Abstract
The heat shock RNA-1 (HSR1) is a noncoding RNA (ncRNA) reported to be involved in mammalian heat shock response. HSR1 was shown to significantly stimulate the heat-shock factor 1 (HSF1) trimerization and DNA binding. The hamster HSR1 sequence was reported to consist of 604 nucleotides (nt) plus a poly(A) tail and to have only a 4-nt difference with the human HSR1. In this study, we present highly convincing evidence for bacterial origin of the HSR1. No HSR1 sequence was found by exhaustive sequence similarity searches of the publicly available eukaryotic nucleotide sequence databases at the NCBI, including the expressed sequence tags, genome survey sequences, and high-throughput genomic sequences divisions of GenBank, as well as the Trace Archive database of whole genome shotgun sequences, and genome assemblies. Instead, a putative open reading frame (ORF) of HSR1 revealed strong similarity to the amino-terminal region of bacterial chloride channel proteins. Furthermore, the 5′ flanking region of the putative HSR1 ORF showed similarity to the 5′ upstream regions of the bacterial protein genes. We propose that the HSR1 was derived from a bacterial genome fragment either by horizontal gene transfer or by bacterial infection of the cells. The most probable source organism of the HSR1 is a species belonging to the order Burkholderiales.
Keywords: heat shock RNA-1, heat shock response, bacterial sequence, horizontal gene transfer
INTRODUCTION
A large number of mRNA-like long noncoding RNAs (ncRNAs) have been reported to be involved in crucial biological processes in mammalian cells including transcriptional regulation and epigenetic gene regulation (Prasanth and Spector 2007; St. Laurent and Wahlestedt 2007; Ponting et al. 2009; Wilusz et al. 2009). For example, ncRNA XIST is associated exclusively with the inactive X chromosome (Brown et al. 1992) and ncRNA H19 is expressed only from the maternal allele of the imprinted IGF2/H19 locus (Brannan et al. 1990; Webber et al. 1998). Many long ncRNAs have been reported to be directly involved in regulation of activities of the associated proteins. The ncRNA Evf-2, for example, forms a stable complex with the Dlx-2 protein to increase the transcriptional activity of the Dlx-5/6 enhancer (Feng et al. 2006). The maternally expressed imprinted ncRNA MEG3 activates p53 and functions as a tumor suppressor (Zhou et al. 2007). Recently it has been shown that large intergenic ncRNAs such as XIST and HOTAIR guide chromatin-modifying complexes to specific genomic loci and act as epigenetic regulators of gene expression (Khalil et al. 2009).
Many of ncRNAs show clear evolutionary conservation, indicating that they are subject to strong purifying selection (Guttman et al. 2009; Ponjavic et al. 2009). Some of these highly conserved ncRNA genes such as HAR1F show significant evolutionary acceleration in the human genome, possibly resulting in emergence of human-specific traits (Pollard et al. 2006). Interestingly, ncRNA XIST evolved from a protein-coding gene and a set of transposable elements in placental mammals after the divergence of placentals and marsupials (Duret et al. 2006; Elisaphenko et al. 2008).
It has been reported that a novel mammalian ncRNA called heat shock RNA-1 (HSR1) activates the heat shock transcription factor 1 (HSF1), which is essential for the induction of expression of heat shock proteins (HSPs) (Shamovsky et al. 2006). The HSR1 was initially isolated from the hamster kidney cell line BHK-21. The hamster HSR1 was detected as an RNA species of ∼2 kb, and it is composed of 604 nucleotides (nt) and a poly(A) tail. The human HSR1 was reported to be almost identical to the hamster HSR1 with a 4-nt difference (Shamovsky et al. 2006). Subsequently, it has been reported by the same group of researchers that noncoding RNAs homologous to mammalian HSR1 are present in other eukaryotic organisms including Xenopus, Drosophila, and Caenorhabditis elegans, and that the homologs are functionally interchangeable (abstract number 2C_03_S at the World Conference of Stress in 2007; available at http://www.stress07.com/051abs2.htm). However, the HSR1 homolog sequences of the latter three species are not publicly available.
The importance of HSR1 was immediately recognized by the ncRNA research field (Kugel and Goodrich 2006; Costa 2007). Involvement of HSR1 in the trimerization of HSF1 was referred as favorable support for existence of noncoding-RNA regulators of other important processes such as RNA polymerase II transcription (Goodrich and Kugel 2006).
In this study, we present that the HSR1 sequence shows strong sequence similarity to the 5′ flanking region and part of the coding region of chloride channel proteins of bacterial genomes. We also discuss possible mechanism for detection of the HSR1 molecules in the mammalian cells.
RESULTS AND DISCUSSION
As an initial attempt to perform functional analysis of the human HSR1 gene, we set to find its sequence information from the publicly available nucleotide sequence databases. Initially, we searched the National Center for Biotechnology Information (NCBI) nonredundant nucleotide sequence database with the BLASTN program using the hamster HSR1 sequence as a query. We could not obtain any match from human or other mammalian genomic clones or mRNA sequences. Instead, we obtained many partial-length hits to bacterial genomes with HSR1 regions 140–210 and 250–310 with the E-values ranging from 10−9 to 10−4. Next, we searched genome assemblies of mammalian species including human, chimpanzee, orangutan, rhesus macaque, marmoset, mouse, rat, guinea pig, cat, dog, horse, cow, opossum, and platypus with the BLAT program available at the University of California Santa Cruz (UCSC) Genome Browser (http://genome.ucsc.edu/). No match was found in these genome assemblies either.
Then we speculated that the HSR1 gene could be embedded in a heterochromatic region which has been known to be refractory to cloning and sequencing (International Human Genome Sequencing Consortium 2004). Heterochromatic regions are reported to contain many transcriptionally active genes (Lyle et al. 2007). Some constitutively heterochromatic regions become transcriptionally active in response to stress stimuli such as heat shock (Rizzi et al. 2004). The HSR1 RNA was reported to be constitutively expressed in the hamster BHK-21 cells and the human HeLa cells (Shamovsky et al. 2006). The HSR1 RNA molecules were polyadenylated and detected in size of ∼2 kb. Therefore, we assumed that cDNA sequences for HSR1 RNA could have been isolated as expressed sequence tags (ESTs) and searched the database for ESTs (dbEST). We also searched other high-throughput genome sequence databases such as genomic survey sequences (GSS), high-throughput genomic sequences (HTGS), and whole-genome shotgun reads (WGS) at the NCBI BLAST website. To find matches to unassembled, raw sequence data for ESTs and genomes, furthermore, we searched the NCBI Trace Archive databases using the Discontiguous MegaBLAST program. The searched trace data included human, chimpanzee, bonobo, rhesus macaque, marmoset, mouse, rat, cow, dog, cat, and horse. However, we were not able to find any significant nucleotide matches from all the high-throughput sequence data, suggesting that no sequence record for the HSR1 has been deposited in the current sequence databases.
Interestingly, however, a BLASTX search of the NCBI nonredundant protein database yielded about 100 meaningful hits to bacterial proteins. Inspection of titles of these proteins revealed that almost all the matches were chloride channel proteins; the rest were labeled as hypothetical proteins. The matches were between a putative open reading frame (ORF) of the 3′ part of the HSR1 sequence (nucleotide positions 390–604) and the amino-terminal regions of the bacterial chloride channel proteins. The predicted HSR1 ORF would encode a total of 71 amino acid residues and runs off at the end of the sequence which was reported to be polyadenylated in the mammalian cells.
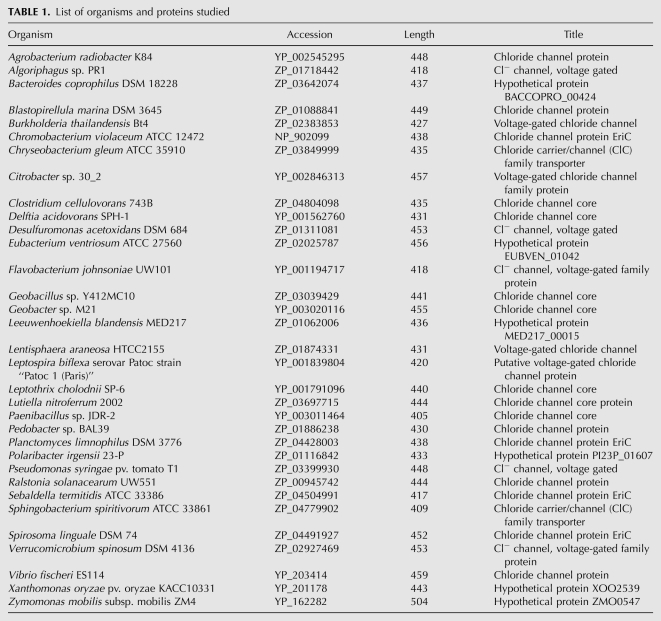
The matched bacterial protein sequences were downloaded and sorted according to the genus name. The protein sequences were from 33 bacterial genera (Table 1). From each genus, only one sequence was selected based on FASTA alignment score between the bacterial protein and the HSR1 ORF sequence. A multiple sequence alignment of the HSR1 ORF and the selected 33 bacterial chloride channel proteins with MUSCLE software revealed many conserved residues (Fig. 1). The HSR1 ORF shares 33 (with Burkholderia, Lutiella, and Pseudomonas proteins) to 18 identical amino acid residues with the bacterial proteins out of its 71 residues. We also found the similar positioning of the translation start codon and the conserved amino acid sequence motifs, which clearly indicates that the HSR1 ORF would encode a truncated version of the chloride channel protein. It is unlikely that the HSR1 sequence shows this high degree of sequence similarity to the bacterial proteins simply by chance. Rather, it suggests that the HSR1 is actually derived from a bacterial protein coding gene.
TABLE 1.
List of organisms and proteins studied
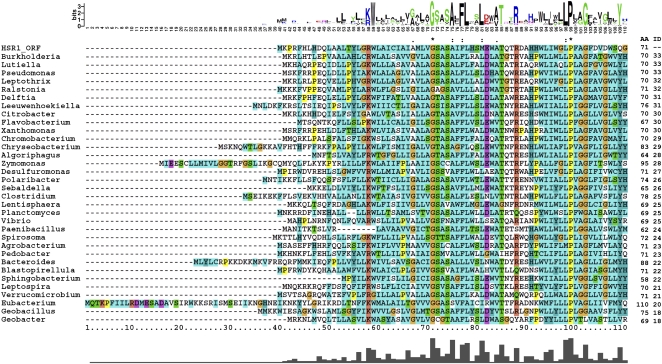
FIGURE 1.
Multiple sequence alignment of predicted HSR ORF and bacterial chloride channel proteins. The protein sequence deduced from the HSR1 ORF (top) and bacterial channel proteins with sequence similarity from 33 bacterial genera were aligned multiply by using the MUSCLE program. A nucleotide was removed to produce the HSR1 ORF (see Fig. 2). Detailed organism names and NCBI accession numbers for the proteins can be found in Table 1. AA, number of amino acid residues aligned; ID, number of identical residues to the HSR1 ORF.
A phylogenetic tree was constructed to infer the possible phylogenetic position of the HSR1 ORF with amino acid sequence alignments. Bootstrap consensus trees were constructed using maximum parsimony (MP) and neighbor-joining (NJ) methods. The result was phylogenetically uninformative. Almost all the branches were not supported by bootstrap values, and the position of HSR1 ORF was ambiguous. This is because the ORF sequences are mutationally saturated and highly diverged except for the functionally important amino acid positions, or because the size of the sequence data is not sufficient for reliable results.
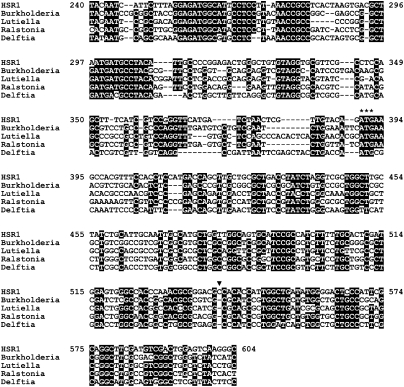
To examine whether the sequence similarity can be found at the nucleotide level, we retrieved genome sequences of the bacteria showing high protein sequence identity to the HSR1 ORF. When the HSR1 nucleotide sequence and the genomic sequences of species of Burkholderia, Lutiella, Ralstonia, and Delftia genera were multiply aligned, the 3′ half of the HSR1 sequence (from 240 to 604) showed a high level of identity with these bacterial genomes (Fig. 2): 57.0% with the Burkholderia species, and 56.7% with the Lutiella and the Ralstonia species, and 55.3% with the Delftia species. The highest pairwise identity among these five sequences was 69.0% obtained between the Burkholderia and the Ralstonia species. The nucleotide sequence conservation extends to the 5′ upstream region (from 240 to 389) of the HSR1 ORF. In fact, sequence conservation is higher in the 5′ upstream region than in the ORF itself, indicating that the region may harbor transcriptional and/or translational regulatory elements. Interestingly, the HSR1 regions 140–210 and 250–310 yielded meaningful hits to many bacterial genomes with significant E-values (up to 10−9) by BLASTN search. Moreover, the region 140–210 showed multiple matches in each genome. Many of them were located within or adjacent to an integrase/transposase of an insertion sequence (IS) element, implying that the 5′ half of the HSR1 may be part of a bacterial IS element. The high nucleotide sequence identity between the HSR1 sequence and the bacterial genomes further supports that the HSR1 is derived from a genomic fragment of currently uncharacterized bacterium. All the four bacterial species above belong to Betaproteobacteria: Burkholderia, Ralstonia, and Delftia species are in the order Burkholderiales and Lutiella in the order Neisseriales.
FIGURE 2.
Multiple alignment of bacterial genome sequences and a part of HSR1. A part of HSR1 sequence from 240 to 604 was aligned with four representative bacterial genomes. The start codons for the chloride channel protein ORFs are indicated by three consecutive asterisks. An additional nucleotide (arrowhead) in the HSR1 ORF, which would be present due to an insertion or perhaps a sequencing error, was removed to deduce the protein sequence. Residues identical in four or five sequences are highlighted on black background. GenBank accession numbers and positions for the nucleotide sequences are: Burkholderia, NZ_ABBH01000052.1:1764-2131; Lutiella, NZ_ACIS01000003.1:194423-194791; Ralstonia, NZ_AAKL01000043.1:37423-37060; Delftia, NC_010002.1: 1926518-1926158.
It is questionable how the bacterial genome sequence-like HSR1 molecule was discovered in the hamster and human cell lines and other eukaryotic cells. One possible mechanism is the horizontal gene transfer. As demonstrated in Wolbachia-insect cases, integration of bacterial genome fragments to eukaryotic nuclear genome is rather common (Nikoh et al. 2008). Some of the integrated Wolbachia genes were even detected by RT-PCR, indicating that they were transcriptionally active in the host insect genome. Similarly, the HSR1 gene might have been horizontally transferred from a bacterium that had infected an early metazoan ancestor. The integrated bacterial genomic fragment could have been exapted as a regulator of the heat shock process. As in the case of XIST, which evolved from a protein-coding gene (Duret et al. 2006; Elisaphenko et al. 2008), the HSR1 sequence might have lost its protein-coding potential.
Another possible mechanism is a simple bacterial infection of the cells examined. Infection of eukaryotic cell lines with microbial organisms such as Mycoplasma species is rather common and problematic during cell culture research (Schmitt and Pawlita 2009). The gene sequences derived from the pathogenic viruses, bacteria, fungi, and protozoa can be detected in the human cells and tissues (Weber et al. 2002). If this is the case, the HSR1 RNA molecules would have been directly derived from some bacterial cells or transcribed from the nuclear genome after the integration. The absence of HSR1 sequence in mammalian genomes and transcripts despite their extensive sequence data deposited in the current sequence databases may justify the simple bacterial infection. Furthermore, the fact that the most similar sequence to HSR1 was found in the intracellular pathogens Ralstonia and Burkholderia (Valvano et al. 2005) could be suggestive of this possibility.
In conclusion, we have found that the mammalian HSR1 sequence shows strong similarity to bacterial genomes, implying that the HSR1 molecules have bacterial origin. Judging from all the data, we propose that the most probable source organism of the HSR1 is a species belonging to the order Burkholderiales. We advise that the origin of the HSR1 should be further scrutinized for better understanding of the evolution and the mechanism of mammalian heat shock process.
MATERIALS AND METHODS
Database search and analysis of the HSR1 sequence
BLAST programs were used to search the NCBI sequence databases (Altschul et al. 1997). All the NCBI nucleotide databases including the nonredundant database, EST database (dbEST), genomic survey sequences (GSS), high-throughput genomic sequences (HTGS), whole-genome shotgun (WGS) reads, and environmental samples (ENV_NT) were searched with the BLASTN program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Discontiguous MegaBLAST searches of the WGS trace databases were carried out at the NCBI Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/). BLASTX search of the NCBI nonredundant protein database was performed at the NCBI website with default parameters. All the sequence similarity searches were conducted on September 24, 2009.
BLAT searches of the mammalian genome assemblies such as human (hg18), chimpanzee (panTro2), orangutan (ponAbe2), rhesus macaque (rheMac2), marmoset (calJac1), mouse (mm9), rat (rn4), guinea pig (cavPor3), cat (felCat3), dog (canFam2), horse (equCab2), cow (bosTau4), opossum (monDom5), and platypus (ornAna1) were performed at the UCSC Genome Browser server (http://genome.ucsc.edu/) (Kent 2002; Kent et al. 2002).
The FASTA program was used to obtain pairwise alignment (Pearson and Lipman 1988). Multiple sequence alignment was generated using MUSCLE (Edgar 2004) and visualized using ClustalX2 (Larkin et al. 2007) or BOXSHADE (http://www.ch.embnet.org/software/BOX_form.html). Sequence Logo representation (Schneider and Stephens 1990) was generated with the WebLogo server (Crooks et al. 2004).
Molecular phylogenetic analysis
Molecular phylogenetic analysis was performed by using MEGA program version 4 (http://www.megasoftware.net/) (Kumar et al. 2008). We used MP and NJ methods to infer phylogenetic trees. Positions with gaps were deleted from the alignment. Bootstrap consensus trees were constructed with 1000 replicates.
We also used PAUP* software (version 4.0b10 for Linux platform) (http://paup.scs.fsu.edu/) (Swofford 2002). We constructed bootstrap 50% majority-rule consensus trees using MP and NJ methods with 100 replicates. The gaps were treated as missing data.
ACKNOWLEDGMENTS
This work was supported in part by the Korea Research Foundation Grant funded by the Korean Government (MOEHRD, Basic Research Promotion Fund) (KRF-2008-331-C00263), and in part by Grant (R01-2008-000-11660-0) from the Korea Science and Engineering Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1879610.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990;10:28–36. doi: 10.1128/mcb.10.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Costa FF. Noncoding RNAs: Lost in translation? Gene. 2007;386:1–10. doi: 10.1016/j.gene.2006.09.028. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312:1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elisaphenko EA, Kolesnikov NN, Shevchenko AI, Rogozin IB, Nesterova TB, Brockdorff N, Zakian SM. A dual origin of the Xist gene from a protein-coding gene and a set of transposable elements. PLoS One. 2008;3:e2521. doi: 10.1371/journal.pone.0002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes & Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JA, Kugel JF. Non-coding-RNA regulators of RNA polymerase II transcription. Nat Rev Mol Cell Biol. 2006;7:612–616. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large noncoding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT: The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel JF, Goodrich JA. Beating the heat: A translation factor and an RNA mobilize the heat shock transcription factor HSF1. Mol Cell. 2006;22:153–154. doi: 10.1016/j.molcel.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lyle R, Prandini P, Osoegawa K, ten Hallers B, Humphray S, Zhu B, Eyras E, Castelo R, Bird CP, Gagos S, et al. Islands of euchromatin-like sequence and expressed polymorphic sequences within the short arm of human chromosome 21. Genome Res. 2007;17:1690–1696. doi: 10.1101/gr.6675307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoh N, Tanaka K, Shibata F, Kondo N, Hizume M, Shimada M, Fukatsu T. Wolbachia genome integrated in an insect chromosome: Evolution and fate of laterally transferred endosymbiont genes. Genome Res. 2008;18:272–280. doi: 10.1101/gr.7144908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long noncoding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal/pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: An answer to the ‘genome complexity’ conundrum. Genes & Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Rizzi N, Denegri M, Chiodi I, Corioni M, Valgardsdottir R, Cobianchi F, Riva S, Biamonti G. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell. 2004;15:543–551. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M, Pawlita M. High-throughput detection and multiplex identification of cell contaminations. Nucleic Acids Res. 2009;37:e119. doi: 10.1093/nar/gkp581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- St Laurent G, 3rd, Wahlestedt C. Noncoding RNAs: Couplers of analog and digital information in nervous system function? Trends Neurosci. 2007;30:612–621. doi: 10.1016/j.tins.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Swofford DL. In Sinaur Associates; Sunderland, MA: 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods), Version 4. [Google Scholar]
- Valvano MA, Keith KE, Cardona ST. Survival and persistence of opportunistic Burkholderia species in host cells. Curr Opin Microbiol. 2005;8:99–105. doi: 10.1016/j.mib.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Webber AL, Ingram RS, Levorse JM, Tilghman SM. Location of enhancers is essential for the imprinting of H19 and Igf2 genes. Nature. 1998;391:711–715. doi: 10.1038/35655. [DOI] [PubMed] [Google Scholar]
- Weber G, Shendure J, Tanenbaum DM, Church GM, Meyerson M. Identification of foreign gene sequences by transcript filtering against the human genome. Nat Genet. 2002;30:141–142. doi: 10.1038/ng818. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes & Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A. Activation of p53 by MEG3 noncoding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]