Abstract
Proper 3′ end processing of a nascent transcript is critical for the functionality of the mature RNA. Although it has long been thought that virtually all long RNA polymerase II transcripts terminate in a poly(A) tail that is generated by endonucleolytic cleavage followed by polyadenylation, noncanonical 3′ end processing mechanisms have recently been identified at several gene loci. Unexpectedly, enzymes with well-characterized roles in other RNA processing events, such as tRNA biogenesis and pre-mRNA splicing, cleave these nascent transcripts to generate their mature 3′ ends despite the presence of nearby polyadenylation signals. In fact, the presence of multiple potential 3′ end cleavage sites is the norm at many human genes, and recent work suggests that the choice among sites is regulated during development and in response to cellular cues. It is, therefore, becoming increasing clear that the selection of a proper 3′ end cleavage site represents an important step in the regulation of gene expression and that the mature 3′ ends of RNA polymerase II transcripts can be generated via multiple mechanisms.
Keywords: polyadenylation, post-transcriptional regulation of gene expression, RNA splicing, tRNA biogenesis, RNA processing, RNA polymerase II
INTRODUCTION
3′ End processing of a nascent transcript is critical for allowing the release of RNA polymerase from its template and for ensuring the proper functionality of the mature RNA. By varying the 3′ end cleavage site, additional sequence motifs are included (or excluded) at the 3′ end of the mature RNA, which can, for example, affect the transcript's stability or subcellular localization (for review, see Moore 2005). In addition, the choice of an alternative 3′ end processing site can result in a mature messenger RNA that encodes a protein with very different domains and function, as exemplified by the immunoglobulin M (IgM) heavy chain gene locus, where the choice between 3′ end cleavage sites determines if the mature RNA encodes a protein that is secreted or localized to the membrane (Takagaki et al. 1996). As there are two or more functional polyadenylation signals at more than half of all human genes (Tian et al. 2005; Yan and Marr 2005), the selection of each 3′ end cleavage site must be tightly regulated. In fact, mutations in polyadenylation signals or in 3′ end processing factors have been linked to numerous human diseases, including cancer and thalassemia, highlighting the impact that inappropriate 3′ end formation can have on human health (for review, see Danckwardt et al. 2008).
It has long been thought that the mature 3′ end of nearly all long RNA polymerase II transcripts, besides many histone mRNAs, is generated in a two-step reaction that involves endonucleolytic cleavage followed by the addition of adenosine (A) residues in a nontemplated fashion. However, large-scale studies of the human transcriptome now indicate that transcription is pervasive throughout the human genome (for review, see Kapranov et al. 2007; Wilusz et al. 2009) and suggest that a significant fraction (>25%) of long transcripts present in cells lack a classical poly(A) tail (Cheng et al. 2005; Wu et al. 2008), indicating that additional 3′ end processing mechanisms likely exist in vivo. In this review, we discuss the known mechanisms by which the mature 3′ ends of long RNA polymerase II (Pol II) transcripts are generated. Canonical cleavage/polyadenylation and histone 3′ end formation are only briefly addressed as these mechanisms have been thoroughly reviewed elsewhere (Colgan and Manley 1997; Zhao et al. 1999; Proudfoot 2004; Marzluff et al. 2008). Instead, we focus on two recently identified 3′ end processing mechanisms that use enzymes with well-characterized roles in other RNA processing events. As there are often multiple potential cleavage sites at the 3′ ends of human genes, we then highlight recent genome-wide studies that have begun to reveal how the choice of a 3′ end cleavage site is systematically regulated and the effects that this has on the fate of the mature transcripts.
CANONICAL CLEAVAGE/POLYADENYLATION
A poly(A) tail is thought to be added post-transcriptionally to the 3′ end of almost all eukaryotic mRNAs, which influences the transcript's stability, translational efficiency, and export to the cytoplasm (for review, see Colgan and Manley 1997; Zhao et al. 1999; Proudfoot 2004). Core polyadenylation sequence elements present in nascent transcripts, including the hexanucleotide AAUAAA (or a close variant) and a downstream G/U-rich sequence, recruit the cleavage/polyadenylation machinery, resulting in endonucleolytic cleavage of the RNA by CPSF-73 (Mandel et al. 2006). A poly(A) tail (up to 200–250 adenosines in mammalian cells) is then added by poly(A) polymerase in a nontemplated fashion to the 3′ end of the transcript. Interestingly, the length of the poly(A) tail can be regulated to control translation of the mRNA, as exemplified by the regulation of many maternal mRNAs in developing oocytes (for review, see Richter 1999).
Although the mechanism by which a poly(A) tail is added seems fairly simple, proteomic analysis has revealed that the human cleavage/polyadenylation complex may be composed of up to ∼85 proteins (Shi et al. 2009). Interestingly, many of these protein components may not play a direct role in 3′ end formation, but instead function to closely couple cleavage/polyadenylation to multiple other cellular processes, including transcription, especially transcriptional termination, and pre-mRNA splicing (for review, see Shatkin and Manley 2000; Maniatis and Reed 2002; Proudfoot et al. 2002; Bentley 2005; Richard and Manley 2009). Coupling aids in maximizing the efficiency of each step in gene expression and likely contributes to the specificity of poly(A) site choice. In particular, some transcripts have, in addition to the core polyadenylation sequence elements, auxiliary sequence motifs located upstream of or downstream from the cleavage site, which affect the usage of the nearby poly(A) site by binding various accessory proteins (Lutz 2008, and references therein).
Unexpectedly, several recent reports indicate that proteins other than CPSF-73 can generate the 3′ end of an mRNA prior to polyadenylation. For example, at the yeast CTH2 locus, co-transcriptional cleavage at the poly(A) signal does not normally occur, resulting in a readthrough transcript that extends ∼1.8 kilobases (kb) beyond the poly(A) signal (Ciais et al. 2008). However, this primary transcript is then progressively degraded by the 3′-5′ exonuclease activity of the nuclear exosome/TRAMP complex until the complex pauses at a G/U-rich sequence, allowing post-transcriptional polyadenylation and production of the mature polyadenylated mRNA. The RNase III type endonuclease Rnt1 has also now been shown in yeast to be able to cleave and generate the 3′ end of some mRNAs, which are subsequently polyadenylated (Rondón et al. 2009).
HISTONE AND snRNA 3′ END FORMATION
Although nearly all mRNAs are thought to have a poly(A) tail at their 3′ end, it is not an absolute rule. It has long been known that the mature 3′ end of replication-dependent histone mRNAs is generated by U7 snRNA base pairing to a highly conserved element known as the histone downstream element (HDE), followed by endonucleolytic cleavage but no polyadenylation (for review, see Marzluff et al. 2008). Instead of having a poly(A) tail, histone mRNAs have a highly conserved stem–loop structure in their 3′ UTRs that binds the stem–loop binding protein (SLBP) and is functionally analogous to a poly(A) tail as it ensures RNA stability and enhances translational efficiency. Interestingly, despite the obvious mechanistic differences between canonical cleavage/polyadenylation and histone 3′ end formation, CPSF-73 is the endonuclease involved in both mechanisms (Dominski et al. 2005; Mandel et al. 2006).
In addition to protein-coding mRNAs, RNA polymerase II transcribes numerous small noncoding RNAs, including the majority of snRNAs (small nuclear RNAs), which also lack a poly(A) tail at their 3′ ends. At these snRNA loci, phosphorylation of serine 7 of the carboxy-terminal domain (CTD) of RNA polymerase II recruits a complex of at least 12 proteins known as the Integrator, which mediates 3′ end processing of snRNAs near the 3′ box sequence (Baillat et al. 2005; Egloff et al. 2007). Perhaps not surprisingly, two of the Integrator subunits are similar to the subunits of the cleavage and polyadenylation specificity factor (CPSF) complex (Baillat et al. 2005).
RNase P GENERATES THE MATURE 3′ END OF TWO LONG NONCODING RNAS
In its well-characterized role, RNase P endonucleolytically cleaves tRNA precursors to produce the mature 5′ termini of functional tRNAs (for review, see Kirsebom 2007). Importantly, it does this by structural, not sequence-dependent, recognition of tRNAs, allowing RNase P to recognize similar features present in other RNA transcripts. For example, RNase P has been shown to cleave the Saccharomyces cerevisiae noncoding RNA HRA1 (Yang and Altman 2007) and several bacterial riboswitches (Altman et al. 2005), as well as been proposed to cleave several intron-encoded box C/D small nucleolar RNAs (snoRNAs) as part of their maturation process in yeast (Coughlin et al. 2008). A connection between RNase P and 3′ end formation of mRNAs has long been known in mitochondria, where tRNAs generally flank protein-coding sequences in the mitochondrial genome (Ojala et al. 1981). Unexpectedly, we recently showed that RNase P also generates the 3′ ends of two long RNA polymerase II transcripts, MALAT1 and MEN β (Wilusz et al. 2008; Sunwoo et al. 2009).
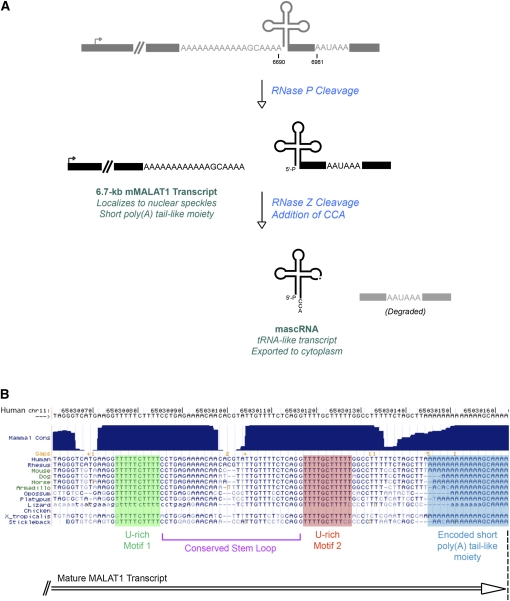
MALAT1 (metastasis-associated lung adenocarcinoma transcript 1), also known as NEAT2 (Hutchinson et al. 2007), is a long noncoding RNA that is misregulated in many cancers (Ji et al. 2003; Lin et al. 2007) and specifically retained in the nucleus in nuclear speckles (Hutchinson et al. 2007), domains that are thought to be involved in the assembly, modification, and/or storage of the pre-mRNA processing machinery (for review, see Lamond and Spector 2003). At the 3′ end of the MALAT1 locus is a cleavage/polyadenylation signal, which can be used to generate a polyadenylated MALAT1 transcript that is ∼7 kb in mouse (∼7.4 kb in human). Unexpectedly, we found this polyadenylated isoform of MALAT1 to be a very minor isoform in vivo (<1% of the MALAT1 transcripts present in the cell). Instead, the mature 3′ end of MALAT1 is almost always generated several hundred nucleotides upstream of the poly(A) site at a region that folds into a tRNA-like structure (Fig. 1A; Wilusz et al. 2008). This upstream region is the most highly evolutionarily conserved portion of the MALAT1 locus (with conservation extending to stickleback) and, like canonical tRNAs, folds into a cloverleaf secondary structure. RNase P recognizes the tRNA-like structure and cleaves to simultaneously generate the mature 3′ end of the abundant MALAT1 transcript and the 5′ end of a small 61-nucleotide (nt) tRNA-like transcript. Additional enzymes involved in tRNA biogenesis, including RNase Z and the CCA-adding enzyme, further process the small RNA, which we have named mascRNA (MALAT1-associated small cytoplasmic RNA), prior to its export to the cytoplasm (Wilusz et al. 2008). Therefore, by using RNase P to generate the mature 3′ end of MALAT1, the cell is able to process a single nascent transcript into two mature transcripts that localize to distinct subcellular locations and likely have unique functions.
FIGURE 1.
MALAT1 is processed at its 3′ end by the tRNA processing machinery. (A) Although there is a canonical polyadenylation signal at the 3′ end of the mouse MALAT1 locus, MALAT1 is primarily processed via an upstream cleavage mechanism, which yields a mature ∼6.7-kb transcript with a short poly(A) tail-like moiety at its 3′ end (Wilusz et al. 2008). Endonucleolytic cleavage by RNase P simultaneously generates the mature 3′ end of MALAT1 and the 5′ end of mascRNA. The small RNA is subsequently cleaved by RNase Z and subjected to CCA addition to generate the mature 61-nt tRNA-like transcript. (B) The MALAT1 poly(A) tail-like moiety (shaded in blue) is highly conserved and encoded in the genome. Further upstream are two nearly perfectly conserved U-rich motifs (shaded in green and orange) separated by a conserved predicted stem–loop.
Despite the 3′ end of MALAT1 being generated via a mechanism very distinct from canonical cleavage/polyadenylation, the mature MALAT1 transcript still has a short (<20 nt) poly(A)-rich tract on its 3′ end (Wilusz et al. 2008). Interestingly, rather than being added on post-transcriptionally, as occurs during polyadenylation, the MALAT1 poly(A) tail-like moiety is actually encoded in the genome and thus part of the nascent transcript (Fig. 1B). RNase P simply cleaves immediately downstream from the A-rich motif (Fig. 1A), providing a twist on how a poly(A) tract can be generated on the 3′ end of a mature Pol II transcript. Considering the short length of the A-rich motif and the fact that it is interrupted by nucleotides other than A, it is perhaps not too surprising that there are additional sequence motifs that stabilize the 3′ end of MALAT1. Upstream of the A-rich motif are two highly conserved U-rich motifs that are predicted to be able to base pair with the poly(A) tail-like moiety (Fig. 1B). Indeed, when we mutated these U-rich motifs to disrupt the base pairing, MALAT1 was deadenylated in vitro, implicating these upstream motifs in stabilizing the MALAT1 transcript (Wilusz et al. 2008). Short upstream U-rich motifs have been shown to stabilize other transcripts by interacting with their poly(A) tails (Muhlrad and Parker 2005; Conrad et al. 2006, 2007), suggesting that this mechanism may be more common than we currently appreciate. In addition, it is tempting to speculate that there are additional transcripts with genomically encoded poly(A) tail-like moieties at their 3′ ends.
Upon searching the mouse and human genomes for sequences similar to the 3′ end of MALAT1, we found that MEN β, a >20-kb noncoding RNA that is, curiously, also retained in the nucleus, is processed at its 3′ end via a very similar mechanism involving RNase P (Sunwoo et al. 2009). In contrast to MALAT1, which localizes to nuclear speckles, MEN β serves as a key structural component of paraspeckles (Sasaki et al. 2009; Sunwoo et al. 2009). Considering that both MALAT1 and MEN β are nuclear retained, a simple model could be envisioned in which 3′ end processing of a long RNA polymerase II transcript by RNase P is a signal to retain the transcript in the nucleus. However, the MALAT1 and MEN β loci both make additional RNA isoforms whose 3′ ends are generated by the canonical cleavage/polyadenylation mechanism and are retained in the nucleus (Wilusz et al. 2008; Sunwoo et al. 2009). Therefore, instead of RNase P cleavage serving as the nuclear retention signal, there are likely sequence motifs within the transcripts that designate them for nuclear retention, perhaps via specific protein interactions. It should be noted that a number of other long nuclear-retained noncoding RNAs, such as Xist (Memili et al. 2001), Hsr-omega-n (Hogan et al. 1994), Airn (Seidl et al. 2006), and Kcnq1ot1 (Redrup et al. 2009), are also polyadenylated, indicating that not all polyadenylated transcripts are exported to the cytoplasm.
THE SPLICEOSOME GENERATES THE MATURE 3′ END OF SCHIZOSACCHAROMYCES POMBE TELOMERASE RNA
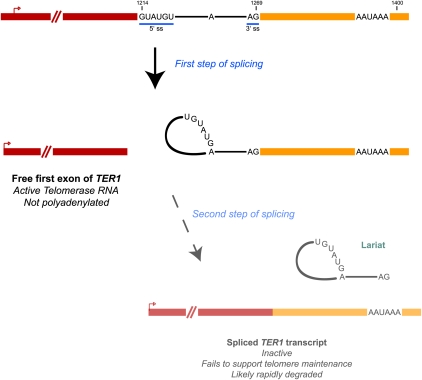
The complete replication of telomeric DNA at the ends of eukaryotic chromosomes requires telomerase, a ribonucleoprotein complex that copies a short template sequence within its intrinsic RNA moiety onto chromosome ends (for review, see Blackburn 2005). In S. pombe, although there is a poly(A) site at the 3′ end of the telomerase RNA (TER1) locus, 95% of the TER1 RNA in the cell ends upstream of this site and is not polyadenylated (Fig. 2; Leonardi et al. 2008; Webb and Zakian 2008). In fact, the longer polyadenylated TER1 isoforms are inactive in vivo as they fail to form a complex with the catalytic protein subunit of telomerase (Box et al. 2008; Leonardi et al. 2008), emphasizing the significance of proper 3′ end site choice for the generation of a functional transcript. How then is the 3′ end of the shorter, functional S. pombe telomerase RNA transcript generated? Recent work indicates that the answer, unexpectedly, is the spliceosome as the mature 3′ end of TER1 precisely maps to a 5′ splice site (Box et al. 2008).
FIGURE 2.
Incomplete splicing generates the functional S. pombe TER1 transcript. A short intron, with canonical 5′ and 3′ splice sites (denoted 5′ ss and 3′ ss, respectively) and branch point sequences, is located upstream of a canonical polyadenylation signal at the 3′ end of the S. pombe TER1 locus. Although the two transesterification steps of splicing are normally tightly coupled, they somehow become uncoupled at the TER1 locus such that only the first step is normally completed. The 5′ exon is released and subsequently functions as the active RNA component of telomerase (Box et al. 2008). If both splicing steps are completed or if the mature 3′ end of TER1 is generated by cleavage/polyadenylation, the resulting TER1 transcripts are inactive and fail to support telomere maintenance.
In its well-characterized role, the spliceosome removes each noncoding intron from pre-messenger RNAs via two transesterification reactions to generate mature messenger RNAs that can be translated (for review, see Wahl et al. 2009). In the first step of RNA splicing, the phosphodiester bond at the 5′ splice site is attacked by the 2′ hydroxyl of an adenosine at the branch point in the intron, forming the lariat intermediate. The 3′ hydroxyl of the free 5′ exon then attacks the phosphodiester bond at the 3′ splice site, resulting in ligation of the exons and excision of the lariat intron. These two steps of RNA splicing are tightly coupled to prevent the release of intermediates and ensure that the exons are properly joined.
However, at the S. pombe TER1 locus, these two splicing steps become uncoupled and only the first transesterification reaction occurs, resulting in the release of the 5′ exon (corresponding to the active TER1 RNA) without exon ligation (Fig. 2; Box et al. 2008). Supporting this model, mutations in the 5′ splice site or the branch point, which affect the first step of splicing, were found to cause a stark decrease in the levels of mature active TER1 RNA and resulted in telomere shortening. In contrast, mutating the 3′ splice site, which only affects the second step in splicing, had no effect on mature TER1 levels or telomere length. Therefore, in a single-step reaction, the spliceosome can generate the mature 3′ end of TER1 RNA in S. pombe (Box et al. 2008) and likely other yeast species (Gunisova et al. 2009). Curiously, if both splicing steps are completed, the resulting TER1 transcript is not active, likely because it is rapidly degraded (Fig. 2). Exactly how the first and second steps of splicing become uncoupled is still a mystery, although the presence of suboptimal splicing motifs in the TER1 transcript appears to play a role.
As with the MALAT1 locus, the mature 3′ end of the S. pombe TER1 transcript is nearly exclusively generated by a noncanonical 3′ end processing mechanism, despite the presence of a downstream poly(A) site. It is possible that these transcripts may be polyadenylated prior to the upstream noncanonical cleavage event, but further experiments are needed to clarify this point and determine if sequential 3′ end cleavage reactions occur or if the noncanonical cleavage event can occur in the absence of downstream polyadenylation. Nevertheless, although there are currently only a handful of known genetic loci that use these noncanonical mechanisms, it is important to specifically point out that they are the rule, rather than the exception, for how the mature 3′ ends of these particular RNAs are generated. In addition, as these noncanonical 3′ end processing mechanisms use enzymes with well-established roles in other RNA processing events, they underscore the previously suggested idea that all of the various RNA processing pathways are interconnected (Shatkin and Manley 2000; Maniatis and Reed 2002; Proudfoot et al. 2002) and provide additional connections that were unexpected.
POST-TRANSCRIPTIONAL CLEAVAGE OF AN mRNA TO ALLOW TRAFFICKING TO THE CYTOPLASM
Although 3′ end formation is generally thought to occur co-transcriptionally (Bentley 2005), some nuclear-retained transcripts are cleaved post-transcriptionally to generate a shorter mRNA isoform that is subsequently exported to the cytoplasm and translated. For example, CTN-RNA is transcribed from the protein-coding mouse cationic amino acid transporter 2 (mCAT2) locus, but the use of a distal poly(A) site causes the ∼8-kb transcript to have a long 3′ UTR that is subjected to RNA editing, resulting in nuclear retention of the transcript (Prasanth et al. 2005). When cells are later subjected to stress, CTN-RNA is somehow post-transcriptionally cleaved in its 3′ UTR to generate a ∼4.2-kb transcript that can traffic to the cytoplasm to be translated. As RNA editing has been observed in the 3′ UTRs of many genes (Kim et al. 2004; Levanon et al. 2004; Chen et al. 2008), it is perhaps not too surprising that a similar regulatory mechanism appears to be employed at the migration stimulating factor (MSF) gene locus (Kay et al. 2005) and proposed to occur at many other genes (Chen et al. 2008). However, the molecular mechanism of this post-transcriptional cleavage process is unclear, as neither the enzymes responsible nor the exact RNA cleavage sites are known. In one possible model, post-transcriptional cleavage would simply generate a new mature 3′ end on the shorter transcript (which may or may not be subsequently polyadenylated). However, a recent bioinformatics study has suggested the intriguing possibility that CTN-RNA and other edited transcripts instead may undergo a noncanonical splicing mechanism that removes the edited region (Osenberg et al. 2009). The resulting noncanonically spliced transcript thus would lack the sequence elements responsible for nuclear retention, but maintain the same mature 3′ end as the precursor transcript. Further experiments are required to distinguish between these potential mechanisms and determine the extent that alternative poly(A) site usage combined with RNA editing and post-transcriptional cleavage is used to regulate protein expression.
THE SELECTION OF 3′ END CLEAVAGE SITES IS REGULATED DURING DEVELOPMENT
Although we are only beginning to appreciate the extent to which noncanonical 3′ end processing mechanisms are used throughout the genome, it will be of considerable interest to determine how the various 3′ end processing machineries are selectively recruited and used to generate the proper mature 3′ end of each Pol II transcript. Currently, the most is known about alternative polyadenylation, as it has been estimated that more than half of all human genes have the choice between two or more 3′ end cleavage sites (Tian et al. 2005; Lutz 2008). Many of these alternative polyadenylation signals are evolutionarily conserved (Ara et al. 2006; Lee et al. 2007) and their use systematically changes during development (Zhang et al. 2005; Liu et al. 2007; Ji et al. 2009). Interestingly, upstream poly(A) sites are preferentially used in rapidly proliferating cells, especially in cancer cells, resulting in mRNAs with shorter 3′ UTRs (Sandberg et al. 2008; Mayr and Bartel 2009). In some cases, these mRNAs with truncated 3′ UTRs have increased stability or are translated more efficiently due to the lack of microRNA binding sites, showing that even relatively subtle changes to the mRNA caused by alternative poly(A) site usage, such as a change in the length of the 3′ UTR, can have drastic effects on gene expression. In contrast, as mouse embryonic development progresses, downstream poly(A) sites are preferentially used, resulting in mRNAs with longer 3′ UTRs, likely increasing the post-transcriptional regulation of these transcripts (Ji et al. 2009).
How then is the choice between different 3′ end cleavage sites regulated? Although we do not yet know how noncanonical 3′ end processing mechanisms are selected over canonical cleavage/polyadenylation sites, patterns of alternative splicing and alternative cleavage/polyadenylation are strongly correlated, suggesting that splicing and 3′ end site selection are likely regulated in a coordinated manner (Wang et al. 2008). Interestingly, recent reports suggest that nucleosome positioning (Spies et al. 2009), as well as epigenetic modifications, namely DNA methylation, may play a role in 3′ end site selection (Wood et al. 2008). Additionally, neuronal activity has been shown to affect poly(A) site selection at many genes and a short motif was found to be enriched near these activity-regulated poly(A) sites (Flavell et al. 2008). Considering the importance of 3′ end site selection for the fate of the mature transcript, it is likely that we are only beginning to appreciate how the choice of sites is regulated.
SUMMARY AND PERSPECTIVES
Although canonical cleavage/polyadenylation is the mechanism by which the mature 3′ end of many, if not the vast majority, of long RNA polymerase II transcripts is generated, recent studies have uncovered several gene loci that use other endonucleases, including RNase P and the spliceosome, to cleave the nascent transcript. Interestingly, at the MALAT1 and S. pombe TER1 loci, despite the presence of nearby poly(A) sites, mechanisms other than canonical cleavage/polyadenylation are nearly exclusively employed to generate the mature 3′ end of the transcripts. It is not yet clear how broadly these noncanonical 3′ end processing mechanisms are used throughout the genome, but the complexity of the transcriptome and the presence of many long transcripts that lack a poly(A) tail (Cheng et al. 2005; Wu et al. 2008) suggest that they may be much more common than we currently appreciate.
ACKNOWLEDGMENTS
J.E.W. was supported by a Beckman Graduate Studentship while at the Watson School of Biological Sciences. Research in the Spector laboratory is supported by NIGMS42694, 5P01CA013106-38, and NIH/EY18244.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1907510.
REFERENCES
- Altman S, Wesolowski D, Guerrier-Takada C, Li Y. RNase P cleaves transient structures in some riboswitches. Proc Natl Acad Sci. 2005;102:11284–11289. doi: 10.1073/pnas.0505271102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ara T, Lopez F, Ritchie W, Benech P, Gautheret D. Conservation of alternative polyadenylation patterns in mammalian genes. BMC Genomics. 2006;7:189. doi: 10.1186/1471-2164-7-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Näär AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Bentley DL. Rules of engagement: Co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Blackburn EH. Telomeres and telomerase: Their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005;579:859–862. doi: 10.1016/j.febslet.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Box JA, Bunch JT, Tang W, Baumann P. Spliceosomal cleavage generates the 3′ end of telomerase RNA. Nature. 2008;456:910–914. doi: 10.1038/nature07584. [DOI] [PubMed] [Google Scholar]
- Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Ciais D, Bohnsack MT, Tollervey D. The mRNA encoding the yeast ARE-binding protein Cth2 is generated by a novel 3′ processing pathway. Nucleic Acids Res. 2008;36:3075–3084. doi: 10.1093/nar/gkn160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes & Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Conrad NK, Mili S, Marshall EL, Shu MD, Steitz JA. Identification of a rapid mammalian deadenylation-dependent decay pathway and its inhibition by a viral RNA element. Mol Cell. 2006;24:943–953. doi: 10.1016/j.molcel.2006.10.029. [DOI] [PubMed] [Google Scholar]
- Conrad NK, Shu MD, Uyhazi KE, Steitz JA. Mutational analysis of a viral RNA element that counteracts rapid RNA decay by interaction with the polyadenylate tail. Proc Natl Acad Sci. 2007;104:10412–10417. doi: 10.1073/pnas.0704187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin DJ, Pleiss JA, Walker SC, Whitworth GB, Engelke DR. Genome-wide search for yeast RNase P substrates reveals role in maturation of intron-encoded box C/D small nucleolar RNAs. Proc Natl Acad Sci. 2008;105:12218–12223. doi: 10.1073/pnas.0801906105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Hentze MW, Kulozik AE. 3′ end mRNA processing: Molecular mechanisms and implications for health and disease. EMBO J. 2008;27:482–498. doi: 10.1038/sj.emboj.7601932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Marzluff WF. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science. 2007;318:1777–1779. doi: 10.1126/science.1145989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunisova S, Elboher E, Nosek J, Gorkovoy V, Brown Y, Lucier JF, Laterreur N, Wellinger RJ, Tzfati Y, Tomaska L. Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA. 2009;15:546–559. doi: 10.1261/rna.1194009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan NC, Traverse KL, Sullivan DE, Pardue M. The nucleus-limited Hsr-ω-n transcript is a polyadenylated RNA with a regulated intranuclear turnover. J Cell Biol. 1994;125:21–30. doi: 10.1083/jcb.125.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage nonsmall cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Kay RA, Ellis IR, Jones SJ, Perrier S, Florence MM, Schor AM, Schor SL. The expression of migration stimulating factor, a potent oncofetal cytokine, is uniquely controlled by 3′-untranslated region-dependent nuclear sequestration of its precursor messenger RNA. Cancer Res. 2005;65:10742–10749. doi: 10.1158/0008-5472.CAN-05-2038. [DOI] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsebom LA. RNase P RNA mediated cleavage: Substrate recognition and catalysis. Biochimie. 2007;89:1183–1194. doi: 10.1016/j.biochi.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: A model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Lee JY, Yeh I, Park JY, Tian B. PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res. 2007;35:D165–D168. doi: 10.1093/nar/gkl870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardi J, Box JA, Bunch JT, Baumann P. TER1, the RNA subunit of fission yeast telomerase. Nat Struct Mol Biol. 2008;15:26–33. doi: 10.1038/nsmb1343. [DOI] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- Liu D, Brockman JM, Dass B, Hutchins LN, Singh P, McCarrey JR, MacDonald CC, Graber JH. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic Acids Res. 2007;35:234–246. doi: 10.1093/nar/gkl919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS. Alternative polyadenylation: A twist on mRNA 3′ end formation. ACS Chem Biol. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memili E, Hong YK, Kim DH, Ontiveros SD, Strauss WM. Murine Xist RNA isoforms are different at their 3′ ends: A role for differential polyadenylation. Gene. 2001;266:131–137. doi: 10.1016/s0378-1119(01)00353-5. [DOI] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: The complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 2005;24:1033–1045. doi: 10.1038/sj.emboj.7600560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- Osenberg S, Dominissini D, Rechavi G, Eisenberg E. Widespread cleavage of A-to-I hyperediting substrates. RNA. 2009;15:1632–1639. doi: 10.1261/rna.1581809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Redrup L, Branco MR, Perdeaux ER, Krueger C, Lewis A, Santos F, Nagano T, Cobb BS, Fraser P, Reik W. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development. 2009;136:525–530. doi: 10.1242/dev.031328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes & Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD. Cytoplasmic polyadenylation in development and beyond. Microbiol Mol Biol Rev. 1999;63:446–456. doi: 10.1128/mmbr.63.2.446-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondón AG, Mischo HE, Kawauchi J, Proudfoot NJ. Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol Cell. 2009;36:88–98. doi: 10.1016/j.molcel.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENɛ/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl CI, Stricker SH, Barlow DP. The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. EMBO J. 2006;25:3565–3575. doi: 10.1038/sj.emboj.7601245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkin AJ, Manley JL. The ends of the affair: Capping and polyadenylation. Nat Struct Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, 3rd, Frank J, Manley JL. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN ɛ/β nuclear-retained noncoding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/s0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Luhrmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CJ, Zakian VA. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat Struct Mol Biol. 2008;15:34–42. doi: 10.1038/nsmb1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes & Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, Bourc'his D, Oakey RJ. Regulation of alternative polyadenylation by genomic imprinting. Genes & Dev. 2008;22:1141–1146. doi: 10.1101/gad.473408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Kim YC, Lu J, Xuan Z, Chen J, Zheng Y, Zhou T, Zhang MQ, Wu CI, Wang SM. Poly A- transcripts expressed in HeLa cells. PLoS One. 2008;3:e2803. doi: 10.1371/journal.pone.0002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Marr TG. Computational analysis of 3′-ends of ESTs shows four classes of alternative polyadenylation in human, mouse, and rat. Genome Res. 2005;15:369–375. doi: 10.1101/gr.3109605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Altman S. A noncoding RNA in Saccharomyces cerevisiae is an RNase P substrate. RNA. 2007;13:682–690. doi: 10.1261/rna.460607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Lee JY, Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]