaging, encompassing a larger imaging window, can be used for the localization of molecules over larger tissue areas (for example biopsy sections) or even sections of entire organs. Imaging SIMS technologies have been applied to a wide range of biological tissues including whole brain sections (as shown in Fig. 9) (84), aorta (85), liver (86), kidney (87), muscle (88), and skin (89).
Fig. 9.
Negative ion SIMS images obtained from a rat brain section using a Bi3+ primary ion source. Analysis window is 256 × 256 pixels, pixel size 70 × 70 µm. Color overlays show the different localizations of the following negative ions: a m/z (255 + 283, red), m/z 892 (green) and m/z 771 (blue); b m/z 255 (blue) and m/z 283 (red). (Adapted with permission from Ref. 19)
Lipids remain the major analyte class investigated with larger window SIMS imaging, and one such example is the localization of globotriaosylceramide and digalactosylceramide as biomarkers of Fabry disease in skin and kidney biopsies of patients (89). Clear accumulations of both lipids were observed in patients in comparison to healthy volunteers as well as colocalization with the distribution of vitamin E and cholesterol.
Sample Optimization
As previously discussed, one main advantage of SIMS is the acquisition of images direct from tissue with a minimum of handling or sample preparation steps. However, sample coating methods can be utilized which have the potential to significantly increase the secondary ion yield and thus the sensitivity of the analysis as well as enabling the ionization of larger lipids and peptides (90). Two described methods are matrix-enhanced SIMS (ME-SIMS) and metal-assisted SIMS (MetA SIMS).
The application of an organic acid matrix has been shown to significantly increase the signal intensity of larger analytes directly from tissue (68). However, for larger molecules (>5 kDa), the signal to noise ratio of ME-SIMS drops dramatically in comparison to MALDI (67). Intact proteins have been observed up to 17 kDa but only when present at high concentrations and purified prior to analysis. Sample preparation is of key importance particularly concerning the choice of matrix and application method with regard to the ultimate crystal size, which may become the limiting step for high spatial resolution imaging analysis. It is important to note that, for ME-SIMS, the ion yields will be determined by the matrix application process (i.e., tissue extraction and cocrystallization), and thus, there is a significant loss of surface specificity in relation to the regular “surface sputtering” SIMS.
Studies have shown that coating the sample with a thin layer of metal (typically in the region of 1–5 nm) such as silver or gold increases the secondary ion yield of intact molecular ions (91). Conclusive reasoning for this is as of yet unconfirmed, although the migration of analytes onto metal “nano islands” and the ability of the metal to act as a catonizing species (92–94) have been proposed. As the metal is sputter coated onto the tissue, there is no risk of analyte spreading and thus loss of spatial information which may occur with the application of a wet organic matrix. The main problem encountered with MetA SIMS is the observation of ions in the resulting spectra in the form of metal adducts which may prove difficult to assign to specific analytes and analyte fragments.
Recent Advancements
A particularly exciting development in the SIMS imaging field is the development of true MS/MS SIMS instrumentation. The Winograd laboratory has adapted an AB Sciex Q-Star to create a SIMS quadrupole time of flight instrument (95). Although the spatial resolution is currently 10 µm and thus has limitations in its application to single-cell-imaging experiments, there is significant potential to provide information used in conjunction with submicrometer resolution SIMS instrumentation. The use of this novel MS/MS methodology in this way has enabled new diagnostic fragments for cholesterol to be determined which could not be conclusively identified in single MS mode. Another novel MS/MS SIMS instrument has been developed around a J105 3D chemical imager platform and has been applied to the imaging of whole Xenopus blastoma sections where the MS/MS product spectrum was used to confirm the identities of phospholipid species (96).
Ion microscopy imaging by TOF-SIMS has been described, whereby the spatial resolution is independent of the spot size of the primary ion beam (96). This decoupling enables a larger area to be imaged from a primary ion pulse without moving the beam or sample. The stigmatic ion optics transport magnified images of the ion distributions to the detector while retaining lateral position information, thus making the ion optic quality and spatial resolution of the detector limiting factors for spatial resolution. Using a related methodology, Luxembourg et al. have imaged the distribution of peptides and proteins in prepared mixtures (97) and whole rat brain sections (67) by stigmatic MALDI imaging coupled to an established TOF-SIMS instrument. Significant increases in spatial resolution (∼4 µm) and acquisition speed were reported in comparison to standard MALDI microprobe imaging.
Conclusion
SIMS MSI has shown significant potential for more frequent application to the spatial analysis of compounds and biomolecules in tissue and cells. However, its use is still only having limited impact in biomedical research, and it is relatively underused in comparison to the more accepted MALDI-MSI methodology. This may be due to limitations in mass ranges which can be successfully ionized and a lack of understanding of the fundamental ionization process (67). It has gained a much larger acceptance in the field of material science, and the majority of SIMS instruments are located in physics-focused institutes where inorganic species are primarily analyzed.
SIMS-MSI offers additional, complementary information to MALDI and so may be used in collaboration to provide a dual MSI approach enabling both high mass and high spatial resolution analysis of biological tissues. Recent advances in instrument technology such as the ability to perform MS/MS analysis on an imaging SIMS instrument may further enhance its acceptance as a method for imaging drug distribution (where absolute confirmation of the compound is required) and the identification and structural confirmation of metabolites. It is clear that, with the recent developments and publications featuring applications to valid biomedical analysis, SIMS imaging has the potential to fill a niche for the analysis of low-molecular weight compounds and biomolecules at high spatial resolution for biomedical research purposes.
THE FUTURE OF EX VIVO/IN SITU IMAGING IN DRUG DEVELOPMENT
Molecular imaging has grown dramatically over the last 50 years, and most recently, in vivo imaging modalities, such as PET (98) and fluorescence (99) compounds, have captured the attention of scientists in drug discovery and development owing to the ability to visualize organ distribution in living lab animals. However, these new in vivo modalities have their limitations, such as relying on the use of short-lived isotopes as tags as in PET imaging, which limits the ability to study tissue pharmacokinetics over days or weeks, or due to problems of reliable quantitation due to background fluorescence and poor resolution, which occurs when imaging fluorescent tags in vivo. Additionally, the positron-emitting isotopes for PET and the fluorescent labels used can alter the structure and/or binding capabilities of small molecules, thus rendering them ineffective at providing the needed information. These modalities are also limited in that they are imaging a radioactive or fluorescent label, and like QWBA, they do not provide information on metabolites. To this end, it seems that ex vivo/in situ imaging modalities, such as QWBA, MARG, MALDI-MS, and SIMS, will undoubtedly be relied upon for many years to come to provide the kind of detailed tissue distribution information required for drug discovery and development programs.
Of course, QWBA also has its limitations especially regarding sampling processing and analysis, but that technology may help improve in the future. Improvements made to the freezing process may provide sections with improved tissue morphology, and if improvements can be made on the transfer of whole-body sections from tape to glass slides and/or other matrices, then whole-body sections may be used for things such as whole-body histological examination. For example, far into the future, the whole-body sectioning technique may be transferred into the pathology lab, where a histopathologist could review all tissues from the same animal in situ instead of using slides of individual tissue sections. In fact, this may already be accomplished if whole animals are infused with fixatives. The trouble comes when attempting to stain the sections while they are on the adhesive tape used for their collection. Additionally, whole-body sections transferred to glass would be more easily stained by immunohistochemistry for known target receptors, thus enabling the colocalization of radiolabeled test articles when autoradioluminographic images are combined with an image of the stained sections.
Improvements made to the cryomicrotome, such as robotic section collection system that incorporates an anatomy recognition software to control section collection, would greatly reduce technician operation time. Something similar to this has been developed at Case Western University in the labs of by Dr. David Wilson. Dr. Wilson has developed a cryo-imaging system that utilizes a large cryomicrotome that is outfitted with an automated camera capable of imaging serial photographic and fluorescence images from the surface of the frozen, sectioned block face of whole-body preparations at different levels through the specimen (David Wilson, Case Western University, Keynote speech at the 2007 Meeting of the Society for Whole-Body Autoradiography, Charleston, SC). The imaging software is also capable of 3D reconstructions and localization of such things as biomarkers (using green fluorescent protein) and fluorescent-labeled drugs. Whole-body sections may also be collected and used for QWBA and images integrated to show drug localization and biomarker activation.
Image analysis software has improved slowly over the last 10 years, and most improvements have pertained to data management and regulatory compliance. More recently, improvements have been made to imaging software to provide the ability to coregister autoradiographic images with color images of the matching whole-body sections. Raytest Inc. offers a software known as AIDA, which makes this process easy to use. This helps autoradiographers improve on the identification of tissue areas for quantification during image analysis. This feature is especially useful when tissue concentrations are very low and identification on the autoradioluminograph may be difficult. This is also most helpful when new technicians are learning to do image analysis.
But perhaps the biggest limitation faced by all autoradiographers (including in vivo imaging techniques) is the fact that radioactive or fluorescent labels are being imaged, and investigators do not know if it reflects the parent molecule and/or its metabolites. This is where a combination of standard PK studies with MALDI-MSI or SIMS-MSI can be very useful. By combining PK and MALDI-MSI, scientists can gain quantitative tissue concentration and metabolite information, completely label free and therefore earlier in the drug discovery process. So in the next 5–10 years, pharmaceutical scientists can expect to see improvements in MALDI-MS and SIMS, potentially allowing a direct quantification based on the MS images. As more collaborations between autoradiographers and bioanalytical scientists occur, improvements will be made, but in order to maintain the high resolution, it will still require ex vivo samples. This is because of the low, nonpenetrating energy of the isotopes (mostly 14C and 3H), which can be stably labeled on most small molecules and must be in close contact with the detector and/or a focused camera typically used for these studies. In 10–15 years, it is possible that MALDI-MSI may be developed to an extent that is robustly quantifiable. In order to accomplish that, many improvements will be needed to process and analyze samples more efficiently. Currently, it is estimated that it would take a MALDI-MS expert approximately 1 week to analyze all the tissues routinely measured by QWBA for a single rat (30–40 tissues and bodily fluids), and so, analyzing all the tissues in ten or more rats on a study could prove to be a daunting task. This would be a particular issue for SIMS imaging during which smaller images are acquired at higher spatial resolutions, and thus, the acquisition duration for imaging larger tissues, or even whole bodies, would be extremely long and therefore impractical for larger population studies. Hence, for such investigations, SIMS-MSI is envisaged as being complimentary to MALDI-MSI technologies by enabling increasingly detailed molecular localization information to be determined from small, specific tissue regions.
Ideally, a pharmaceutical scientist would like to be able to dose an animal with a nonlabeled test article and follow the distribution of the parent drug and its metabolites throughout the body of a living animal over a long period of time. However, to do that, one would need to overcome all the hurdles of individual tissue effects on identification and quantification of a xenobiotic and/or its metabolites among the milieu of endogenous compounds in animals. Finally, the use of ex vivo animal models where investigators can literally freeze an animal in time and obtain high-resolution quantitative data on parent drugs and metabolites will remain the state-of-the-art method to determine the distribution of xenobiotics for many years to come.
Footnotes
This paper presents an overview of these techniques, history, study designs, and considerations for use in support of drug discovery and development.
An erratum to this article can be found at http://dx.doi.org/10.1208/s12248-009-9167-3
References
- 1.Hahn EJ. Autoradiography: a review of basic principles. Am Lab. 1983;15:64–71. [Google Scholar]
- 2.Luckey G. US Patent. 1975;3,859,527.
- 3.Niepse de Saint Victor, Compt Rend. 1867;65:505-7. In: Rogers AW, editor. Techniques of autoradiography. Elsevier Scientific Publishing Company; 1973.
- 4.London ES. O fiziologopatologicheskrom znachenii emanastii radya (Physiopathological importance of radium emanation) Russki Vrach (St. Petersburg) 1904;3:869–72. [Google Scholar]
- 5.Dziewaitkovski DD. Sulfate-sulfur metabolism in the rat fetus as indicated by sulfur-35. J Exp Med. 1953;98:119. doi: 10.1084/jem.98.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ullberg S. Studies on the distribution and fate of 35S-labelled benzylpenicillin in the body. Acta Radiol Suppl. 1954;118:1–110. [PubMed] [Google Scholar]
- 7.Cohen Y, Epierre JW. Methode d'etude autoradiographique de substances marquées volatiles, Rapport C.E.A. 2071; 1961.
- 8.Pellerin P. La technique d'autoradiographie anatomique à la température de l'azote liquide. Pathol Biol Sent Hôp. 1961;9:233. [PubMed] [Google Scholar]
- 9.Martin LE, Harrison C, Bates CM. A simple low-temperature radioautographic technique. Biochem J. 1962;82:17P. [Google Scholar]
- 10.Kalberer F. A new method of macroautoradiography. Adv Tracer Methodol. 1966;3:139. doi: 10.1007/978-1-4684-8625-4_15. [DOI] [PubMed] [Google Scholar]
- 11.Ullberg S. The technique of whole-body autoradiography: cryosectioning of large specimens. In: Elvefeldt O, editor. Special issue on whole-body autoradiography LKB Instr J. Science Tools. Bromma Sweden; 1977.
- 12.Berlin M, Ullberg S. Accumulation and retention of mercury in the mouse. Arch Environ Health. 1963;6:589. doi: 10.1080/00039896.1963.10663447. [DOI] [PubMed] [Google Scholar]
- 13.Kutzim H. The quantitative determination of the distribution of S35-sulfate in mice using autoradiography. Nucl Med (Stuttg) 1962;15(3):39–50. [PubMed] [Google Scholar]
- 14.Cross SAM, Groves AD, Hesselbo T. A quantitative method for measuring radioactivity in tissues sectioned for whole body radiography. Int J Appl Radiat Isot. 1974;25:381–6. doi: 10.1016/0020-708X(74)90017-9. [DOI] [PubMed] [Google Scholar]
- 15.Longshaw S, Fowler JSL. A poly (methy l4C) methacrylate source for use in whole-body autoradiography and beta-radiography. Xenobiotica. 1978;8:289–95. doi: 10.3109/00498257809060952. [DOI] [PubMed] [Google Scholar]
- 16.Coe RAJ. An evaluation of X-ray films suitable for autoradiographs using ß14C radiation. Int J Appl Radiat Isot. 1982;36:93–6. [Google Scholar]
- 17.Franklin ER. The use of measurements of radiographic film response of X-ray film in quantitative and semi-quantitative autoradiography. Int J Appl Radiat Isot. 1985;36:193–6. doi: 10.1016/0020-708X(85)90066-3. [DOI] [PubMed] [Google Scholar]
- 18.Geary WA, II, Toga AW, Wooten GF. Quantitative film autoradiography for tritium: methodological considerations. Brain Res. 1985;337:99–118. doi: 10.1016/0006-8993(85)91613-0. [DOI] [PubMed] [Google Scholar]
- 19.Steinke W, Archimbaud Y, Becka M, Binder R, Busch U, Dupont P, et al. Quantitative distribution studies in animals: cross-validation of radioluminography versus liquid-scintillation measurement. Regul Toxicol Pharmacol. 2000;31:S33–43. doi: 10.1006/rtph.2000.1384. [DOI] [PubMed] [Google Scholar]
- 20.Schweitzer A, Fahr A, Niederberger W. A simple method for quantitation of 14C-whole-body autoradiograms. Appl Radiat Isotopes. 1975;33:329–33. doi: 10.1016/0883-2889(87)90019-0. [DOI] [PubMed] [Google Scholar]
- 21.Coulson F, Carr CJ, editors. The validation of radioluminography for sue in quantitative distribution studies. Regul Toxicol Pharmacol (Special Edition). 2000;31(2), part 2 of 2 parts.
- 22.Sonada M, Takana M, Miyahara J, Kato H. Computed radiography utilising scanning laser stimulated luminescence. Radiology. 1988;148:833–8. doi: 10.1148/radiology.148.3.6878707. [DOI] [PubMed] [Google Scholar]
- 23.Miyahara J. The imaging plate: a new radiation image sensor. Chem Today. 1989;223:29–36. [Google Scholar]
- 24.Shigematsu A, Motoji N, Hatori A, Satoh T. Progressive application of autoradiography in pharmaceutical and metabolic studies for development of new drugs. Regul Toxicol Pharmacol. 1990;22:122–42. doi: 10.1006/rtph.1995.1078. [DOI] [PubMed] [Google Scholar]
- 25.Mori K, Hamaoka T. Protein, nucleic acid enzyme. 39:181–91. In: Kolbe H, Dietzel G, editors. Techincal validation of radioluminography systems. Regul Toxicol Pharmacol (Special Edition). 2000;31:S5–14. [DOI] [PubMed]
- 26.Motoji N, Hayama E, Shigematsu A. Radioluminography for quantitative autoradiography of 14C. Eur J Drug Metab Pharmacokinet. 1995;20:89–105. doi: 10.1007/BF03226362. [DOI] [PubMed] [Google Scholar]
- 27.Potchioba MJ, Tensfeldt TG, Nocerini MR, Silber BM. A novel quantitative method for determining the biodistribution of radiolabeled xenobiotics using whole-body cryosectioning. J Pharmacol Exp Ther. 1995;272:953–62. [PubMed] [Google Scholar]
- 28.Tanaka M. Collaborative studies on the distribution and metabolism of radiolabelled drugs using radioluminography. Xenobiot Metabol Dispos. 1994;9:393–407. [Google Scholar]
- 29.Jeavons AP, Hood K, Herlin O, et al. The high density avalanche chamber for positron emission tomography. IEEE Trans Nucl Sci. 1983;30:640–5. doi: 10.1109/TNS.1983.4332347. [DOI] [Google Scholar]
- 30.Charpak G, Imrie D, Jeanjean J, Miné P, Nguyen H, Scigocki D, et al. A new approach to positron emission tomography. Eur J Nucl Med. 1989;15:690–3. doi: 10.1007/BF00631758. [DOI] [PubMed] [Google Scholar]
- 31.d’Argy R, Sundwall A. Quantitative whole-body autoradioluminography—future strategy for balance and tissue distribution studies. Regul Toxicol Pharmacol. 2000;31(2):S57–62. doi: 10.1006/rtph.2000.1387. [DOI] [PubMed] [Google Scholar]
- 32.Solon EG, Kraus L. Quantitative whole-body autoradiography in the pharmaceutical. Survey results on study design, methods and regulatory compliance. J Pharmacol Toxicol Methods. 2002;43:73–81. doi: 10.1016/s1056-8719(02)00161-2. [DOI] [PubMed] [Google Scholar]
- 33.Kaim A, Weber B, Kurrer M, Westera G, Schweitzer A, Gottschalk J, et al. 18F-FDG and 18F-FET uptake in experimental soft tissue infection. Eur J Nucl Med. 2002;29:648–54. doi: 10.1007/s00259-002-0780-y. [DOI] [PubMed] [Google Scholar]
- 34.Schweitzer A, Hasler-Nguyen N, Zijlstra J. Preferential uptake of the non steroid anti-inflammatory drug diclofenac into inflamed tissues after a single oral dose in rats. BMC Pharmacol. 2009;9:5. doi: 10.1186/1471-2210-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solon E, Balani SK, Lee FW. Whole-body autoradiography in drug discovery. Curr Drug Metab. 2002;3:451–62. doi: 10.2174/1389200023337207. [DOI] [PubMed] [Google Scholar]
- 36.Foster C, Howard L, Schweitzer A, Persohn E, Hiestand P, Balatoni B, et al. Brain penetration of the oral immunomodulatory drug FTY720 and its phosphorylation in the central nervous system during experimental autoimmune encephalomyelitis: consequences for mode of action in multiple sclerosis. JPET. 2007;323:469–76. doi: 10.1124/jpet.107.127183. [DOI] [PubMed] [Google Scholar]
- 37.Weiss H, Pfaar U, Schweitzer A, Wiegand H, Skerjanec A, Schran H. Biodistribution and plasma protein binding of zoledronic acid. Drug Metab Dispos. 2008;36:2043–9. doi: 10.1124/dmd.108.021071. [DOI] [PubMed] [Google Scholar]
- 38.Potchioba MJ, Nocerini MR. Utility of whole-body autoradiography in drug discovery for the quantification of tritium-labeled drug candidates. Drug Metab Dispos. 2004;32(10):1190–8. doi: 10.1124/dmd.32.10.. [DOI] [PubMed] [Google Scholar]
- 39.Bruin G, Faller T, Wiegand H, Schweitzer A, Nick H, Schneider J, et al. Pharmacokinetics, distribution, metabolism, and excretion of deferasinox and its iron complex in rats. Drug Metab Dispos. 2008;36:2523–38. doi: 10.1124/dmd.108.022962. [DOI] [PubMed] [Google Scholar]
- 40.Solon E. Autoradiography: high-resolution molecular imaging in pharmaceutical discovery and development. Ex Opin Drug Discov. 2007;2(4):503–14. doi: 10.1517/17460441.2.4.503. [DOI] [PubMed] [Google Scholar]
- 41.Rico A, Benard P, Braun JP, Burgat-Sacaze V. Application of macroscopic autoradiography to large animals in veterinary pharmacokinetics: the distribution of sodium selenite labelled with 75Se in the pig. Ann Rech Vét. 1978;9:25–32. [PubMed] [Google Scholar]
- 42.Baker JRJ. Autoradiography: a comprehensive review. Royal microscopical society, microscopy handbooks 18. Oxford Science Publications; 1989. pp. 30–32.
- 43.Stumpf WE. Drug localization in tissues and cells. IDDC Press. Library of Congress Control Number 2003105179; 2003.
- 44.Stumpf WE, Roth LJ. Vacuum freeze drying of frozen sections for dry-mounting high resolution autoradiography. Stain Technol. 1964;39:219–23. doi: 10.3109/10520296409061233. [DOI] [PubMed] [Google Scholar]
- 45.Lacassagne A, Lattes J. R’éparitiondu polonium (injecté sous la peau) dans l’organisme de rats porteurs de griffes cancereuses. C R Séance Soc Biol. 1924;90:352–3. [Google Scholar]
- 46.Bélanger LF, Leblond CP. A method for locating radioactive elements in tissues by covering histological sections with a photographic emulsion. Endocrinology. 1946;39:8. doi: 10.1210/endo-39-1-8. [DOI] [PubMed] [Google Scholar]
- 47.Joftes DL, Warren S. Simplified liquid emulsion radioautography. J Biol Photogr Assoc. 1955;23(4):145–50. [PubMed] [Google Scholar]
- 48.Appleton TC. Autoradiography of soluble labeled compounds. J R Microsc Soc. 1964;83:277–81. doi: 10.2307/3224737. [DOI] [PubMed] [Google Scholar]
- 49.Caro LG. Electron microscopic radiography of thin sections—golgi zone as a site of protein concentration in pancreatic acinar cells. J Biophys Biochem Cytol. 1961;10:37. doi: 10.1083/jcb.10.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagata T. Techniques and application of microscopic autoradiography. Histol Histopathol. 1997;12:1091–124. [PubMed] [Google Scholar]
- 51.Rauvast V, Mavon A. Transfollicular delivery of linoleic acid in human scalp skin: permeation study and microautoradiographic analysis. Int J Cosmet Sci. 2006;28(2):117–23. doi: 10.1111/j.1467-2494.2006.00303.x. [DOI] [PubMed] [Google Scholar]
- 52.Hayakawa N, Kubota N, Imai N, Stumpf WE. Receptor microscopic autoradiography for the study of percutaneous absorption, in vivo skin penetration, and cellular–intercellular deposition. J Pharmacol Toxicol Methods. 2004;50(2):131–7. doi: 10.1016/j.vascn.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 53.Young LS, Regan MC, Sweeney P, Barry KM, Ryan MP, Fitzpatrick JM. Changes in regional renal blood flow after unilateral nephrectomy using the techniques of autoradiography and microautoradiography. J Urol. 1998;160:926–31. doi: 10.1016/S0022-5347(01)62834-9. [DOI] [PubMed] [Google Scholar]
- 54.Ogura T, Toshio N, Asano N, Katayama E, Oishi T, Mimura Y, et al. In vitro micro-autoradiography of atrial natriuretic peptide in biopsy specimens from patients with renal diseases. J Med. 1994;25:203–17. [PubMed] [Google Scholar]
- 55.Yamato M, Kataoka Y, Mizuma H, Wada Y, Watanabe Y. PET and macro- and microautoradiographic studies combined with immunohistochemistry for monitoring rat intestinal ulceration and healing processes. J Nucl Med. 2009;50:266–73. doi: 10.2967/jnumed.108.057943. [DOI] [PubMed] [Google Scholar]
- 56.Coe RAJ. Quantitative whole-body autoradiography. Regul Toxicol Pharmacol. 2000;31.2:S1–3. doi: 10.1006/rtph.2000.1379. [DOI] [PubMed] [Google Scholar]
- 57.McDonnell LA, Heeren RMA. Imaging mass spectrometry. Mass Spectrom Rev. 2007;26.4:606–43. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 58.Rohner TC, Staab D, Stoeckli M. MALDI mass spectrometric imaging of biological tissue sections. Mech Ageing Dev. 2005;126:177–85. doi: 10.1016/j.mad.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 59.Stoeckli M, Staab D, Schweitzer A. Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole-body tissue sections. Int J Mass Spectrom. 2007;260(2–3):195–202. [Google Scholar]
- 60.Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Anal Chem. 2006;78(18):6448–56. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- 61.Karas M, Bachmann D, Bahr U, Hillenkamp F. Matrix-assisted ultraviolet laser desorption of non-volatile compounds. Int J Mass Spectrom Ion Process. 1987;78:53–68. doi: 10.1016/0168-1176(87)87041-6. [DOI] [Google Scholar]
- 62.Van Berkel GJ, et al. Liquid microjunction surface sampling probe electrospray mass spectrometry for detection of drugs and metabolites in thin tissue sections. J Mass Spectrom. 2008;43(4):500–8. doi: 10.1002/jms.1340. [DOI] [PubMed] [Google Scholar]
- 63.Wiseman JM, et al. Desorption electrospray ionization mass spectrometry: imaging drugs and metabolites in tissues. Proc Natl Acad Sci USA. 2008;105(47):18120–5. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burns MS. Applications of secondary ion mass-spectrometry (Sims) in biological-research—a review. J Micros-Oxford. 1982;127:237–58. doi: 10.1111/j.1365-2818.1982.tb00419.x. [DOI] [PubMed] [Google Scholar]
- 65.Taira S, et al. Nanoparticle-assisted laser desorption/ionization based mass imaging with cellular resolution. Anal Chem. 2008;80:4761–6. doi: 10.1021/ac800081z. [DOI] [PubMed] [Google Scholar]
- 66.Teng C, Hsun H, Kun C, Lin YS, Chen Y-C. Gold nanoparticles as selective and concentrating probes for samples in MALDI MS analysis. Anal Chem. 2004;76(15):4337–42. doi: 10.1021/ac049963x. [DOI] [PubMed] [Google Scholar]
- 67.Heeren RMA, McDonnell LA, Amstalden E, Luxembourg SL, Altelaar AFM, Piersma SR. Why don’t biologists use SIMS? A critical evaluation of imaging MS. Appl Surf Sci. 2006;252:6827–35. doi: 10.1016/j.apsusc.2006.02.134. [DOI] [Google Scholar]
- 68.McDonnell LA, Heeren RMA, de Lange RPJ, Fletcher IW. Higher sensitivity secondary ion mass spectrometry of biological molecules for high resolution, chemically specific imaging. J Am Soc Mass Spectrom. 2006;17:1195–202. doi: 10.1016/j.jasms.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 69.Liebl HJ. Ion microprobe mass analyzer. J Appl Phys. 1967;38:5277–80. doi: 10.1063/1.1709314. [DOI] [Google Scholar]
- 70.Chait BT, Standing KG. Secondary ion mass spectrometry of oligopeptides. Int J Mass Spectrom Ion Phys. 1981;40:185–93. doi: 10.1016/0020-7381(81)80041-1. [DOI] [Google Scholar]
- 71.Levi-Setti R, Hallegot P, Girod C, Chabala JM, Li J, Sadonis A, et al. Critical issues in the application of a gallium probe to high resolution secondary ion imaging. Surf Sci. 1991;246:94–106. doi: 10.1016/0039-6028(91)90399-D. [DOI] [Google Scholar]
- 72.Walker AV. Why is SIMS underused in chemical and biological analysis? Challenges and opportunities. Anal Chem. 2008;80:8865–70. doi: 10.1021/ac8013687. [DOI] [PubMed] [Google Scholar]
-
73.Nagy G, Gelb LD, Walker AV. An investigation of enhanced secondary ion emission under
 bombardment. J Am Soc Mass Spectrom. 2005;16:733–42. doi: 10.1016/j.jasms.2005.01.024. [DOI] [PubMed] [Google Scholar]
bombardment. J Am Soc Mass Spectrom. 2005;16:733–42. doi: 10.1016/j.jasms.2005.01.024. [DOI] [PubMed] [Google Scholar] - 74.Todd PJ, Schaff TG, Chaurand P, Caprioli RM. Organic ion imaging of biological tissue with MALDI and SIMS. J Mass Spectrom. 2001;36:355–69. doi: 10.1002/jms.153. [DOI] [PubMed] [Google Scholar]
- 75.Jones EA, Lockyer NP, Vickerman JC. Depth profiling brain tissue sections with a 40 keV C60+ primary ion beam. Int J Mass Spectrom. 2007;260:146–57. doi: 10.1016/j.ijms.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 76.Debois D, Brunelle A, Laprevote O. Attempts for molecular depth profiling directly on a rat brain tissue section using fullerene and bismuth cluster ion beams. Int J Mass Spectrom. 2007;260:115–20. doi: 10.1016/j.ijms.2006.09.026. [DOI] [Google Scholar]
- 77.Maier O, Oberle V, Hoekstra D. Fluorescent lipid probes: some properties and applications (a review) Chem Phys Lipids. 2002;116:3–18. doi: 10.1016/S0009-3084(02)00017-8. [DOI] [PubMed] [Google Scholar]
- 78.Chandra S, Smith DR, Morrison GH. Subcellular imaging by dynamic SIMS ion microscopy. Anal Chem. 2000;202:217–29. doi: 10.1021/ac002716i. [DOI] [PubMed] [Google Scholar]
- 79.Strick R, Strissel PL, Gavrilov K, Levi-Setti R. Cation-chromatin binding as shown by ion microscopy is essential for the structural integrity of chromosomes. J Cell Biol. 2002;155:899–910. doi: 10.1083/jcb.200105026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ostrowski SG, Kurczy ME, Roddy TP, Winograd N, Ewing AG. SIMS imaging of cholesterol in membranes of fluorescently identified single cells. Anal Chem. 2007;79:3554–60. doi: 10.1021/ac061825f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McQuaw CM, Zheng L, Ewing AG, Winograd N. Localization of sphingomyelin in cholesterol domains by imaging mass spectrometry. Langmuir. 2007;23:5645–50. doi: 10.1021/la063251f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fletcher JS, Rabbani S, Henderson A, Blenkinsopp P, Thompson SP, Lockyer NP, et al. A new dynamic in mass spectral imaging of single biological cells. Anal Chem. 2008;80:9058–64. doi: 10.1021/ac8015278. [DOI] [PubMed] [Google Scholar]
- 83.Parry S, Winograd N. High-resolution TOF-SIMS imaging of eukaryotic cells preserved in a trehalose matrix. Anal Chem. 2005;77:7950–7. doi: 10.1021/ac051263k. [DOI] [PubMed] [Google Scholar]
- 84.Altelaar AFM, Klinkert I, Jalink K, de Lange RPJ, Adan RAH, Heeren RMA, et al. Gold-enhanced biomolecular surface imaging of cells and tissue by SIMS and MALDI mass spectrometry. Anal Chem. 2006;78:734–42. doi: 10.1021/ac0513111. [DOI] [PubMed] [Google Scholar]
- 85.Touboul D, Kollmer F, Niehuis E, Brunelle A, Laprevote O. Improvement of biological time-of-flight-secondary ion mass spectrometry imaging with a bismuth cluster ion source. J Am Soc Mass Spectrom. 2005;16:1608–18. doi: 10.1016/j.jasms.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 86.Malmberg P, Börner K, Yun C, Friberg P, Hagenhoff B, Mansson JE, et al. Localization of lipids in the aortic wall with imaging TOF-SIMS. Biochim Biophys Acta. 2007;1771:185–95. doi: 10.1016/j.bbalip.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 87.Debois D, Bralet MP, Le Naour F, Brunelle A, Laprevote O. In situ lipidomic analysis of nonalcoholic fatty liver by cluster TOF-SIMS imaging. Anal Chem. 2009;81:2823–31. doi: 10.1021/ac900045m. [DOI] [PubMed] [Google Scholar]
- 88.Nygren H, Börner K, Malmberg P, Tallarek E, Hagenhoff B. Imaging TOF-SIMS of rat kidney prepared by high-pressure freezing. Microsc Res Tech. 2005;68:329–34. doi: 10.1002/jemt.20258. [DOI] [PubMed] [Google Scholar]
- 89.Magnusson Y, Friberg P, Sjövall P, Dangardt F, Malmberg P, Chen Y. Lipid imaging of human skeletal muscle using TOF-SIMS with bismuth cluster ion as a primary ion source. Physiol Funct Imaging. 2008;28:202–9. doi: 10.1111/j.1475-097X.2008.00796.x. [DOI] [PubMed] [Google Scholar]
- 90.Touboul D, Roy S, Germain DP, Chaminade P, Brunelle A, Laprevote O. MALDI-TOF and cluster-TOF-SIMS imaging of Fabry disease biomarkers. Int J Mass Spectrom. 2007;260:158–65. doi: 10.1016/j.ijms.2006.09.027. [DOI] [Google Scholar]
- 91.McDonnell LA, Piersma SR, Altelaar AFM, Mize TH, Luxembourg SL, Verhaert PDEM, et al. Subcellular imaging mass spectrometry of brain tissue. J Mass Spectrom. 2005;40:160–8. doi: 10.1002/jms.735. [DOI] [PubMed] [Google Scholar]
- 92.Delcourte A, Bertrand P. Interest of silver and gold metallization for molecular SIMS and SIMS imaging. Appl Surf Sci. 2004;231:250–5. doi: 10.1016/j.apsusc.2004.03.029. [DOI] [Google Scholar]
- 93.Keune K, Boon JJ. Enhancement of the static SIMS secondary ion yields of lipid moieties by ultrathin gold coating of aged oil paint surfaces. Surf Interface Anal. 2004;36:1620–8. doi: 10.1002/sia.1996. [DOI] [Google Scholar]
- 94.Delecorte A, Bour J, Aubriet F, Muller JF, Bertrand P. Sample metallization for performance improvement in desorption/ionization of kilodalton molecules: quantitative evaluation, imaging secondary ion MS, and laser ablation. Anal Chem. 2003;75:6875–85. doi: 10.1021/ac0302105. [DOI] [PubMed] [Google Scholar]
- 95.Piehowski PD, Carado AJ, Kurczy ME, Ostrowski SG, Heien ML, Winograd N, et al. MS/MS methodology to improve subcellular mapping of cholesterol using TOF-SIMS. Anal Chem. 2008;80:8662–7. doi: 10.1021/ac801591r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schueler B. Microscope imaging by time-of-flight secondary ion mass spectrometry. Microsc Microanal Microstruct. 1992;3:119–39. doi: 10.1051/mmm:0199200302-3011900. [DOI] [Google Scholar]
- 97.Luxembourg SL, Mize TH, McDonnell LA, Heeren RMA. High-spatial resolution mass spectrometric imaging of peptide and protein distributions on a surface. Anal Chem. 2004;76:5339–44. doi: 10.1021/ac049692q. [DOI] [PubMed] [Google Scholar]
- 98.Vaalburg W. Preclinical pharmacokinetic PET studies with radiolabeled potential new drugs in man. Drug Inf J. 1997;31:1015–8. [Google Scholar]
- 99.Rao J, Dragulescu A, Yao H. Fluorescence imaging in vivo: recent advances. Curr Opin Biotechnol. 2007;18(1):17–25. doi: 10.1016/j.copbio.2007.01.003. [DOI] [PubMed] [Google Scholar]