Abstract
Identification of small nucleolar RNAs (snoRNAs) in genomic sequences has been challenging due to the relative paucity of sequence features. Many current prediction algorithms rely on detection of snoRNA motifs complementary to target sites in snRNAs and rRNAs. However, recent discovery of snoRNAs without apparent targets requires development of alternative prediction methods. We present an approach that combines rule-based filters and a Bayesian Classifier to identify a class of snoRNAs (H/ACA) without requiring target sequence information. It takes advantage of unique attributes of their genomic organization and improved species-specific motif characterization to predict snoRNAs that may otherwise be difficult to discover. Searches in the genomes of Caenorhabditis elegans and the closely related Caenorhabditis briggsae suggest that our method performs well compared to recent benchmark algorithms. Our results illustrate the benefits of training gene discovery engines on features restricted to particular phylogenetic groups and the utility of incorporating diverse data types in gene prediction.
Keywords: H/ACA snoRNA, computational prediction, nested genes, naive Bayesian Classifier
INTRODUCTION
Interest in small non-protein-coding RNAs (ncRNA), including tRNAs, snRNAs, snoRNAs, and miRNAs, has burgeoned in recent years, following a growing recognition of their roles in a wide range of biological processes (Eddy 2001; Hüttenhofer et al. 2005; Mattick and Makunin 2005). Their discovery has been aided by high-throughput methods such as genome sequencing projects and DNA microarrays (Hüttenhofer et al. 2005; He et al. 2007; Matera et al. 2007). Computational discovery of these small RNAs, however, has been lagging, particularly for snoRNAs and miRNAs, due to the paucity of sequence features. We sought to address this problem for box-H/ACA genes, a major class of the snoRNAs.
Early screens for snoRNAs used experimental methods and yielded relatively few genes (Ni et al. 1997; Liang-Hu et al. 2001; Higa et al. 2002; Wachi et al. 2004; Yang et al. 2005) because experimental methods tend to be biased in favor of highly expressed sequences (Hüttenhofer et al. 2001; Gu et al. 2005). More recent efforts combined bioinformatics methods with sequencing of cDNA libraries or microarray data to carry out genome-wide scans—Caenorhabditis elegans (Deng et al. 2006; Zemann et al. 2006; Huang et al. 2007); other genomes: mouse (Hüttenhofer et al. 2001), Arabidopsis thaliana (Marker et al. 2002), and Drosophila melanogaster (Yuan et al. 2003). These studies, however, often rely on homology-based (using BLAST or similar algorithms) or target site searches, snoGPS (Schattner et al. 2004) and Snoscan (Lowe and Eddy 1999). This complicates the discovery of highly diverged or novel sequences, or sequences (orphan snoRNAs) that apparently lack target sites in snRNAs and rRNAs (Hüttenhofer et al. 2001; Bachellerie et al. 2002; Huang et al. 2005; Yang et al. 2006). This discovery bias may be a concern in light of recent reports that some snoRNAs may guide modifications of messenger RNAs (Kishore and Stamm 2006; Bazeley et al. 2008) or function as precursors to other short RNAs (Saraiya and Wang 2008; Taft et al. 2009).
Two recently developed snoRNA prediction algorithms aim to address these problems: SnoReport (Hertel et al. 2008) and SnoSeeker (Yang et al. 2006). SnoReport eschews target site searches entirely; instead, it uses a support vector machine approach trained on folding energies and distance constraints of a set of known snoRNAs. SnoSeeker was designed to search for both guide and orphan snoRNA genes, using a probabilistic model with conserved primary and secondary sequence motifs. SnoReport was designed to be a universal snoRNA predictor, whereas SnoSeeker was designed specifically for mammalian sequences.
While a universal predictor may have wide applicability, the published results (Yang et al. 2006; Hertel et al. 2008) suggest that a clade-specific predictor may perform better than a universal one. We therefore sought to develop a method for identifying H/ACA snoRNAs in the genomes of Caenorhabditis species, by defining nematode-specific versions of previously described sequence motifs and combining them with additional features to achieve higher prediction accuracy.
RESULTS AND DISCUSSION
The canonical structure of the H/ACA snoRNA is defined by the presence of an H-box (AnAnnA) and an ACA-box (ACA), as well as two hairpins of approximately equal lengths (Supplemental Fig. S1). However, as currently defined, the information content of these sequence motifs is quite low, and the hairpins are common enough (Rivas and Eddy 2000) that the mere presence of these features does not provide sufficient basis for identifying H/ACA snoRNAs. To identify other sequence features that may help in the prediction of nematode H/ACA snoRNAs, we assembled a comprehensive set (Supplemental Table S1) of all known H/ACA snoRNAs in Caenorhabditis elegans from a number of previous studies (Wachi et al. 2004; Deng et al. 2006; Zemann et al. 2006; Huang et al. 2007).
Distinctive genomic organization of C. elegans H/ACAs
Like many ncRNAs, snoRNAs are often found nested within the introns of protein-coding genes (He et al. 2006). Our survey of the C. elegans ncRNAs (including 608 tRNAs, 81 snRNAs, 87 C/D snoRNAs, and 62 H/ACAs) showed that H/ACAs have distinct host-nested gene organization (Supplemental Table S2). It may be worth noting that all features described here pertain to the host genes rather than the RNA itself.
H/ACAs are parallel-nested in introns
Of the 62 currently annotated C. elegans H/ACAs, 56 (90%) are found nested within the introns of protein-coding genes, and all but two (i.e., 54) are in the same orientation (“parallel”) as the host gene. The propensity toward parallel nesting is significantly higher than for other small RNAs (tRNAs: 144/608 [24%], snRNAs: 25/81 [30%], C/D snoRNAs: 43/87 [49%]) (Supplemental Table S2).
Host genes preferentially reside in operons
The 54 parallel-nested H/ACAs are located within 40 distinct host genes, 23 (58%) of which are contained within operons. This is significantly higher than the fraction of operon-contained genes among all genes (2871/20,084 = 14%), compact genes (see below) (17%), or host genes of other ncRNAs (tRNA: 9.4%; snRNA: 11%, C/D snoRNA: 15%) (Supplemental Table S2).
Size range of host introns
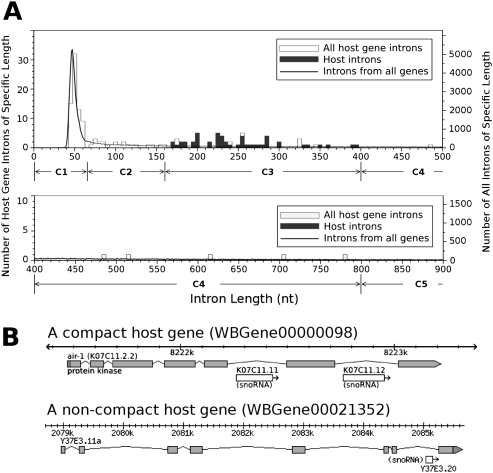
We found that in C. elegans most host introns (introns that harbor H/ACA RNAs) range between 161 and 400 nucleotides (nt) (Fig. 1A). The nested H/ACAs display no discernable positional bias within the host introns, although they never reside closer than 5 nt to the 3′ splice site or 19 nt to the 5′ splice site, likely due to the presence of the splicing signals (data not shown). Given that most H/ACAs range between 120 and 160 nt, the limited range of host intron lengths implies that nematode H/ACAs do not share host introns. This is consistent with their processing being mechanistically linked to splicing, in a manner similar to snoRNA processing in vertebrates and higher plants (Tollervey and Kiss 1997; Bachellerie et al. 2002).
FIGURE 1.
Genomic organization of C. elegans H/ACA snoRNAs. (A) Intron length distribution of H/ACA host genes compared to all C. elegans protein-coding genes. Note that the left scale refers to the counts of host gene introns, while the right scale refers to the counts of all introns. (C1–C5) The intron length categories (see the text for details). (B) Examples showing structure of a compact gene and a noncompact gene (from WormBase).
Host genes are compact
Most host genes containing nested H/ACAs have a distinct exon–intron structure. Specifically, all but the host introns tend to be short (≤65 nt) regardless of the total number of introns or the length of exons (Fig. 1A). We designated these as “compact genes” (Fig. 1B). We also observed a correlation between compactness of a host gene and its residence within an operon. Specifically, host genes residing in operons are less compact than other host genes (see below).
Previous studies have observed that introns harboring snoRNAs are often restricted in length (Fedorov et al. 2005; Yang et al. 2006; Zhou and Lin 2008). While host intron length is related to host gene compactness, they are not equivalent, as host gene compactness also requires other (nonhost) introns to be short. Currently no direct link is established between host gene compactness and snoRNA biogenesis. However, it is known that (A) many snoRNAs reside in ribosomal protein genes (Yoshihama et al. 2002; Zemann et al. 2006), which comprise a subset of housekeeping genes; and (B) housekeeping genes in some species (including C. elegans) are compact, according to a definition similar to the one used here (Duret and Mouchiroud 1999; Eisenberg and Levanon 2003; Vinogradov 2004). These data and our observations suggest that compactness of host genes may have biological relevance, possibly due to the processing of the H/ACA snoRNAs from the host introns, or the overall level and ubiquity of expression. We tested whether annotations of host genes were over-represented for particular functional categories, but other than many being housekeeping and broadly expressed, did not discover any particularly strong trends (Supplemental Table S2).
Empirical rule describing host gene compactness
To utilize host gene compactness for predicting likely genomic locations of the H/ACA snoRNAs, we divided the intron length spectrum into five categories: (C1) ≤65 nt, (C2) 66–160 nt, (C3) 161–400 nt, (C4) 401–800 nt, and (C5) ≥801 nt (Fig. 1A). Category 1 is the default intron size for a compact gene. Category 3 introns (C3) are host intron-sized; indeed, most C3 introns in annotated host genes contain H/ACA snoRNAs. A compact host gene is therefore expected to have mostly C1 and at least one C3 intron, although many host genes also contain several C2 or C4 introns, but none have any C5 introns. The number of C2 or C4 introns in a host gene is operon-dependent: nonoperon host genes have more C2 introns, but no C4 introns (data not shown). Based on 36 of the 40 H/ACA host genes that we considered to be compact (Fig. 1B; Supplemental Table S3), compact host genes satisfy the following criteria:
May contain any number of C1 introns.
Must contain one to four C3 (host-sized) introns.
For a nonoperon gene, C2 introns are limited to a maximum of one or 50% of the total number of introns, whichever is greater; the limit is reduced to 25% if the gene is inside an operon.
C4 introns are only allowed if the gene resides in an operon, and only up to a maximum of one intron or 25% of the total number of introns, whichever is greater.
C5 introns are not allowed.
C. elegans-specific sequence features of H/ACA snoRNAs
One of the major difficulties in computational identification of H/ACA snoRNAs is the low information content of sequence features. Through detailed analysis of the currently annotated C. elegans H/ACA snoRNAs, we discovered several additional features of these genes (Fig. 2).
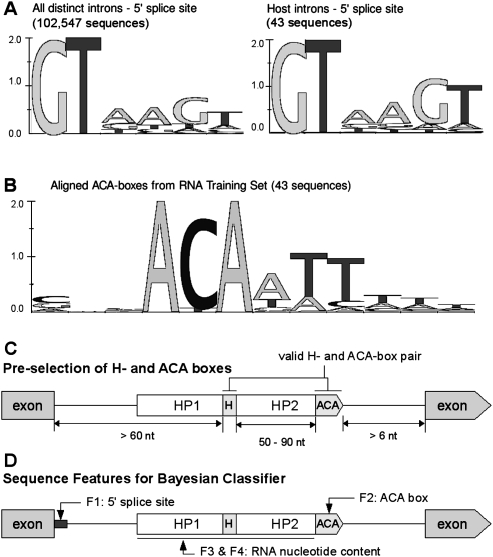
FIGURE 2.
Sequence features of C. elegans H/ACA. (A) Sequence logos showing that GTnnGT usage is higher in host introns than all introns. Sequence logos were generated using in-house-developed software based on literature (Schneider and Stephens 1990). (B) Sequence logo showing the consensus motif of the extended 6-nt ACA-box. (C) Pre-selection of H- and ACA-boxes requires the box motifs to conform to sequence consensus, as well as to be positioned within spatial constraints with respect to each other and the intron boundaries. (D) Sequence features of the H/ACA used by the Naive Bayesian Classifier to evaluate candidate sequences.
Host introns show preference for a specific 5′ splice site signal
Whereas 48% (49,112/102,547) of all C. elegans introns start with GTnnGT, this motif is substantially more prevalent among H/ACA host introns (39/54 = 72%) (Fig. 2A). This trend is not seen among short (≤65 nt) introns (22,111/51,657 = 43%), introns of operon genes (6002/14,903 = 40%), nonhost introns of host genes (102/210 = 49%), or all host-sized (161–400 nt) introns (8514/15,719 = 54%). A preference for a specific 5′ splice site motif (GTnnGT) may reflect a link between splicing and intronic RNA processing (Bachellerie et al. 2002).
Refining consensus motifs of H-boxes and ACA-boxes
All H/ACA snoRNAs are characterized by two A-rich motifs, the H-box (AnAnnA) and the ACA-box (ACA); these are common in the A/T-rich introns of C. elegans. Alignment of the currently annotated C. elegans H/ACAs showed that the ACA-box extends to the three downstream nucleotides, with an overall consensus of “ACAATT” (Fig. 2B). We thus refined the sequence of the 3′-end of C. elegans H/ACAs, as the ACA-box is located 3 nt upstream of the cleavage site (Bachellerie et al. 2002). Refining the H-box sequence was more challenging because we frequently found multiple valid motifs between the two hairpins, as well as numerous H-box-like motifs, such as AnAnA or AnAnnnA. We found, however, that all C. elegans H-box motifs always contain at least one non-A nucleotide.
Nucleotide content in hairpin sequences
We found that C. elegans H/ACAs exhibit substantially different nucleotide usage than intronic sequences, including significant differences (Kolmogorov–Smirnov tests) (data not shown) in the frequency of certain dinucleotides, particularly, “AA,” “CG,” “GC,” “GT,” and “TG.” This sequence composition difference may be imposed by the demands of higher thermodynamic stability of hairpins (Rivas and Eddy 2000). We did not find any additional primary sequence motifs in the currently annotated H/ACA sequences using MEME (Bailey et al. 2006).
Combining host gene and H/ACA sequence features into a prediction engine
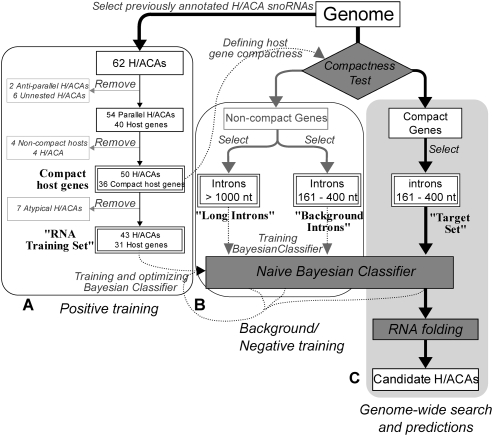
Our prediction engine consists of two separate parts (Fig. 3). The first relies on the compactness rules described above to rapidly select candidate genes and introns likely to contain snoRNAs. The second is a naive Bayesian Classifier that uses newly refined sequence features (Fig. 2C,D) to evaluate the probability that a given candidate intron, indeed, contains an H/ACA snoRNA.
FIGURE 3.
Schematic representation of the workflow. (A) Selection of H/ACA snoRNAs (RNA training set) and host genes used for training the host gene compactness test and the classifier. (B) Selection of sequence sets used for background training (background introns) and negative controls (long and short introns). (C) Selection of candidate host genes and host introns (target set) and subsequent steps to identify candidate H/ACA snoRNAs. (Boxes with double borders) Sequence sets used for training; (solid lines) selection processes; (dashed lines) training processes.
Selection of candidate host genes and introns
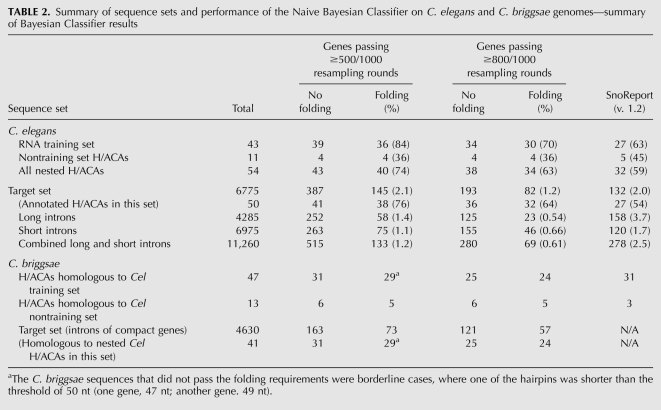
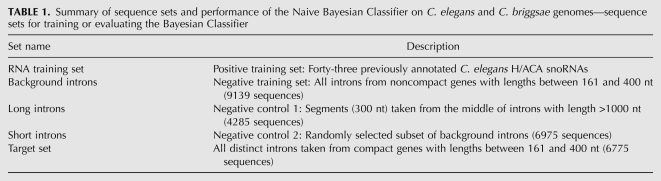
By applying the compactness criteria to the C. elegans genome, 4699 out of 19,543 protein-coding genes were classified as compact; these contained 6775 distinct host-sized (161–400 nt) introns. The 54 parallel-nested C. elegans H/ACA snoRNAs were found within 40 distinct host genes, and 36 of these host genes (containing 50 H/ACAs) were classified as compact. Although this classification of host gene compactness is ad hoc and deterministic, it enabled a rapid and sensitive scan of the whole genome, reducing the search space for H/ACA snoRNAs to 6775 introns, spanning just 2 Mb in total. This is a substantial reduction of the sequence space compared to the combined intronic and intergenic regions on both strands, totaling ∼150 Mb (Stein et al. 2003). Importantly, this selection retained most of the annotated H/ACA sequences: 81% (50/62) of all H/ACA snoRNAs and 93% (50/54) of all parallel-nested H/ACAs (Table 2, see below).
TABLE 2.
Summary of sequence sets and performance of the Naive Bayesian Classifier on C. elegans and C. briggsae genomes—summary of Bayesian Classifier results
Construction of the naive Bayesian Classifier
Having reduced the search space to 6775 introns (dubbed the “target set”) using the host gene compactness criteria, we used the sequence features described above to construct a naive Bayesian Classifier to search the target set introns for H/ACA snoRNAs. The Bayesian Classifier is a probabilistic approach that calculates the likelihood that a given sequence belongs to one class (H/ACAs) or another (Table 1, “background introns”). The classifier, therefore, has to be trained on two sequence sets representing the two different classes (for detailed descriptions, see Materials and Methods and Supplemental Material). For each candidate intron, we first performed a fast scan for the presence of valid H-boxes and ACA-boxes within specific distance constraints (Fig. 2C), and then evaluated the likelihood of them belonging to a genuine H/ACA snoRNA (Supplemental Material; Materials and Methods). To estimate the confidence of each prediction, we used multiple rounds of random re-sampling of the training sets (Materials and Methods). We decided that being classified as H/ACA snoRNA by the Bayesian Classifier at least 500 out of 1000 re-sampling rounds provided a reasonable compromise between sensitivity and specificity. As the Bayesian Classifier did not use any information on RNA secondary structure, we added further stringency to our predictions by calculating the RNA secondary structure of the two hairpin regions using RNAfold (Hofacker et al. 1994; Hofacker 2003).
TABLE 1.
Summary of sequence sets and performance of the Naive Bayesian Classifier on C. elegans and C. briggsae genomes—sequence sets for training or evaluating the Bayesian Classifier
Classifier sensitivity and specificity
To determine the quality of our predictions, we applied the Bayesian Classifier to the control sequence sets. We tested the classifier on the 43 introns of the “RNA training set” (Table 1; Materials and Methods). For negative control we used the “long introns” and “short introns” sets (Table 1; Materials and Methods) because most of these are not expected to contain H/ACA snoRNAs (Table 1). The classifier alone provided a relatively high recall rate (Table 2), with 39 out of the 43 (91%) RNA training set sequences passing at least 500/1000 re-sampling rounds. The estimates of the corresponding false-positive rates (predicted H/ACAs in long and short introns sets) were 4.6% (515/11,260) and 2.5% (280/11,260). After applying the RNA folding step, the number of recovered RNA training set sequences was reduced to 36 (84%), but the false-positive rate was substantially reduced to 1.2% (133/11,260, average of the two negative control sets) (Table 2). When more stringent conditions were applied (i.e., requiring the passing of 800, not 500, rounds), the false-positive rate was reduced by half, while the loss of annotated H/ACAs was fairly modest (30/43 = 70%) (Table 2).
As mentioned above, several H/ACA sequences were excluded from the RNA training set because they resided in noncompact host genes or possessed noncanonical sequence features (see conditions for exclusion in Materials and Methods and Supplemental Material). We tested whether the classifier could recover any of these excluded genes. The classifier identified four of 11 such sequences (Table 1; Supplemental Table S3). Because none of them were homologous to any of the RNA training set sequences, they could not have been discovered using homology-based searches.
Search within C. elegans genome
The results from applying the classifier to the host-sized (160–400 nt) introns selected from the compact genes in the C. elegans genome are shown in Table 2. At the default level of confidence (requiring that a given sequence is predicted in at least 500/1000 re-sampling rounds), 145 H/ACAs were predicted in the target set, including 38 previously annotated H/ACAs. The estimated false-positive rate of 1.2%, and therefore 81 (6775 × 0.012) false-positive predictions, suggests the presence of 26 (145 − 38 − 81) additional, currently not annotated H/ACAs. A more stringent approach (requiring the passing of 800/1000 re-sampling rounds) resulted in 82 predicted H/ACAs, of which 32 were previously annotated. At this level of stringency the false-positive rate is 0.61%. Therefore, most (41 = 6775 × 0.0061) of the remaining 50 sequences are expected to be false-positive predictions, suggesting that few, if any, additional H/ACAs remain to be discovered in the genome of C. elegans.
Comparison of algorithm performance
Two currently available prediction engines, SnoReport (Hertel et al. 2008) and SnoSeeker (Yang et al. 2006), do not rely on target-site matching, making them comparable to our approach. However, we only used SnoReport for comparison because SnoSeeker was developed specifically for mammalian sequences. We applied the program to the same C. elegans sequence sets used to evaluate the Bayesian Classifier (Table 2).
At the default confidence level (passing at least 500/1000 re-sampling rounds), our classifier achieved a higher recall rate, 91% (39/43), than SnoReport, 63% (27/43), while recovering similar numbers of H/ACAs not included in the RNA training set, four versus five (Table 2). Furthermore, our method gains additional specificity, without considerable loss of recovery, using folding (Table 2). When applied to 6775 host-sized (161–400 nt) introns of the target set, our method and SnoReport predicted similar numbers of candidate H/ACAs (145 and 132, respectively). However, we have achieved a somewhat higher recovery of annotated H/ACAs (38 versus 27), with a twofold lower estimated rate of false positives (133 versus 278).
It seems important to underscore that this comparison was actually biased against our method, because it was carried out on a pre-selected set of host-sized (161–400 nt) introns, not the entirety of intergenic and intronic sequences. As mentioned above, the former represents a 75-fold reduction of the search space (2 Mb versus 150 Mb), greatly reducing the number of false-positive predictions.
Search within C. briggsae genome
To test whether our method, trained on the C. elegans genome, could be applied to other nematodes, we tested it on Caenorhabditis briggsae, a related species with an assembled (although incompletely annotated) genome (Stein et al. 2003; Hillier et al. 2007). As a benchmark for algorithm performance, we first identified C. briggsae loci that were homologous to previously annotated C. elegans H/ACAs. There were 60 such parallel-nested genes, of which 47 were homologous to the genes in the RNA training set.
First, we sought to establish that a classifier trained on the C. elegans data was capable of discovering H/ACA snoRNAs of C. briggsae. At the default confidence level (passing at least 500/1000 re-sampling rounds) and imposing folding requirements, 62% (29/47) of homologs of the RNA training set sequences were recovered. This rate was comparable to that of SnoReport (66% = 31/47) (Table 2). Homologs of nontraining set genes cannot be discovered via BLAST searches using the RNA training set sequences, because there is no sequence similarity between the two sets. Yet both our method and SnoReport recovered some such sequences (five and three, respectively, of 13 total), suggesting that rules inferred from C. elegans can be used to discover H/ACA genes in C. briggsae.
We then applied compactness rules inferred from the C. elegans RNA training set to identify compact genes in C. briggsae. These contained 4630 host-sized introns (C. briggsae target set). Among these sequences, the Bayesian Classifier identified 73 candidate sequences (passing at least 500/1000 re-sampling rounds) that satisfied folding requirements (Table 2), including 29 that were homologous to known C. elegans H/ACAs, implying that compactness rules were sufficiently similar between the two species.
Validation of candidate C. elegans H/ACA snoRNAs
Genuine H/ACA snoRNAs are expected to be expressed and possibly conserved in C. briggsae. We used whole-genome tiling microarray data for C. elegans (He et al. 2007) to determine whether candidate H/ACA genes were expressed. As a reference, 53 of the 54 currently annotated parallel-nested H/ACAs were expressed at sufficiently high levels to be detected, suggesting a false-negative (FN) rate of 2% (1/54). We carried out BLAST searches against the assembled C. briggsae genome (Stein et al. 2003) to determine whether our candidate sequences were conserved.
Within the set of 145 candidate C. elegans H/ACAs (passing at least 500/1000 re-sampling rounds), 38 were annotated, leaving 107 sequences of unknown status. Of these, 31 sequences either had a significant match (E < 1e-5) in the C. briggsae genome or were expressed at a sufficiently high level to be detected (Supplemental Table S4). This number is consistent with our estimate (see above) that 26 genuine H/ACA snoRNAs remain to be discovered in the genome of C. elegans. Eight predicted sequences are particularly noteworthy because they are either expressed and conserved between C. elegans and C. briggsae, or they are conserved in orthologous introns of the two species (Supplemental Table S4). We therefore consider them likely genuine snoRNA genes. Three other sequences were predicted by both our method and SnoReport; however, unlike the genes above, these were found in multiple copies throughout the genome and are thus less likely to be genuine snoRNAs. Taken together, our results suggest that nearly the entire complement of C. elegans H/ACA genes has now been discovered.
Conclusions
We developed a two-part engine to predict H/ACA snoRNAs. First, utilizing the knowledge of genomic architecture of host genes, we achieved a 75-fold reduction of the search space to include only the introns likely to contain H/ACAs. Next, we used a Bayesian Classifier trained on nematode-specific sequence features to make gene predictions that were further verified by RNA folding. Our results suggest that clade-specific features allow accurate predictions of new genes to be made even in this extensively studied gene category. The validity of our predictions is supported by expression data and sequence conservation. Finally, we see this work as an addition, not an alternative, to current methods. Many genes have few informative features and are therefore recalcitrant to computational discovery. Prediction engines containing separate modules, each exploiting a different set of clade-specific features, may prove to be a useful approach to solving this problem.
MATERIALS AND METHODS
For a detailed description of the methods used, see Supplemental Materials.
Genomic sequences and annotated ncRNA data
Genomic sequence data were collected from WormBase release WS170 (http://www.wormbase.org) (Chen et al. 2005). Additional snoRNA sequences (both H/ACA and C/D) were curated from relevant literature (Higa et al. 2002; Wachi et al. 2004; Deng et al. 2006; Zemann et al. 2006; Huang et al. 2007), resulting in a set of 62 H/ACA genes (Supplemental Table S1). We refer to C. elegans genes by their WormBase ID (e.g., “WBGene00012345”), except in those cases where the snoRNA has not yet been annotated by WormBase.
Analysis of genomic organization
Chromosomal positions and exon–intron information were based on WormBase WS170 annotations. As the annotation of the untranslated regions was incomplete, we used the start and stop codons to mark the boundaries of the coding genes. For genes with multiple transcription start sites and splice variants, we considered each transcript separately. There are a few discrepancies in the annotation of the ncRNAs between different sources (see Supplemental Material).
Identification of candidate host introns within compact host genes
Host gene compactness was initially noted by comparing the exon–intron structures of the 40 host genes of parallel-nested H/ACA snoRNAs (Fig. 3). Once we found that 90% (36/40) of these genes were, indeed, compact, we constructed a set of empirical rules to describe the compactness property. These rules were applied to classify genes as either compact or not compact. From the introns of compact genes, we selected host-sized (161–400 nt) introns that could harbor H/ACA snoRNAs. There exist many compact genes containing only Category I introns (≤65 nt), but these are likely too short to harbor any snoRNAs in their introns.
Analysis of H/ACA sequence features
As our set of H/ACA snoRNAs was assembled from several published sources, there were occasional discrepancies in the annotated locations of the H-boxes and ACA-boxes. The selection of H-boxes was further complicated by the presence of multiple (and frequently overlapping) candidate motifs. For each H/ACA snoRNA, we manually selected the H-boxes and ACA-boxes, ensuring that the position of the predicted hairpin structures (calculated using mfold) (Matthews et al. 1999; Zuker 2003) conformed with previous annotations.
Selection and evaluation of candidate H/ACA sequences
For each candidate host-sized (161–400 nt) intron, we first performed a fast scan for the presence of valid H-boxes and ACA-boxes within certain distance constraints (Fig. 2C). A valid H-box had to conform to the consensus, AnAnnA, and contain at least one non-A residue. An ACA-box was considered valid if it scored above the minimum threshold using the ACA-box position weight matrix (see below). Valid ACA-boxes were situated >120 nt downstream from the 5′ splice site and >6 nt upstream of the 3′ splice site. Valid H-boxes were located >60 nt downstream from the 5′ splice site and 50–90 nt upstream of an ACA-box (Fig. 2C). If both motifs were found within these distance constraints, we consider them position markers for a candidate H/ACA. A candidate host intron could contain multiple pairs of H-boxes and ACA-boxes. Once all possible pairs of H-boxes and ACA-boxes within an intron were identified, we evaluated them using the Bayesian Classifier and selected the most likely candidates.
Position weight matrix for scoring the ACA-box
A position–weight matrix (PWM) was generated for the ACA-box motif using the nucleotide frequencies in the set of ACA-boxes from the RNA training set. The PWM covers the ACA trinucleotide as well as the three downstream nucleotides. A threshold value, used for candidate ACA-box selection in subsequent genome-wide screens, was set at 95% of the lowest score from the RNA training set.
Construction of the naive Bayesian Classifier
Our Bayesian Classifier was constructed using four sequence features (Fig. 2D), which distinguished H/ACA snoRNAs from intronic sequences:
Starting hexamer of the host intron (5′ splice site). This feature checked whether the splice site of the host intron conformed to GTnnGT.
The expanded 6-nt ACA-box (ACAnnn). This feature determined whether the ACA-box of the candidate sequence was similar to the ACA-box of currently annotated H/ACAs. Five alternative sequences (in decreasing order of preference) were deemed acceptable: ACAATT, ACAnTT, ACAAAn, ACAnnn, and ATAnTT.
“In-class” tetramer usage. This feature calculated the likelihood of finding a particular tetramer in H/ACAs versus background introns (regardless of the number of times it appeared in the sequence).
“In-sequence” dinucleotide and trinucleotide frequency distribution. This feature examined how frequently a particular di- or trinucleotide was present in either an H/ACA or intron sequence.
Features 3 and 4 employed a classifier-within-classifier approach, which was a simplified version of the word-based naive Bayesian Classifier used in the Ribosomal Database Project Classifier (Wang et al. 2007). Data files and binary and source code for the Bayesian Classifier are available from http://www.sourceforge.net/projects/snobac.
Sequence sets for training and testing the naive Bayesian Classifier
Two sequence sets were used to train the Bayesian Classifier: the “RNA training set” and the “background introns,” which, respectively, consisted of introns harboring, or devoid of, annotated H/ACA snoRNAs. The RNA training set, containing 43 sequences, was derived by removing 11 sequences from the 54 parallel-nested snoRNAs. The removed sequences included four nested in the introns of noncompact host genes, three with noncanonical H-box motifs, and four that were significantly longer than other annotated H/ACAs (Fig. 3A; Supplemental Table S3). For the background introns, we used a set of 9139 host-sized (161–400 nt) introns from all noncompact genes.
We also generated two negative control sets, “long introns” and “short introns” (Table 1), for testing and evaluation purposes. The long introns contained 300-nt segments selected from the middle of introns longer than 1000 nt, as well as their own 5′ splice site hexamers. The short introns were a randomly selected subset of the background introns. As these sequences were all selected from noncompact genes, they are unlikely to contain H/ACA snoRNAs.
Confidence estimation using random re-sampling
The use of the entire RNA training set for predicting candidate H/ACA sequences could result in overtraining. To avoid this problem and to estimate the level of confidence in individual H/ACA predictions, we conducted 1000 rounds of training on subsets of the training and background introns. In each round, the classifier was trained on approximately two-thirds (0.66 × 42 + 1 = 30) of the sequences randomly selected from the RNA training set and an equal number of randomly selected background introns. Candidate sequences classified as an H/ACA in all 1000 rounds could be considered highly confident predictions. Prediction confidence, therefore, can be represented by the number of times out of 1000 in which a candidate sequence was classified as an H/ACA. We further evaluated the classifier using a “leave-1-out” cross-validation method (Supplemental Material; Supplemental Table S3).
Prediction of RNA secondary structures
Predictions of RNA secondary structures were made using RNAfold, a part of the Vienna RNA Package (Hofacker et al. 1994). For HP2, we folded the entire region between the candidate H-boxes and ACA-boxes. To ensure that this sequence folds into a single, rather than a number of smaller, hairpins, we also folded all sequences representing 5′ and 3′ truncations (in steps of 2 nt) between the original sequence and 50 nt. As HP1 is not bound by a defined upstream motif, we folded the sequence extending 90 nt upstream of the candidate H-box and all of its truncated variants. For each candidate, we reported the longest hairpin length for HP1 and HP2.
Comparison with SnoReport and validation of predicted sequences by sequence homology and expression
SnoReport version 1.2 was downloaded from http://www.bioinf.uni-leipzig.de/∼jana/software/SnoReport.html. We used WU-BLAST (W Gish. 1996-2004. WU-BLAST. http://blast.wustl.edu/)to search for sequences homologous between C. elegans and C. briggsae. We matched our candidate sequences against the NPA (nonpolyadenylated) and SNPA (small NPA, <500 nt) sets, derived from the published C. elegans tiling array data (He et al. 2007).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Andrey Rzhetsky, Yoav Gilad, and Rob Knight for thoughtful advice and critical reading of the manuscript.
Footnotes
Article published ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1876210.
REFERENCES
- Bachellerie JP, Cavaillé J, Hüttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazeley PS, Shepelev V, Talebizadeh Z, Butler MG, Fedorova L, Filatov V, Fedorov A. SnoTARGET shows that human orphan snoRNA targets locate close to alternative splice junctions. Gene. 2008;408:172–179. doi: 10.1016/j.gene.2007.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Harris TW, Antoshechkin I, Bastiani C, Bieri T, Blasiar D, Bradnam K, Canaran P, Chan J, Chen C-K, et al. WormBase: A comprehensive data resource for Caenorhabditis biology and genomics. Nucleic Acids Res. 2005;33:D383–D389. doi: 10.1093/nar/gki066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Zhu X, Skogerbø G, Zhao Y, Fu Z, Wang Y, He H, Cai L, Sun H, Liu C, et al. Organization of the Caenorhabditis elegans small noncoding transcriptome: Genomic features, biogenesis, and expression. Genome Res. 2006;16:20–29. doi: 10.1101/gr.4139206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci. 1999;96:4482–4487. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Noncoding RNA genes and the modern RNA world. Nat Rev Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends Genet. 2003;19:362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- Fedorov A, Stombaugh J, Harr MW, Yu S, Nasalean L, Shepelev V. Computer identification of snoRNA genes using a Mammalian Orthologous Intron Database. Nucleic Acids Res. 2005;33:4578–4583. doi: 10.1093/nar/gki754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu AD, Zhou H, Yu CH, Qu LH. A novel experimental approach for systematic identification of box H/ACA snoRNAs from eukaryotes. Nucleic Acids Res. 2005;33:e194. doi: 10.1093/nar/gni185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Cai L, Skogerbø G, Deng W, Liu T, Zhu X, Wang Y, Jia D, Zhang Z, Tao Y, et al. Profiling Caenorhabditis elegans noncoding RNA expression with a combined microarray. Nucleic Acids Res. 2006;34:2976–2983. doi: 10.1093/nar/gkl371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Wang J, Liu T, Liu XS, Li T, Wang Y, Qian Z, Zheng H, Zhu X, Wu T, et al. Mapping the C. elegans noncoding transcriptome with a whole-genome tiling microarray. Genome Res. 2007;17:1471–1477. doi: 10.1101/gr.6611807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel J, Hofacker IL, Stadler PF. SnoReport: Computational identification of snoRNAs with unknown targets. Bioinformatics. 2008;24:158–164. doi: 10.1093/bioinformatics/btm464. [DOI] [PubMed] [Google Scholar]
- Higa S, Maeda N, Kenmochi N, Tanaka T. Location of 2-O-methyl nucleotides in 26S rRNA and methylation guide snoRNAs in Caenorhabditis elegans. Biochem Biophys Res Commun. 2002;297:1344–1349. doi: 10.1016/s0006-291x(02)02377-x. [DOI] [PubMed] [Google Scholar]
- Hillier LW, Miller RD, Baird SE, Chinwalla A, Fulton LA, Koboldt DC, Waterston RH. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 2007;5:e167. doi: 10.1371/journal.pbio.0050167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh Chem. 1994;125:167–188. [Google Scholar]
- Huang ZP, Zhou H, He HL, Chen CL, Liang D, Qu LH. Genome-wide analyses of two families of snoRNA genes from Drosophila melanogaster, demonstrating the extensive utilization of introns for coding of snoRNAs. RNA. 2005;11:1303–1316. doi: 10.1261/rna.2380905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZP, Chen CJ, Zhou H, Li BB, Qu LH. A combined computational and experimental analysis of two families of snoRNA genes from Caenorhabditis elegans, revealing the expression and evolution pattern of snoRNAs in nematodes. Genomics. 2007;89:490–501. doi: 10.1016/j.ygeno.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Hüttenhofer A, Kiefmann M, Meier-Ewert S, O'Brien J, Lehrach H, Bachellerie JP, Brosius J. RNomics: An experimental approach that identifies 201 candidates for novel, small, nonmessenger RNAs in mouse. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A, Schattner P, Polacek N. Noncoding RNAs: Hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- Liang-Hu Q, Qing M, Hui Z, Yue-Qin C. Identification of 10 novel snoRNA gene clusters from Arabidopsis thaliana. Nucleic Acids Res. 2001;29:1623–1630. doi: 10.1093/nar/29.7.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- Marker C, Zemann A, Terhörst T, Kiefmann M, Kastenmayer JP, Green P, Bachellerie JP, Brosius J, Hüttenhofer A. Experimental RNomics: identification of 140 candidates for small nonmessenger RNAs in the plant Arabidopsis thaliana. Curr Biol. 2002;12:2002–2013. doi: 10.1016/s0960-9822(02)01304-0. [DOI] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP. Noncoding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Small regulatory RNAs in mammals. Hum Mol Genet. 2005;14:R121–R132. doi: 10.1093/hmg/ddi101. [DOI] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89:565–573. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- Rivas E, Eddy SR. Secondary structure alone is generally not statistically significant for the detection of noncoding RNAs. Bioinformatics. 2000;16:583–605. doi: 10.1093/bioinformatics/16.7.583. [DOI] [PubMed] [Google Scholar]
- Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Decatur WA, Davis CA, Ares M, Jr, Fournier MJ, Lowe TM. Genome-wide searching for pseudouridylation guide snoRNAs: Analysis of the Saccharomyces cerevisiae genome. Nucleic Acids Res. 2004;32:4281–4296. doi: 10.1093/nar/gkh768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T, Stephens R. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, Chinwalla A, Clarke L, Clee C, Coghlan A, et al. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol. 2003;1:166–192. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- Vinogradov AE. Compactness of human housekeeping genes: Selection for economy or genomic design? Trends Genet. 2004;20:248–253. doi: 10.1016/j.tig.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Wachi M, Ogawa T, Yokoyama K, Hokii Y, Shimoyama M, Muto A, Ushida C. Isolation of eight novel Caenorhabditis elegans small RNAs. Gene. 2004;335:47–56. doi: 10.1016/j.gene.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CY, Zhou H, Luo J, Qu LH. Identification of 20 snoRNA-like RNAs from the primitive eukaryote, Giardia lamblia. Biochem Biophys Res Commun. 2005;328:1224–1231. doi: 10.1016/j.bbrc.2005.01.077. [DOI] [PubMed] [Google Scholar]
- Yang JH, Zhang XC, Huang ZP, Zhou H, Huang MB, Zhang S, Chen YQ, Qu LH. SnoSeeker: An advanced computational package for screening of guide and orphan snoRNA genes in the human genome. Nucleic Acids Res. 2006;34:5112–5123. doi: 10.1093/nar/gkl672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihama M, Uechi T, Asakawa S, Kawasaki K, Kato S, Higa S, Maeda N, Minoshima S, Tanaka T, Shimizu N, et al. The human ribosomal protein genes: Sequencing and comparative analysis of 73 genes. Genome Res. 2002;12:379–390. doi: 10.1101/gr.214202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan G, Klämbt C, Bachellerie JP, Brosius J, Hüttenhofer A. RNomics in Drosophila melanogaster: Identification of 66 candidates for novel nonmessenger RNAs. Nucleic Acids Res. 2003;31:2495–2507. doi: 10.1093/nar/gkg361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemann A, op de Bekke A, Kiefmann M, Brosius J, Schmitz J. Evolution of small nucleolar RNAs in nematodes. Nucleic Acids Res. 2006;34:2676–2685. doi: 10.1093/nar/gkl359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Lin K. Excess of microRNAs in large and very 5′ biased introns. Biochem Biophys Res Commun. 2008;368:709–715. doi: 10.1016/j.bbrc.2008.01.117. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]