Abstract
The yeast three-hybrid system (Y3H) is a powerful tool to select or confirm RNA–protein interactions. Target protein recognition of an RNA insert within a test transcript depends on at least three factors: intrinsic protein affinity for the properly folded insert, retention of RNA insert tertiary structure within a longer RNA transcript, and accessibility of the RNA insert to the target protein. Y3H reporter gene readout reflects the combination of these factors. Here, we discuss RNA insert tertiary structure and accessibility in the Y3H as “RNA display.” We review evidence that RNA display can sometimes be optimized during Y3H selections that do not increase the intrinsic affinity of an RNA insert for a target protein. This situation is more likely when a library of RNA inserts and heterogeneous flanking sequences is subjected to selection, and is less likely when point mutations are targeted to the insert in a fixed context. An RNA display vector with enhanced modularity has been developed to minimize sequence context effects in the Y3H.
Keywords: RNA folding, aptamer, genetic selection, yeast three-hybrid system
INTRODUCTION
In 1996, SenGupta et al. (1996) introduced the yeast three-hybrid system (Y3H) (Fig. 1), an ingenious method to assay RNA–protein interactions in vivo. Identifying RNA–protein interactions involves transformation of a yeast strain with RNA and protein expression plasmids, followed by assays of HIS3 and lacZ reporter genes (SenGupta et al. 1996, 1999). An experimental RNA insert (“bait”) is cloned into an RNA expression vector, for example, pIIIA-MS2-2, which provides the molecular recognition required for tethering to yeast chromatin upstream of HIS3 and lacZ reporter genes (Fig. 1). An MS2 RNA hairpin–MS2 protein interaction is exploited for this purpose in the original design. A protein (“prey”) is expressed as a fusion with the Gal4 transcription activation domain. The Y3H has been successfully used to confirm natural or artificial RNA–protein interactions and to identify novel RNA partners for target proteins by selection from RNA or protein libraries (SenGupta et al. 1996; Zhang et al. 1999, 2000; Kraemer et al. 2000; Cassiday and Maher 2001, 2003; van Zon et al. 2001; Bernstein et al. 2002, 2005; Sokolowski et al. 2003; Spiridonov et al. 2003; Klimek-Tomczak et al. 2004; Hook et al. 2005; Riley et al. 2006; Seay et al. 2006; Konig et al. 2007; Stumpf et al. 2008; Koh et al. 2009; Wurster et al. 2009).
FIGURE 1.
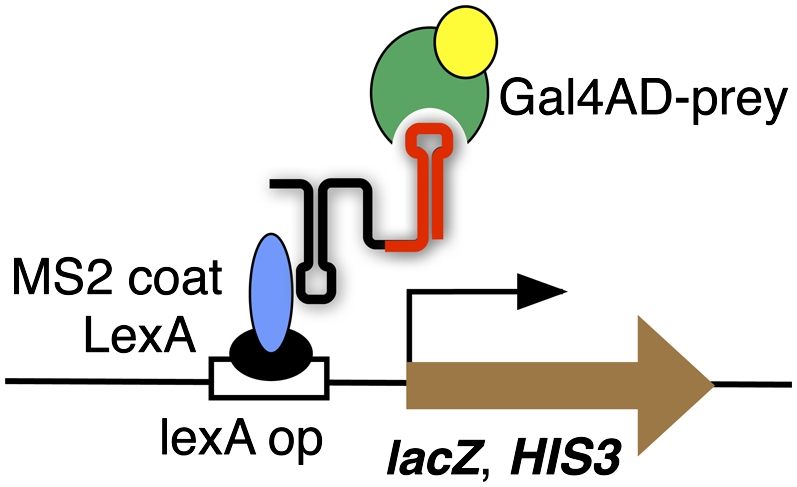
Schematic illustration of Y3H. (A) The RNA insert (red) is expressed in the context of RNA vector sequences (black) tethered upstream of lacZ (brown) and HIS3 reporter genes via a MS2 coat–LexA fusion protein (blue and black). Gene activation depends on binding of the Gal4 activation domain (yellow) –prey fusion protein (green).
Our laboratory has implemented the Y3H to confirm the ability of in vitro selected anti-NF-κB RNA aptamers to bind their transcription factor targets, and to optimize the RNA aptamers by in vivo selection (Cassiday and Maher 2003; Wurster and Maher 2008; Wurster et al. 2009). We have routinely selected aptamer variants with improved Y3H reporter gene readout, but frequently observe that the identified RNA aptamer variants do not display increased affinity for the target protein when isolated from the display template and tested by in vitro assay. Here we discuss the basis for these findings and suggest a Y3H RNA display vector with improved modularity.
RNA DISPLAY
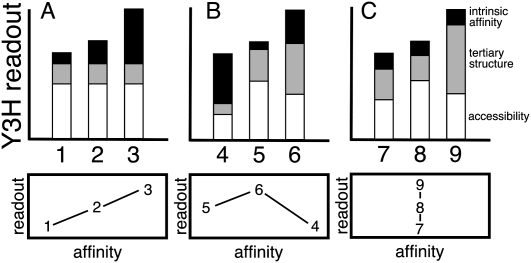
When assaying protein recognition of RNA inserts, in vivo interaction depends on at least three factors: intrinsic affinity of the isolated RNA insert for the target protein, retention of required RNA insert tertiary structure within a longer RNA transcript, and accessibility of the insert to the target protein. The combination of these three factors dictates the fitness of an RNA insert for target protein interaction and reporter gene readout. We use the term “RNA display” to describe the tertiary structure and accessibility of an RNA insert in the Y3H. Figure 2 illustrates how intrinsic affinity, tertiary structure, and accessibility can be considered as additive factors in determining Y3H readout for strains carrying different RNA inserts in different sequence contexts in the presence of a single target protein. If display factors (Fig. 2, white and gray bars) are constant, Y3H readout will be correlated with RNA–protein affinity (Fig. 2, black bars) measured in biochemical experiments with the isolated RNA insert. This ideal situation is most likely if RNA inserts are similar in sequence and carry mutations targeted to sites of RNA–protein interaction (Hook et al. 2005; Koh et al. 2009).
FIGURE 2.
Theoretical contributions to Y3H readout. Y3H readout reflects the combination of the intrinsic affinity of an isolated RNA insert for a protein target as measured in a biochemical experiment (black), RNA insert tertiary structure in the context of the Y3H transcript (gray), and accessibility of the RNA insert at the reporter gene promoter in vivo (white). (A) Correlation between intrinsic affinity and Y3H readout for RNAs 1–3 because display factors are constant. (B) Lack of correlation between intrinsic target affinity and Y3H readout for RNAs 4–6 because of differing display. (C) Different Y3H readouts for RNAs 7–9 with comparable intrinsic target affinities because of differing display.
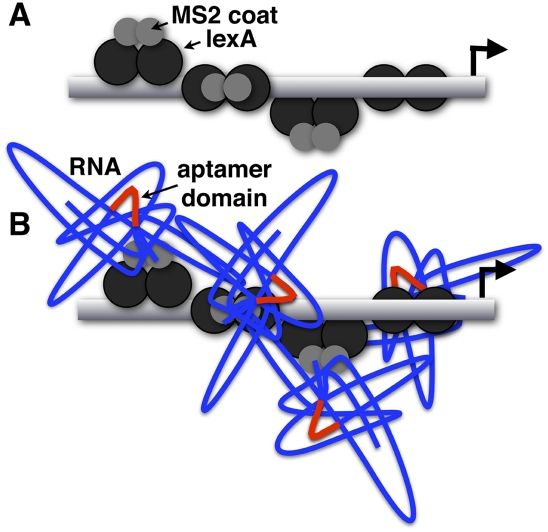
Alternatively, we have commonly observed selection for increased Y3H reporter readout without a corresponding increase in biochemical affinity (Fig. 2B,C). This kind of display optimization can be dramatic, such that a well-displayed RNA insert of moderate intrinsic biochemical affinity can give a higher Y3H readout than a higher-affinity RNA insert displayed sub-optimally (Fig. 2B). Likewise, RNA inserts with comparable biochemical affinities can give different Y3H readouts because of differences in display (Fig. 2C). Unintentional selection for improved context (RNA display) is therefore possible when a functional RNA insert is subjected to genetic selection (HIS3 readout) in a library of different sequence contexts (Fig. 3). The significance of accessibility is compounded by the fact that four or eight LexA operators are present upstream of Y3H reporter genes (Benton et al. 1990). Each LexA operator site may be bound by a LexA–MS2 fusion protein dimer (Valegard et al. 1994; Butala et al. 2009), and each bound protein dimer has the potential to display an RNA (typically >300 nt in length). Thus molecular crowding may also contribute to the accessibility of the RNA insert (Fig. 4B).
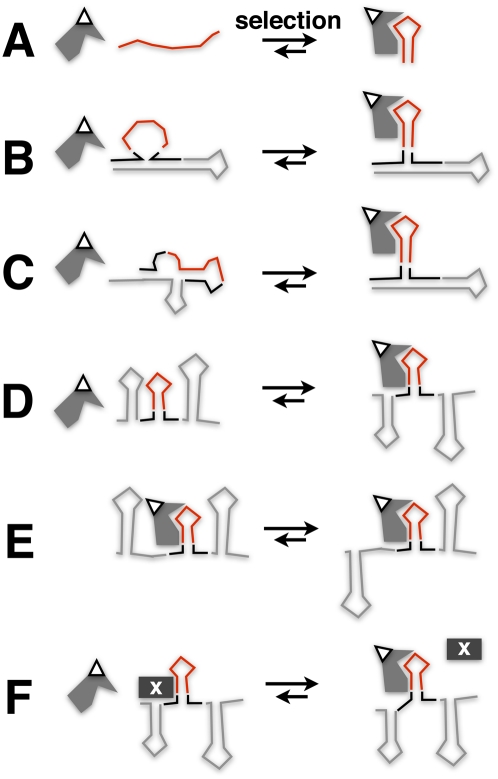
FIGURE 3.
Optimization of Y3H readout by selection. (A) An RNA insert (red) selected in isolation for proper folding to interact with target (gray polygon)/activation domain (white triangle) fusion protein. (B) Selection for flanking RNA sequences (black) that enhance proper local tertiary structure context (gray) for the RNA insert. (C) Selection for flanking RNA sequences that enhance proper global folding of display RNA. (D) Selection for flanking RNA sequences that promote accessibility of the RNA insert to target the protein-binding domain. (E) Selection for flanking RNA sequences that enhance access of bound target protein to transcriptional cofactors. (F) Selection for flanking RNA sequences that destabilize a competing complex with yeast protein “X” (black rectangle), enhancing accessibility of RNA insert.
FIGURE 4.
Steric considerations in the Y3H system. (A) Multiple LexA (black circles)/MS2 coat (gray circles) dimers are bound to yeast promoter DNA near lacZ and HIS3 reporter genes. (B) The RNA insert is displayed in the context of multiple dynamic RNA–protein complexes. Y3H readout reflects the intrinsic affinity of the target protein for the RNA insert, but also contributions of RNA display (RNA tertiary structure and accessibility).
The Y3H has been widely successful in detecting expected RNA–protein interactions, so the faithfulness and sensitivity of the assay are not at issue. The focus here is on quantitative experiments designed to extrapolate Y3H readout to the intrinsic affinity of isolated RNA insert and target protein in biochemical experiments. We provide examples from the literature to illustrate certain cases in which Y3H reporter gene readout and biochemical affinity measurements are and are not consonant.
RELATING Y3H REPORTER GENE READOUT TO IN VITRO RNA/PROTEIN AFFINITY
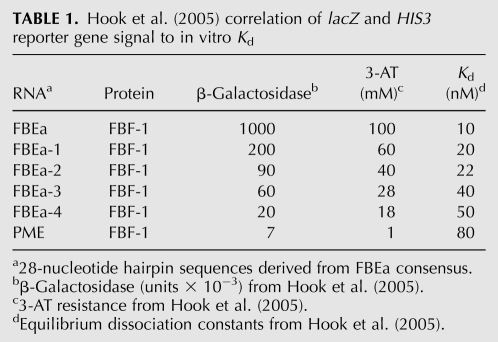
In a systematic analysis, Hook et al. (2005) compared Y3H HIS3 and lacZ reporter gene readout and in vitro equilibrium dissociation constant (Kd) values. The comparison involved PUF family member FBF-1 binding to the wild-type PME RNA hairpin and derivatives of a 28-nt FBEa RNA. PUF family proteins regulate stem cell maintenance by translational repression of factors involved in cell differentiation. PUF proteins bind 3′-untranslated region (UTR) hairpins in target messages (Wickens et al. 2002). FBEa and PME RNAs are naturally occurring hairpin structures bound by FBF-1 (Hook et al. 2005). By introducing point mutations into the 11-nt consensus sequence necessary for FBF-1 binding within the 28-nt wild-type hairpin sequence of FBEa (Bernstein et al. 2005), the authors created a set of RNAs with in vitro binding affinities for FBF-1 over a 10–100 nM range (Bernstein et al. 2005; Hook et al. 2005). For Y3H testing, FBEa and PME RNA sequences were cloned into the pIIIA-MS2-2 display cassette and transformed into the Y3H assay strain in the presence of FBF-1. The investigators reported that both HIS3 activation (3-AT resistance) and the logarithm of the β-galactosidase activity demonstrated linear correlation with the in vitro Kd values measured for isolated RNA hairpins binding to the FBF-1 complex (Table 1). A similar correspondence between in vitro and in vivo data was reported by Koh et al. (2009).
TABLE 1.
Hook et al. (2005) correlation of lacZ and HIS3 reporter gene signal to in vitro Kd
Correlation between in vitro affinity and Y3H readout is robust in these cases. Three aspects of the experimental design may have played important roles. First, insertion of the target RNA sequences into the expressed Y3H transcript adequately preserved RNA display (native tertiary structure presented to PUF proteins in 3′ UTRs and accessibility within the larger RNA). Second, the investigators compared nearly identical sequences. Third, mutations were targeted to the core consensus sequence within the insert.
ANTI-P50 Y3H SELECTIONS DO NOT IMPROVE INTRINSIC TARGET AFFINITY MEASURED IN VITRO
In 2003, our laboratory reported two different Y3H RNA selections to identify point mutants of an in vitro selected anti-p50 RNA aptamer for the purpose of improving its affinity for the target NF-κB transcription factor p502 (Cassiday and Maher 2003). An RNA library encoded aptamers from an early round of the in vitro selection experiment that eventually yielded the anti-p50 aptamer.
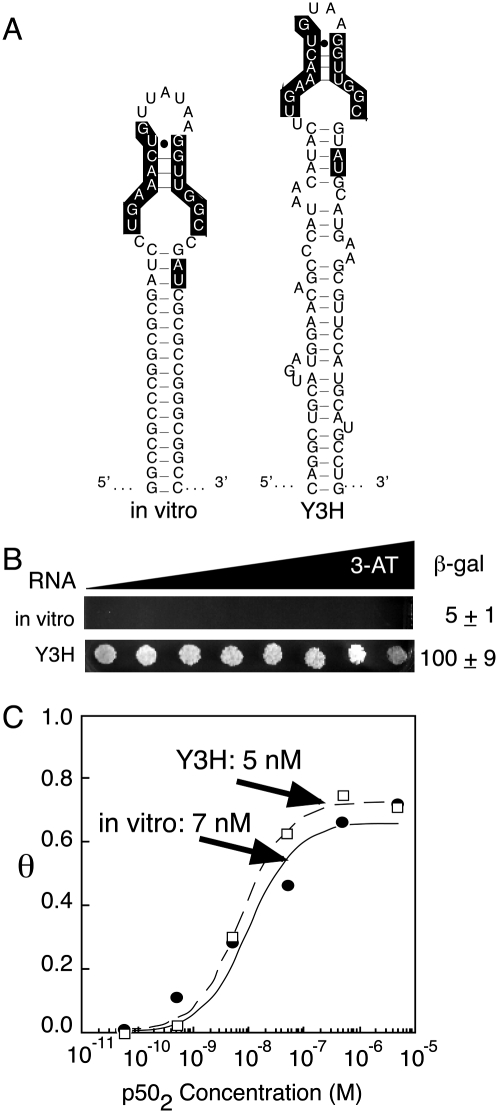
Figure 5A depicts the original anti-p50 RNA aptamer identified by in vitro selection. Subsequent crystallographic analysis of the anti-p50:p502 complex (Huang et al. 2003) revealed that conserved nucleotides are located in the central stem and internal bulge of the hairpin (Fig. 5A, highlighted). Y3H selection yielded RNAs with threefold to 20-fold improved reporter gene readouts (Fig. 5). Because the RNAs were shown to accumulate equally in yeast, it was assumed that improved Y3H activity was due to enhanced target protein binding. In fact, when isolated from the context of the RNA expression cassette sequences, the Y3H-selected RNAs displayed in vitro affinities for NF-κB p502 indistinguishable from that of the original SELEX-derived 31-nt anti-p50 RNA (Fig. 5C). Varying sequence context can evidently influence the display of a single RNA insert, yielding a range of improved Y3H reporter gene readouts after selection. Had the selections been performed in a fixed sequence context with mutations targeted only to positions required for high-affinity complex formation with p502, we predict that correlation between Y3H readout and in vitro affinity would have been more likely.
FIGURE 5.
Anti-p50 Y3H selections and affinity analysis in vitro (Cassiday and Maher 2003). (A) Predicted secondary structure of in vitro selected anti-p50 RNA aptamer (left) compared to predicted secondary structure Y3H-selected variant from early in vitro selection RNA library (right). (B) Comparison of aptamer activities in Y3H assays. 3-AT gradient plate yeast growth image (left) and liquid β-galactosidase assays values (right) compare HIS3 and lacZ reporter gene signals. (C) Binding isotherms for isolated RNA aptamer complexes with p502 showing similar affinities despite different Y3H readouts.
P53 STUDIES DEMONSTRATE IMPORTANCE OF RNA DISPLAY IN Y3H
Our laboratory also reported a study of nonspecific RNA–p53 interactions using the Y3H (Riley et al. 2006). RNA selections were performed seeking an RNA consensus sequence within a 60-nt random region for high-affinity binding by p53. A large fraction of the tested library RNAs bound p53 and produced lacZ and HIS3 signals above background. Signals were sequence-dependent. Efforts were made to correlate predicted RNA secondary structure with Y3H reporter gene activity, but no consensus could be reliably derived. Differences in RNA abundance were ruled out by Northern analysis. Subsequent biochemical experiments revealed that p53 bound all RNA insert sequences that emerged from Y3H selections with equal affinity in vitro (Riley et al. 2006). The Y3H therefore has the ability to select RNAs from heterogeneous libraries on the basis of display to a sequence-non-specific RNA binding protein.
Y3H SELECTIONS FROM GENOMIC SEQUENCES IDENTIFY RNA INSERTS WHERE REPORTER GENE READOUT DOES NOT CORRELATE WITH IN VITRO TARGET AFFINITY
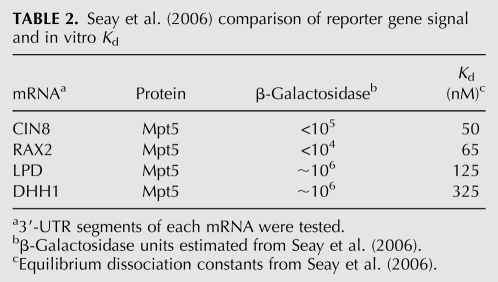
In 2006, Seay et al. (2006) reported a Y3H selection to identify 3′-UTR sequences recognized by the PUF protein Mpt5. The investigators ligated ∼50–150-bp fragments of yeast chromosomal DNA into pIIIA-MS2-2 for expression of test transcripts. Y3H assay strain YBZ-1 was transformed with this library in the presence of plasmid Gal4AD-Mpt5. The authors screened 12 million transformants for HIS3 reporter gene activation. Sixty-nine unique RNAs were identified in the selection, with 36% representing candidate 3′-UTR sequences. Seay and colleagues ranked the selected sequences on the basis of Y3H reporter gene readout and compared these results with equilibrium dissociation constants measured in vitro for each RNA/Mpt5 complex. No correlation between reporter gene readout and in vitro Kd is evident (Table 2). For example, Y3H readouts ranked candidate 3′-UTR sequences in the order DHH1/LPD > CIN8 > RAX2 when tested with Mpt5, whereas the corresponding in vitro analysis yielded the ranking CIN8 > RAX2 > DHH1/LPD. We suggest that DHH1 (best RNA insert by Y3H) benefits from superior RNA display compared to CIN8 since in vitro Kd values were 325 nM for DHH1-Mpt5 and 50 nM for CIN8-Mpt5. This study illustrates both the power of Y3H selections to identify RNA sequences bound by a target protein, and an instance where Y3H readout and biochemical affinity are not correlated.
TABLE 2.
Seay et al. (2006) comparison of reporter gene signal and in vitro Kd
Y3H SELECTIONS FOR ANTI-NF-κB P65 RNA APTAMERS IDENTIFY RNA INSERTS WHERE REPORTER GENE READOUT POORLY CORRELATES WITH IN VITRO TARGET AFFINITY
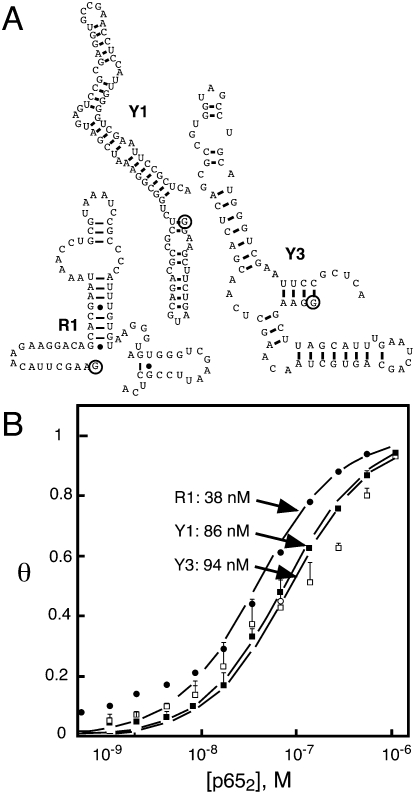
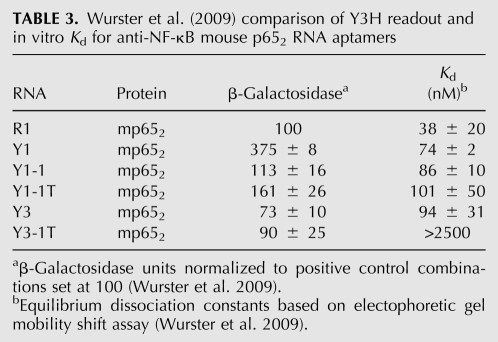
Our laboratory has described in vitro selection of an anti-p65 RNA aptamer termed R1. This RNA is characterized by 5′ and 3′ terminal sequences that are required for p652 binding (low nanomolar range) in vitro (Wurster and Maher 2008). Y3H analysis suggested that R1 has a high specificity for murine NF-κB p652 in the Y3H. Therefore, we undertook Y3H RNA selections to identify candidate anti-p65 RNA aptamers with broad specificities for NF-κB p652 orthologs (Wurster et al. 2009). We transformed the Y3H assay strain with plasmids encoding the Rel homology region of NF-κB zebrafish p652. We included a plasmid expressing an RNA library derived from an early round of the in vitro selection experiment that originally yielded the anti-p65 R1 aptamer (Wurster and Maher 2008). Transformants were selected for HIS3 activation. This procedure identified two new anti-p65 RNA aptamers termed Y1 and Y3. These RNAs are predicted (Jaeger et al. 1989; Zuker 1989) to have much more compact secondary structures compared to the anti-p65 R1 RNA aptamer (Fig. 6). Truncation analysis created variants Y1-1, Y1-1T, and Y3-1/1T (Table 3). In the presence of p652, reporter gene readout for Y1 was approximately fourfold stronger than for the corresponding yeast strain expressing the R1 RNA. In contrast, in vitro analysis of the Y1–p652 complex yielded a Kd value twofold weaker than the R1–p652 complex. Thus, a correlation was not observed between Y3H reporter gene readout and the equilibrium dissociation constant measured for the isolated RNA aptamer in vitro (Table 3).
FIGURE 6.
Anti-p65 Y3H selections and affinity analysis in vitro (Wurster et al. 2009). (A) Comparison of predicted secondary structures for R1, Y1, and Y3 anti-p65 RNA aptamers. (Circles) 5′ RNA termini. (B) In vitro binding isotherms for isolated RNA aptamer complexes with p652.
TABLE 3.
Wurster et al. (2009) comparison of Y3H readout and in vitro Kd for anti-NF-κB mouse p652 RNA aptamers
RNA EXPRESSION VECTOR DESIGN TO MINIMIZE VARIATION IN Y3H RNA DISPLAY
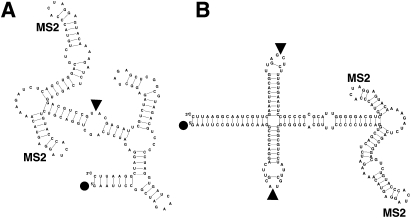
What steps might be taken to increase the correlation between Y3H readout and intrinsic RNA–protein affinity measured in biochemical experiments? One approach (described above) is to limit comparison to targeted mutations within a strictly defined RNA insert tested in a fixed sequence context. Another approach is to sequester the RNA insert in an autonomous domain within the test transcript. Ideally, such a domain would fold independently and project away from the large test transcript. We have studied three approaches to this goal. First, we included complementary G/C-rich sequences flanking the RNA insert to generate a stable stem structure (Cassiday and Maher 2003). This approach permitted strong RNA insert interaction with the target protein but did not prevent inconsistent RNA display when inserts of various sizes were tested. Second, we have attempted systematic optimization of flanking sequences to maximize autonomy (Wurster et al. 2009). Computational approaches were used to predict the folding autonomy of specified or random RNA inserts within the standard Y3H display transcript for all possible 6-bp (4096) or 10-bp (1,048,576) complementary sequences flanking the insert. Optimal stem sequences were then tested in the Y3H. In the case of the optimized 6-bp stem, Y3H readout for the R1 aptamer was marginally improved relative to a non-optimal stem, but aptamer function remained sensitive to details of the stem junction. Several optimized 10-bp stems were included flanking a 60-nt random sequence library adjacent to a functional RNA aptamer. When expressed with the target protein, random inserts projecting from optimized stems were twice as likely to produce detectable Y3H readouts. However, even with optimized stems, Y3H readouts in the presence of an adjacent 60-nt random sequence averaged only 3% of the readout without the random sequence. Thus, 6-bp or 10-bp stems do not resolve RNA display issues. Third, we have recently described a redesigned RNA expression vector for the Y3H. This “T-cassette” strategy (Fig. 7) clones RNA inserts in the context of a four-arm junction structure (Wurster et al. 2009). Spatial separation and insert autonomy are enhanced in such transcripts, with similar transcript accumulation and Y3H readouts increased twofold to sixfold. This T-cassette strategy has the potential to increase consistency in RNA display for experiments wherein RNA insert flanking sequences are heterogeneous but there is the desire to correlate Y3H readout with intrinsic affinity measurements.
FIGURE 7.
Comparison of Y3H RNA expression cassettes. (A) Transcript from conventional RNA expression cassette. (B) T-cassette design (Wurster et al. 2009) to enhance modularity (folding autonomy) and accessibility of RNA inserts emanating from a four-way helical junction. (Black dots) Transcript 5′ termini. Transcript 3′ terminal sequences are truncated for clarity. Bacteriophage MS2 hairpins are labeled. (Arrows) Cloning sites.
CONCLUSION
The Y3H serves as an excellent tool for identifying functional variants of an RNA insert, and for selecting RNA–protein interactions. Y3H readout in the presence of a target protein measures the overall fitness of the RNA insert for protein recognition. This fitness, in turn, reflects contributions from both the intrinsic target affinity of the isolated RNA insert and RNA display. We show that RNA display is a selectable parameter that can affect Y3H readout apart from intrinsic RNA insert–protein affinity. Our review of the literature suggests that the Y3H faithfully detects specific RNA/protein interactions. When conservative RNA insert variants are tested in a fixed sequence context, correlation between Y3H reporter gene readout and biochemical protein–RNA affinity measurements is robust. We reviewed exceptional cases in which this correlation is not observed. These examples appear to reflect selection operating on libraries of RNA inserts in heterogeneous flanking sequence contexts. In these cases, selection for improved RNA display can occur without an improvement in the protein affinity for the isolated RNA insert. RNA expression vectors with highly structured domains for RNA insert cloning may be helpful (Wurster et al. 2009).
ACKNOWLEDGMENTS
We thank Y.F. Her, J.P. Bida, and other members of the Maher laboratory for their contributions. This work was supported by the Mayo Foundation and by NIH grant GM68128 (L.J.M.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1880410.
REFERENCES
- Benton BM, Eng WK, Dunn JJ, Studier FW, Sternglanz R, Fisher PA. Signal-mediated import of bacteriophage T7 RNA polymerase into the Saccharomyces cerevisiae nucleus and specific transcription of target genes. Mol Cell Biol. 1990;10:353–360. doi: 10.1128/mcb.10.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DS, Buter N, Stumpf C, Wickens M. Analyzing mRNA–protein complexes using a yeast three-hybrid system. Methods. 2002;26:123–141. doi: 10.1016/S1046-2023(02)00015-4. [DOI] [PubMed] [Google Scholar]
- Bernstein D, Hook B, Hajarnavis A, Opperman L, Wickens M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butala M, Zgur-Bertok D, Busby SJ. The bacterial LexA transcriptional repressor. Cell Mol Life Sci. 2009;66:82–93. doi: 10.1007/s00018-008-8378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassiday LA, Maher LJ., III In vivo recognition of an RNA aptamer by its transcription factor target. Biochemistry. 2001;40:2433–2438. doi: 10.1021/bi002376v. [DOI] [PubMed] [Google Scholar]
- Cassiday LA, Maher LJ., III Yeast genetic selections to optimize RNA decoys for transcription factor NF-κB. Proc Natl Acad Sci. 2003;100:3930–3935. doi: 10.1073/pnas.0736013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook B, Bernstein D, Zhang B, Wickens M. RNA–protein interactions in the yeast three-hybrid system: Affinity, sensitivity, and enhanced library screening. RNA. 2005;11:227–233. doi: 10.1261/rna.7202705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DB, Vu D, Cassiday LA, Zimmerman JM, Maher LJ, III, Ghosh G. Crystal structure of NF-κB (p50)2 complexed to a high-affinity RNA aptamer. Proc Natl Acad Sci. 2003;100:9268–9273. doi: 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger JA, Turner DH, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimek-Tomczak K, Wyrwicz LS, Jain S, Bomsztyk K, Ostrowski J. Characterization of hnRNP K protein–RNA interactions. J Mol Biol. 2004;342:1131–1141. doi: 10.1016/j.jmb.2004.07.099. [DOI] [PubMed] [Google Scholar]
- Koh YY, Opperman L, Stumpf C, Mandan A, Keles S, Wickens M. A single C. elegans PUF protein binds RNA in multiple modes. RNA. 2009;15:1090–1099. doi: 10.1261/rna.1545309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Julius C, Baumann S, Homann M, Goringer HU, Feldbrugge M. Combining SELEX and the yeast three-hybrid system for in vivo selection and classification of RNA aptamers. RNA. 2007;13:614–622. doi: 10.1261/rna.334307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer B, Zhang B, SenGupta D, Fields S, Wickens M. Using the yeast three-hybrid system to detect and analyze RNA–protein interactions. Methods Enzymol. 2000;328:297–321. doi: 10.1016/s0076-6879(00)28404-8. [DOI] [PubMed] [Google Scholar]
- Riley KJ, Cassiday LA, Kumar A, Maher LJ., III Recognition of RNA by the p53 tumor suppressor protein in the yeast three-hybrid system. RNA. 2006;12:620–630. doi: 10.1261/rna.2286706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seay D, Hook B, Evans K, Wickens M. A three-hybrid screen identifies mRNAs controlled by a regulatory protein. RNA. 2006;12:1594–1600. doi: 10.1261/rna.145306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenGupta DJ, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA–protein interactions in vivo. Proc Natl Acad Sci. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenGupta DJ, Wickens M, Fields S. Identification of RNAs that bind to a specific protein using the yeast three-hybrid system. RNA. 1999;5:596–601. doi: 10.1017/s1355838299002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski M, Scott JE, Heaney RP, Patel AH, Clements JB. Identification of herpes simplex virus RNAs that interact specifically with regulatory protein ICP27 in vivo. J Biol Chem. 2003;278:33540–33549. doi: 10.1074/jbc.M302063200. [DOI] [PubMed] [Google Scholar]
- Spiridonov VG, Smirnova SA, Mel'nichuk MD. Use of three-hybrid system to detect RNA-binding activity of alfalfa mosaic virus coat protein. Ukr Biokhim Zh. 2003;75:75–80. [PubMed] [Google Scholar]
- Stumpf CR, Kimble J, Wickens M. A Caenorhabditis elegans PUF protein family with distinct RNA binding specificity. RNA. 2008;14:1550–1557. doi: 10.1261/rna.1095908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valegard K, Murray JB, Stockley PG, Stonehouse NJ, Liljas L. Crystal structure of an RNA bacteriophage coat protein–operator complex. Nature. 1994;371:623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- van Zon A, Mossink MH, Schoester M, Scheffer GL, Scheper RJ, Sonneveld P, Wiemer EA. Multiple human vault RNAs. Expression and association with the vault complex. J Biol Chem. 2001;276:37715–37721. doi: 10.1074/jbc.M106055200. [DOI] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Wurster SE, Maher LJ., III Selection and characterization of anti-NF-κB p65 RNA aptamers. RNA. 2008;14:1037–1047. doi: 10.1261/rna.878908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurster SE, Bida JP, Her YF, Maher LJ., III Characterization of anti-NF-κB RNA aptamer binding specificity in vitro and in the yeast three-hybrid system. Nucleic Acids Res. 2009;37:6214–6224. doi: 10.1093/nar/gkp670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Kraemer B, SenGupta D, Fields S, Wickens M. Yeast three-hybrid system to detect and analyze interactions between RNA and protein. Methods Enzymol. 1999;306:93–113. doi: 10.1016/s0076-6879(99)06007-3. [DOI] [PubMed] [Google Scholar]
- Zhang B, Kraemer B, SenGupta D, Fields S, Wickens M. Yeast three-hybrid system to detect and analyze RNA–protein interactions. Methods Enzymol. 2000;318:399–419. doi: 10.1016/s0076-6879(00)18066-8. [DOI] [PubMed] [Google Scholar]
- Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]