Abstract
In eubacteria, translation of defective messenger RNAs (mRNAs) produces truncated polypeptides that stall on the ribosome. A quality control mechanism referred to as trans-translation is performed by transfer-messenger RNA (tmRNA), a specialized RNA acting as both a tRNA and an mRNA, associated with small protein B (SmpB). So far, a clear view of the structural movements of both the protein and RNA necessary to perform accommodation is still lacking. By using a construct containing the tRNA-like domain as well as the extended helix H2 of tmRNA, we present a cryo-electron microscopy study of the process of accommodation. The structure suggests how tmRNA and SmpB move into the ribosome decoding site after the release of EF-Tu·GDP. While two SmpB molecules are bound per ribosome in a preaccommodated state, our results show that during accommodation the SmpB protein interacting with the small subunit decoding site stays in place while the one interacting with the large subunit moves away. Relative to canonical translation, an additional movement is observed due to the rotation of H2. This suggests that the larger movement required to resume translation on a tmRNA internal open reading frame starts during accommodation.
Keywords: cryo-electron microscopy, ribosome, SmpB, tmRNA, trans-translation
INTRODUCTION
Translation of defective messenger RNAs (mRNAs) caused by transcription errors, mRNA degradation, or recoding events produces truncated polypeptides that stall on the ribosome (Keiler 2008). In eubacteria, a trans-translation surveillance is performed by a ribonucleoprotein complex composed of the small protein B (SmpB) and transfer-messenger RNA (tmRNA), a specialized RNA with properties of both a tRNA and an mRNA, to provide a key component of the translation quality control pathway. The multifunctional roles of SmpB include alanylation enhancement of tmRNA (Shimizu and Ueda 2002) and association with tmRNA entering the empty A-site of the ribosome (Hanawa-Suetsugu et al. 2002). tmRNA–SmpB interacts with translational complexes blocked at the 3′-end of defective mRNAs to release stalled ribosomes and target their nascent polypeptides and mRNAs for degradation. The mRNA is always cleaved before trans-translation starts, avoiding a competition with canonical translation to occur. A quaternary “tmRNA–SmpB–EF-Tu·GTP” complex enters the A-site of the stalled ribosome still carrying the peptidyl-tRNA in the P-site. Recent biochemical and structural data indicate that SmpB provides contacts in place of the tRNA anti-codon stem, being a functional mimic of the pairing between a codon and an anti-codon at the ribosome decoding center (Nonin-Lecomte et al. 2009). At an early stage during trans-translation, two molecules of SmpB are recovered per stalled ribosome, one on each subunit and both contacting the “tRNA-like domain” (TLD) of tmRNA (Hallier et al. 2004; Kaur et al. 2006). After transpeptidation, the TLD moves to the P-site while the encoded tag reading frame of the tmRNA enters the mRNA path. The truncated mRNA is removed and degraded while the stalled ribosome switches the translation process to the tmRNA coding sequence resume codon, adding the tag peptide to the C terminus of the nascent polypeptide. One SmpB molecule is predicted to accompany tmRNA during its passage through the ribosome (Bugaeva et al. 2008).
Three-dimensional structures of SmpB from thermophiles in solution (Dong et al. 2002; Someya et al. 2003) and bound to the TLD (Gutmann et al. 2003; Bessho et al. 2007) have been determined. These findings all indicate that the SmpB-bound tmRNA functionally mimics a canonical tRNA during aminoacylation and entry into the ribosome. The structure of the remaining portion of the RNA relies on solution mapping (Felden et al. 1997) combined with cryo-electron microscopy (cryo-EM) reconstructions of the RNA backbone within the electron density of a preaccommodated complex (Kaur et al. 2006; Gillet et al. 2007). The subsequent stages of trans-translation, such as tmRNA accommodation, however, remain unknown.
During canonical translation, initial selection corresponds to the reversible association of the tRNA anti-codon with the codon in the 30S A-site, while the aminoacyl-acceptor end is still bound to elongation factor Tu (EF-Tu) (Ogle and Ramakrishnan 2005). Recognition between the codon and anti-codon stabilizes the ternary complex on the ribosome and the transmission of a signal to EF-Tu that hydrolyzes GTP (Schuette et al. 2009; Villa et al. 2009). After the release of the inorganic phosphate, EF-Tu rearranges into a GDP-bound conformation that dissociates from the aminoacyl end of the tRNA and from the ribosome. Either the aminoacyl end of tRNA moves into the A-site of the 50S subunit (accommodation of cognate aa-tRNA), which leads to formation of the peptide bond, or the aa-tRNA dissociates from the ribosome (proofreading of near-cognate aminoacyl-tRNA [aa-tRNA]) (Daviter et al. 2006). Accommodation is linked to the dynamic properties of the tRNA, acting as a molecular spring (Frank et al. 2005).
During trans-translation, the fate of the TLD after accommodation is of particular interest because the entry of Ala-tmRNA into the A-site of a stalled ribosome occurs without a codon–anti-codon interaction. The structure of an accommodated “tmRNA–SmpB” complex into stalled ribosomes will allow its comparison with the one of aminoacyl tRNAs that is promoted by codon recognition during canonical translation. Also, the movement of the aminoacyl-end of tmRNA into the A-site upon accommodation of the TLD is expected to produce a steric clash of the molecule of SmpB bound to the 50S with the P-site tRNA (Gutmann et al. 2003). Therefore, we can predict that the molecule of SmpB bound to the 50S subunit has to move away, but the questions are when and how it happens. To address these questions, we used cryo-EM three-dimesional reconstruction to study a ribonucleoprotein complex made of the TLD as well as the extended helix H2 of tmRNA bound to SmpB into stalled ribosomes.
RESULTS AND DISCUSSION
Structural design of the accommodated complexes
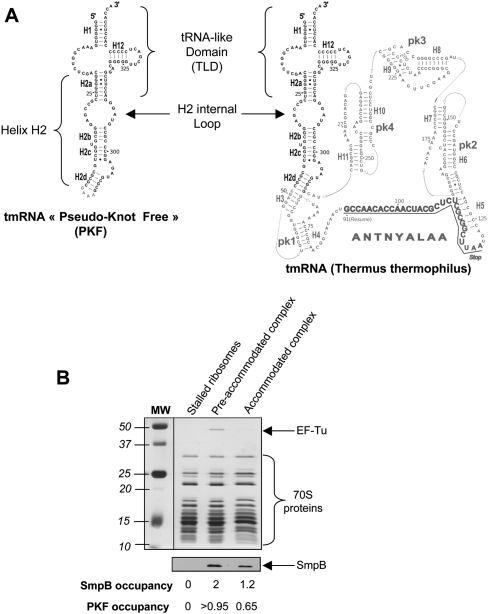
Previously, we carried out a cryo-EM study of full-length tmRNA in an accommodated state (Kaur et al. 2006). However, the recovered density was weak and several portions of tmRNA did not show up, preventing us from visualizing the movements of tmRNA and SmpB when entering the ribosomal A-site. This result can be interpreted as a consequence of (1) the RNA disorganization in and around the pseudoknots PK2–PK4 of the tmRNA and (2) the overall heterogeneity of the accommodated samples, due to the absence of complete purity, low occupancy, and conformational stability of the complexes. Therefore, in order to reduce its flexibility and increase its occupancy in the stalled ribosomes, we decided to work with a truncated version of tmRNA, depleted from its four pseudoknots and internal coding portion. This RNA, named PKF for pseudoknot-free tmRNA, is a 113-nucleotide-long molecule that contains the TLD of Thermus thermophilus tmRNA as well as the extended helix H2 (Fig. 1A). In this construct, H2 was capped by a GAAA tetraloop as used in previous structural studies (Gaudin et al. 2003). We reasoned that this short version of tmRNA might retain the tRNA function of tmRNA and its capability to bind SmpB (Karzai et al. 1999) as well as EF-Tu (Rudinger-Thirion et al. 1999) and AlaRS (Himeno et al. 1997). Indeed, a similar shortened version of tmRNA from Escherichia coli that lacks the messenger region and three of the four pseudoknot structures had been previously shown to be correctly folded and binding SmpB and EF-Tu·GTP simultaneously to its acceptor arm (Barends et al. 2001).
FIGURE 1.
(A) Secondary structure comparisons of PKF and full-length tmRNA from T. thermophilus. The domain in common, comprising the TLD (tRNA-like domain) and the helix H2, is shown in black. The figure is adapted from the tmRDB website (http://rnp.uthct.edu/rnp/tmRDB/tmRDB.html) (Andersen et al. 2006). (B) Biochemical characterization of the ribosomal complexes. After purification by size exclusion chromatography, ribosome complexes were analyzed by SDS-PAGE. The elongation factor EF-Tu was identified on the silver-stained gel, while SmpB and PKF occupancies were measured by Western blot and radiolabeling, respectively.
Biochemical characterization of the accommodated complexes
As for full-length tmRNA (Barends et al. 2001), the yields of aminoacylation of PKF were increased in the presence of EF-Tu·GTP and SmpB. A maximum of 65% alanylation of 1 μM PKF was achieved in the presence of 4 μM AlaRS, 6 μM EF-Tu·GTP, and 2 μM SmpB, after 30-min incubation at 37°C. The accommodated state was obtained by incubating a quaternary complex made of aminoacylated PKF, SmpB, and EF-Tu·GTP in the presence of a stalled ribosome. As a control, the preaccommodated state of the same complex was also prepared by adding 100 μM kirromycin (Stark et al. 1997). After formation, the complexes were loaded on a Superdex 200 gel filtration column (GE Healthcare) and recovered in the void volume while the unbound fraction of PKF could be discarded. The calculated occupancy of the ribosomes by PKF was close to 100% when stalled by the antibiotic kirromycin (preaccommodated state). In the absence of the antibiotic (to allow an accommodated state), the occupancy of ribosomes by PKF drastically reduces to close to 0% (data not shown). This is because the peptidyl transfer reaction lowers the stability of the complex before translocation (Semenkov et al. 2000; Shoji et al. 2009). Therefore, in order to prevent the subsequent ribosome-PKF complex instability, a deacylated P-site tRNAfMet was used to produce accommodated alanyl PKF. In this way, an occupancy of ∼65% could be achieved. This rather high fraction has to be compared to the 45% of occupancy previously obtained by using a full-length accommodated tmRNA (Kaur et al. 2006), confirming the higher stability of the shorter construct PKF. The stoichiometry of SmpB within both complexes was evaluated by Western blot. Band intensity analysis indicates that the amount of SmpB present in the preaccommodated state is almost twice that recovered after accommodation (2 vs. 1.2, respectively) (Fig. 1B), suggesting that accommodation triggers the departure of one ribosome-bound protein. At least one SmpB protein remains on each ribosome, including those lacking PKF (see below, Classification of the Subpopulations of the Complexes), which may account for the higher occupancy of SmpB relative to PKF after accommodation (1.2 vs. 0.65, respectively) (Fig. 1B).
Classification of the subpopulations of the complexes
Images of frozen-hydrated ribosomal complexes were recorded using a LaB6 microscope operating at 200 kV. Micrographs were digitized, and 153,785 individual images of ribosomal particles were extracted semi-automatically. The first iterations of image processing allowed us to discard isolated views of the 30S (5409 particles) and 50S subunits (30,952) as well as ratcheted ribosomes with PKF (41,359). Therefore, 76,035 images were extracted and used for alignment and three-dimensional reconstruction. After initial alignments of images, the first three-dimensional reconstruction clearly indicated heterogeneity in the raw images. This was revealed by the presence of additional densities located in various places of interest inside the ribosome. In addition, the overall ribosome reconstruction was not well defined. These observations indicated that the reconstruction resulted from the back-projection of ribosome images that correspond to the view of ribosomes in different conformational states. Knowing the exact components used to form the complexes, we could easily extrapolate which type of ribosomal conformational state could be present in solution. We then decided to use the local multivariate statistical analysis (MSA) method (Klaholz et al. 2004; Penczek et al. 2006) to separate images with respect to the different conformational states of the ribosome. This method allows the sorting of images into various subsets, depending on the densities present or absent in the area delimited by a mask applied on the projections. The use of a specific mask enabled us to directly sort the projections into two subsets that allowed us to compute two template structures. To eliminate any bias of the splitting procedure and to improve the initial assignment of the projections, the two template structures were projected in quasi-evenly distributed angular directions, and the two sets of reference projections were placed in one set. Then the entire data set of projections was compared with the combined set of reference projections in a standard multireference alignment procedure.
Three conformational states brought to light
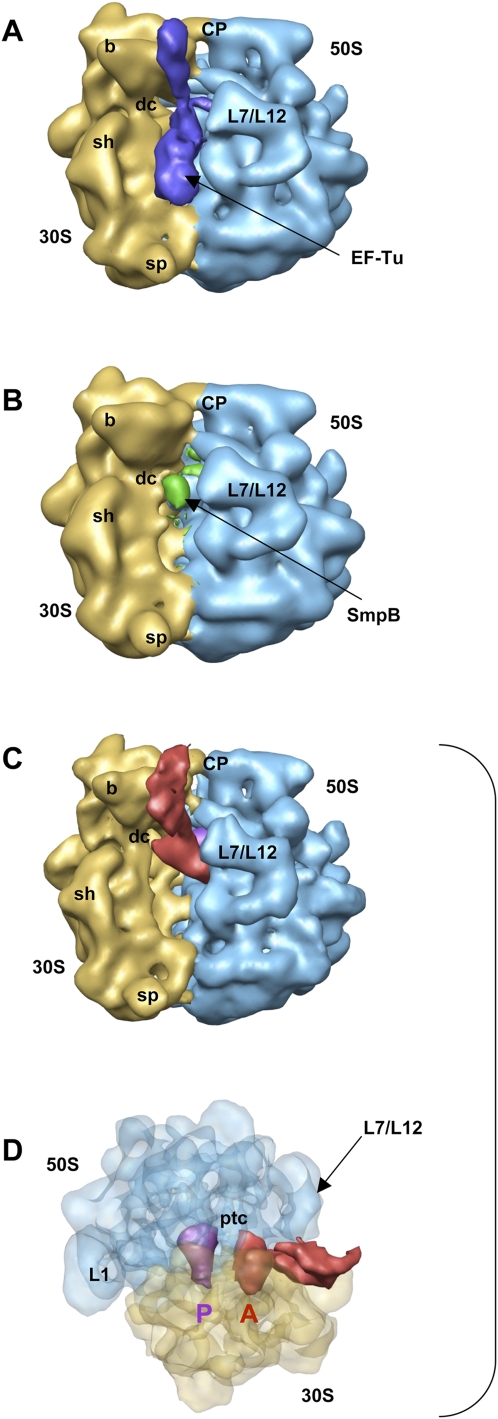
Three consecutive two-dimensional masks were applied on the low-pass filtered images. The first mask was placed on the location of EF-Tu, at the entrance of the ribosome, followed by a second two-dimensional mask located in the A-site, and, finally, a third mask placed toward the peptidyl center. The first two-dimensional mask enabled us to sort 12,557 projections, corresponding to the preaccommodated state (Fig. 2A). As for full-length tmRNA (Valle et al. 2003; Kaur et al. 2006), the TLD of PKF binds EF-Tu at the entrance of the L7/L12 stalk of the ribosome, whereas helix H2 protrudes along the beak of the 30S small subunit. However, the current map was obtained in the absence of kirromycin that immobilizes EF-Tu on the elongating ribosome after GTP hydrolysis. Because of the lack of this antibiotic, some flexibility is recovered on the reconstructed complex, preventing us from determining the exact location of the SmpB proteins or the subtle differences between the conformations of EF-Tu before and after GTP hydrolysis. Nevertheless, the isolation of the preaccommodated state of PKF allowed us (1) to show that the recognition of a ribosome stalled by PKF bound to EF-Tu·GTP occurs as for a full-length tmRNA and (2) to discard this set of particles from the reconstruction of the accommodated state. Subsequently, to discriminate ribosomes containing PKF, the second local-MSA process was applied with a two-dimensional mask located in the A-site, which allowed us to separate 34,144 particles that do not contain PKF. The resulting three-dimensional map showed empty ribosomes with a density present in the decoding site, which was compatible with the presence of a single SmpB molecule (Fig. 2B). This state was already described when incubating free SmpB with stalled ribosomes, in the absence of tmRNA, indicating the high stability of the complex (Gillet et al. 2007; Kurita et al. 2007). From the 29,334 remaining images, a third two-dimensional mask was placed toward the peptidyl center and allowed separation of 15,447 particles containing a homogeneous density in the A-site. This third subset corresponds to the accommodated state of the PKF–SmpB complex after the release of EF-TU·GDP (Fig. 2C,D). To interpret the cryo-EM data, the coordinates of the atomic model of a T. thermophilus 70S (Protein Data Bank [PDB] accession codes 2HGP and 2HGQ) (Yusupova et al. 2006) were fitted into the map. The overall structure of 70S is identical, in a classic, nonratcheted A/A-P/P configuration. The densities at the A- and P-sites were attributed by fitting the X-ray coordinates of the rigid tRNAs from the 70S crystals. Despite the weak density recovered at the expected location of the acceptor branch, which may account for its mobility when using a deacylated tRNA (Selmer et al. 2006; Agirrezabala et al. 2008), the tRNA placed into the P-site fits into the difference map (Fig. 3A,C, blue) and its orientation toward the peptidyl-center is obvious. Next, the orientation of the “PKF–SmpB” complex in the A-site was determined. As for the P-site tRNA, coordinates from the rigid tRNA of the crystal were first docked into the difference map recovered in the A-site, with the acceptor arm directed toward the peptidyl-center and the anti-codon loop interacting with the decoding site (Fig. 3A, red). Then, the X-ray coordinates of the TLD–SmpB complex from T. thermophilus (PDB accession code 2CZJ) (Bessho et al. 2007) were included by superimposing the acceptor arm of the TLD with the one from the A-site tRNA (Fig. 3B, green and black). When substituting the A-site tRNA with the TLD–SmpB structure in the cryo-EM map, a good correlation of the remaining portion of the complex with the experimental density was observed (Fig. 3C). The open L-shaped conformation of the TLD (Stagg et al. 2001; Gutmann et al. 2003) orientates SmpB toward the decoding site, while the rest of the PKF chain (H2) points to the beak of the 30S subunit. Due to the lack of an anti-codon stem-loop, the TLD of tmRNA is connected to the irregular helix H2 that is composed of four helical sections 2a–2d (Fig. 1A) abutting to pseudoknot PK1 (Zwieb et al. 1999). The TLD construct used by Bessho et al. (2007) contains H2a. Therefore, a three-dimensional model of the E. coli tmRNA in the tmRDB database (Andersen et al. 2006) was used to fit the internal loop and H2b-d modules into the nonattributed density from the cryo-EM map (Fig. 3C). The orientation, as well as the size and shape of H2 imposed by the fit of the density of the “TLD–SmpB” complex, convincingly match the remaining density recovered on the cryo-EM map from the accommodated complex (Fig. 3C). H2b-c is in line with H2a along the beak of the 30S subunit. The bulge between H2c and H2d (Fig. 1A) orientates H2d to direct the arc made by the following domains around the beak. This position suggests that a large movement is still necessary for the internal ORF to reach the ribosomal mRNA path.
FIGURE 2.
Reconstruction of PKF–SmpB complex bound to the stalled ribosome. (A) Cryo-EM map generated from the first subset sorted by the local MSA method, corresponding to a preaccommodated state before the release of EF-Tu. (Blue) Density for EF-Tu–PKF–SmpB. (B) Cryo-EM map generated from the second subset and corresponding to a stalled ribosome in the absence of PKF with a molecule of SmpB nested into the decoding site of the small ribosomal subunit. (Green) Density for SmpB. (C) Cryo-EM map generated from the third subset and corresponding to the accommodated state after the release of EF-Tu·GDP (same orientation as in A). (Red) Density for PKF-SmpB; (purple) the P-site tRNA. B adopts the same orientation as in A, while C allows a better view of the positioning of PKF compared to the tRNA stuck in the P-site. (D) View of map shown in B after a rotation by +90° around the vertical axis and −90° around the horizontal axis. The density corresponding to the ribosome is semitransparent. (Blue) The 50S subunit; (yellow) the 30S subunit. Landmarks on the 50S subunit are as follows: CP, central protuberance; L7/L12, stalk formed by proteins L7/L12; ptc, peptidyl transfer center; L1, protein L1. Landmarks on 30S subunit are: b, beak; dc, decoding center; sh, shoulder; sp, spur.
FIGURE 3.
Fitting of the 30S subunit, A- and P-site tRNAs, and PKF–SmpB into the cryo-EM map of the accommodated state. (A) Cryo-EM map of the 30S subunit (yellow), with the densities corresponding to TLD–SmpB complex in red and tRNAfMet in purple. The fitted 30S crystal structure from T. thermophilus with canonical tRNAs in A- and P-sites is shown in ribbon inside the map (yellow, 30S subunit; red, A-site tRNA; blue, P-site tRNA). (B) Superimposition of the acceptor arm from A-site tRNA with the one from the crystal structure of the TLD–SmpB complex. (red, A-site tRNA; light green, TLD; black, SmpB). (C) Substitution of the A-site tRNA by the TLD–SmpB atomic model in the cryo-EM map. The coordinates for helix H2 (dark green) were satisfactorily docked into the unexplained region. Landmarks on 30S subunit are: b, beak; dc, decoding center; sh, shoulder; sp, spur. For clarity, 50S subunit is not shown.
Interactions of accommodated PKF–SmpB complex with the stalled ribosome
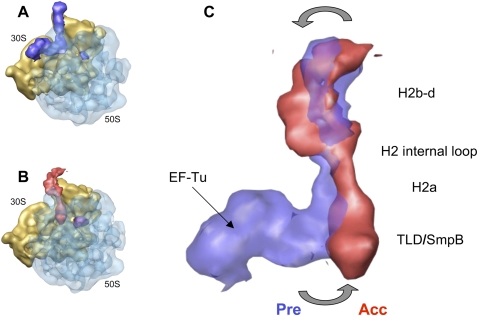
One of the main hurdles during the movement of PKF into the A-site is the expected steric clash between SmpB-1 bound to the large subunit and the P-site tRNA (Gutmann et al. 2003). As depicted on the map (Fig. 3C), the accommodation of PKF leads to the departure of SmpB-1 from the large subunit after the release of EF-Tu·GDP. This result is in agreement with the decrease of SmpB observed by Western blot analysis (Fig. 1B). Despite being absent from the accommodated PKF, the protein could migrate to another tmRNA binding site, located in the pseudoknot string containing the internal ORF, to functionally assist translation resumption (Metzinger et al. 2008). On the other hand, the placement of SmpB-2 confirms interactions with the decoding site during accommodation (Bessho et al. 2007; Kurita et al. 2007; Nonin-Lecomte et al. 2009). The orientation of SmpB-2 agrees with the predicted interactions between the globular body of the protein and the top of helix 44 (comprising decoding bases 1492 and 1493), while its C-terminal tail is directed toward helix 18 (containing decoding base G530). The superimposition of the individual maps of the PKF complex before and after accommodation reveals additional interesting features (Fig. 4). Relative to canonical translation, the process of accommodation of tmRNA involves an additional rotational movement of H2, centered in the conserved internal loop connecting 2a to 2b. This rotation pushes the TLD, including H2a, deep into the ribosomal A-site, while H2b-d moves away from the beak (Fig. 4C, gray arrows). Such a movement is expected to drag the arc made of the following modules around the beak, suggesting that the movement required to resume translation starts during accommodation.
FIGURE 4.
Movements of PKF during accommodation. (A) Cryo-EM map of the preaccommodated state. (B) Accommodated state. For clarity, density for the 50S subunit is shown as semitransparent. (C) Superimposition of the PKF complex before (semitransparent blue) and after (red) accommodation. The orientation is the same as in A and B.
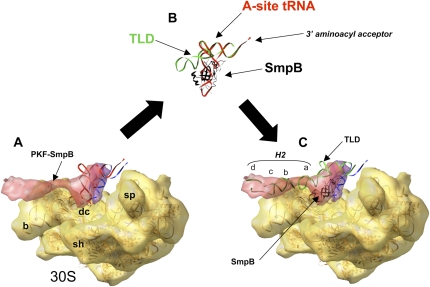
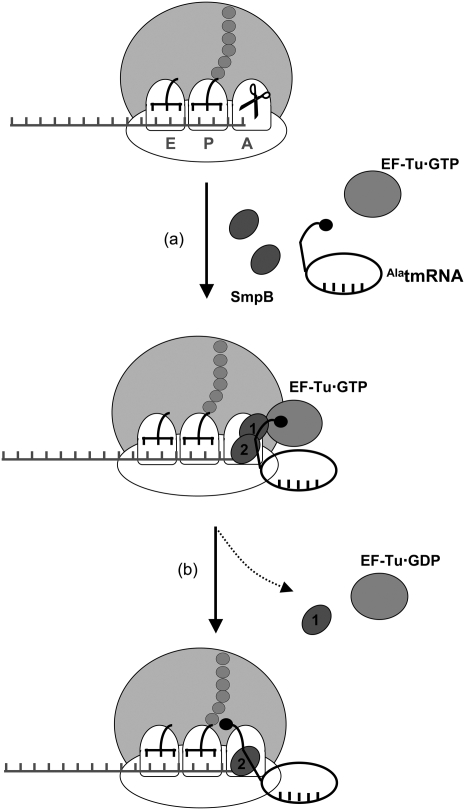
In conclusion, the present cryo-EM reconstruction of tmRNA accommodation suggests how the process takes place in the ribosome decoding site after the release of EF-Tu·GDP. The SmpB protein interacting with the small subunit decoding site stays in place while the one interacting with the large subunit moves away. The rotation movement of H2 triggered by the accommodation prepares the larger shift necessary to switch translation on tmRNA internal ORF (see Fig. 5). Work is now in progress to monitor the fates of SmpB and tmRNA during the EF-G dependent translocation of the complex into the P-site of a stalled ribosome.
FIGURE 5.
Trans-translation model for tmRNA recognition and accommodation. A ribosome stalls on an incomplete message (A), leading to the recruitment of a quaternary “AlatmRNA–SmpB–EF-Tu·GTP” complex to the A-site of the stalled ribosomes. At this step, two molecules of SmpB, one per subunit, are recovered. (B) Then, EF-Tu hydrolyzes the GTP and dissociates from the TLD and from the ribosome; the acceptor arm of TLD moves into the A-site of the 50S subunit (accommodation). The SmpB protein interacting with the 30S decoding site stays in place while the one interacting with the 50S subunit moves away. A movement of H2 triggered by the accommodation prepares the shift on the tmRNA internal ORF.
MATERIALS AND METHODS
Preparation of ribosomal complexes
Salt-washed T. thermophilus 70S ribosomes free of S1 were prepared as previously described (Clemons et al. 2001). His-tagged AlaRS from E. coli, His-tagged SmpB, and EF-Tu from T. thermophilus were overexpressed in E. coli using the T7 expression system and isolated by Ni2+ precharged HisTrap chelating column, according to the manufacturer's procedures (GE Healthcare). The T. thermophilus PKF gene was directly cloned into a pUC19 plasmid, downstream of a T7 RNA polymerase promoter. Plasmids were linearized by BstNI restriction enzyme, and the corresponding RNAs were synthesized for 4 h by using the T7 MEGAscript kit (Ambion). Purification was performed by electrophoresis on denaturing gels followed by electro-elution in a S&S BIOTRAP elution chamber (Schleicher and Schuell). EF-Tu·GDP was activated to EF-Tu·GTP immediately before use, by incubation for 15 min at 37°C in 50 mM HEPES-KOH (pH 7.5), 60 mM NH4Cl, 7 mM MgCl2, 2 mM GTP, 6 mM phosphoenolpyruvate (PEP), and 10 μg/mL pyruvate kinase (PK). The PKF was heated for 2 min at 80°C in a folding buffer (10 mM HEPES-KOH at pH 7.5, 20 mM NH4Cl, 5 mM MgCl2) and cooled for 30 min at room temperature before being aminoacylated. The alanylation was performed by incubation for 30 min at 37°C in 50 mM HEPES-KOH (pH 7.5), 60 mM NH4Cl, 7 mM MgCl2, 2 mM ATP, 6 mM PEP, 10 μg/mL PK, 30 μM alanine, 6 μM EF-Tu·GTP, 2 μM SmpB, 1 μM tmRNA PKF, and 4 μM AlaRS. The rates of alanylation of the PKF were measured by TCA precipitation of radiolabeled alanines, as previously described (Gillet and Felden 2001). After the addition of 0.1 volume of 2 M ice-cold potassium acetate (pH 5.0), the sample was concentrated 10-fold on a 5-kDa cut-off Microcon centrifugal unit (Millipore). Stalled ribosomes were obtained by incubating 70S ribosomes (1 μM) with 2 μM of an mRNA consisting of the GGCAAGGAGGUAAAAAUG sequence, and E. coli deacylated tRNAfMet (2 μM), for 30 min at 37°C in 5 mM HEPES-KOH (pH 7.5), 10 mM NH4Cl, 10 mM MgOAc, 50 mM KCl, and 6 mM β-mercaptoethanol. The final complexes were obtained by mixing 1 μM of stalled ribosomes with 2 μM of SmpB in the presence (preaccommodated complex) or absence (accommodated complex) of 100 μM kirromycin for 5 min at 37°C. The concentrated solution of EF-Tu·GTP–AlaPKF–SmpB complex was then added for 15 min at 37°C.
Purification of ribosomal complexes by size-exclusion chromatography
After filtration through 0.22-μm filters, samples (100 μL) were applied to a Superdex 200 HR 10/30 column (GE Healthcare) pre-equilibrated with 5 mM HEPES-KOH (pH 7.5), 10 mM NH4Cl, 10 mM MgOAc, 50 mM KCl, and 6 mM β-mercaptoethanol at 4°C. The fractions containing the purified 70S ribosomes were pooled and concentrated. 70S concentration was determined by UV absorbance using an extinction coefficient of 35 × 106 M−1cm−1 at 260 nm. To measure its occupancy, a 32P-radiolabeled PKF was used and subsequently quantified within the purified 70S fractions by liquid scintillation counting on a Wallac 1409 (Perkin-Elmer). The rate of occupancy was determined by dividing the amount of PKF by the total amount of ribosomes in the assay. Protein analysis was performed by SDS-PAGE analysis on the same fractions. Gels were stained by using the PlusOne Silver Staining kit (GE Healthcare) or subjected to SmpB immunodetection by Western blot. Detection was performed by using chemiluminescence (GE Healthcare). SmpB occupancy within the ribosomal complexes was obtained by analyzing 2 pmoles of the purified ribosome complexes by Western blot, using anti-SmpB antibodies. Signal intensities were calculated by using ImageJ (Abramoff et al. 2004).
Cryo-EM and image processing of the ribosomal complexes
The accommodated state complexes were diluted to a final concentration of 80 nM. Suspensions were applied to Lacey carbon grids. The excess of solution was blotted, and the grids were flash-frozen in liquid ethane, using the Vitrobot automat (FEI). This resulted in ribosomal complexes embedded in a thin film of vitrified ice. Electron micrographs were recorded under low-dose conditions at liquid nitrogen temperature with a Tecnai Sphera LaB6 200-kV transmission electron microscope. Images were collected at 50,000× magnification with a defocus range of 1–1.8 μm. With the aim of getting the maximal number of complex ribosomal particles and three-dimensional reconstruction in the range of 15–20 Å resolution, good micrographs were digitized on an Epson Perfection V750 Pro with a step size of 10 μm. Single-molecule images were selected semi-automatically from raw micrographs using the BOXER subroutine of EMAN (Ludtke et al. 1999) and were coarsened by a factor of 2, resulting in a pixel size corresponding to 4 Å. The phase-contrast-transfer function was corrected by phase flipping, and all image processing was done in the context of the IMAGIC-V software system (van Heel et al. 1996). The first iterations of image processing (multireference-alignment, multivariate-statistical-analysis, and structure refinement) allowed us to discard ratcheted ribosomes and dissociated 30S and 50S subunits and to compute a first three-dimensional map of the ribosomal complex. As the resulting map displayed some heterogeneity, an approach by local MSA (Klaholz et al. 2004; Penczek et al. 2006) with three consecutive masks delimiting area of interest on the ribosome (location of EF-Tu on the ribosome, 30S decoding site, and 50S A-site) was developed to obtain three three-dimensional structures corresponding to the preaccommodated state with PKF and EF-Tu, stalled ribosomes with SmpB in the decoding site, and the accommodated state without EF-Tu. The maps of the preaccommodated state and stalled ribosome with a molecule of SmpB were low-pass filtered to 20 Å. The estimated resolution, using FSC (Fourier shell correlation) coefficient 0.5 (van Heel et al. 2000), of the map of the accommodated state is 17.9 Å. The visualization and the fitting of the cryo-EM densities with the atomic models were performed using the program Chimera (Pettersen et al. 2004).
ACKNOWLEDGMENTS
We thank V. Ramakrishnan (MRC-LMB, Cambridge, UK) for providing the T. thermophilus ribosomes used in this study, and J.J Lacapère (Inserm U410, Paris, France) for insightful comments on the work. We thank the two anonymous referees for helpful suggestions on the manuscript. This work was supported by grants from ANR08-JCJC-0027-01 to R.G.; Rennes Métropole to D.T.; Ministère de l'Enseignement Supérieur et de la Recherche: “Fonction structure et inactivation d'ARN bactériens”; INSERM: “Fonction structure et inactivation d'ARN bactériens”; ANR606-MIME-016-01 Microbiologie, Immunologie et Maladies émergentes”; Région Bretagne CPER “Agent pathogène et risques et maladies infectieuses” to B.F. F.W. was supported by a PhD fellowship from the Ministère de l'Enseignement Supérieur et de la Recherche.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1757410.
REFERENCES
- Abramoff MD, Magelhaes PJ, Ram SJ. Image pocessing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen ES, Rosenblad MA, Larsen N, Westergaard JC, Burks J, Wower IK, Wower J, Gorodkin J, Samuelsson T, Zwieb C. The tmRDB and SRPDB resources. Nucleic Acids Res. 2006;34:D163–D168. doi: 10.1093/nar/gkj142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barends S, Karzai AW, Sauer RT, Wower J, Kraal B. Simultaneous and functional binding of SmpB and EF-Tu-TP to the alanyl acceptor arm of tmRNA. J Mol Biol. 2001;314:9–21. doi: 10.1006/jmbi.2001.5114. [DOI] [PubMed] [Google Scholar]
- Bessho Y, Shibata R, Sekine S, Murayama K, Higashijima K, Hori-Takemoto C, Shirouzu M, Kuramitsu S, Yokoyama S. Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proc Natl Acad Sci. 2007;104:8293–8298. doi: 10.1073/pnas.0700402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaeva EY, Shpanchenko OV, Felden B, Isaksson LA, Dontsova OA. One SmpB molecule accompanies tmRNA during its passage through the ribosomes. FEBS Lett. 2008;582:1532–1536. doi: 10.1016/j.febslet.2008.03.049. [DOI] [PubMed] [Google Scholar]
- Clemons WM, Jr, Brodersen DE, McCutcheon JP, May JL, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: Purification, crystallization and structure determination. J Mol Biol. 2001;310:827–843. doi: 10.1006/jmbi.2001.4778. [DOI] [PubMed] [Google Scholar]
- Daviter T, Gromadski KB, Rodnina MV. The ribosome's response to codon–anticodon mismatches. Biochimie. 2006;88:1001–1011. doi: 10.1016/j.biochi.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Dong G, Nowakowski J, Hoffman DW. Structure of small protein B: The protein component of the tmRNA-SmpB system for ribosome rescue. EMBO J. 2002;21:1845–1854. doi: 10.1093/emboj/21.7.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felden B, Himeno H, Muto A, McCutcheon JP, Atkins JF, Gesteland RF. Probing the structure of the Escherichia coli 10Sa RNA (tmRNA) RNA. 1997;3:89–103. [PMC free article] [PubMed] [Google Scholar]
- Frank J, Sengupta J, Gao H, Li W, Valle M, Zavialov A, Ehrenberg M. The role of tRNA as a molecular spring in decoding, accommodation, and peptidyl transfer. FEBS Lett. 2005;579:959–962. doi: 10.1016/j.febslet.2004.10.105. [DOI] [PubMed] [Google Scholar]
- Gaudin C, Nonin-Lecomte S, Tisne C, Corvaisier S, Bordeau V, Dardel F, Felden B. The tRNA-like domains of E. coli and A. aeolicus transfer-messenger RNA: Structural and functional studies. J Mol Biol. 2003;331:457–471. doi: 10.1016/s0022-2836(03)00760-5. [DOI] [PubMed] [Google Scholar]
- Gillet R, Felden B. Transfer RNAAla recognizes transfer-messenger RNA with specificity: A functional complex prior to entering the ribosome? EMBO J. 2001;20:2966–2976. doi: 10.1093/emboj/20.11.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet R, Kaur S, Li W, Hallier M, Felden B, Frank J. Scaffolding as an organizing principle in trans-translation. The roles of small protein B and ribosomal protein S1. J Biol Chem. 2007;282:6356–6363. doi: 10.1074/jbc.M609658200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann S, Haebel PW, Metzinger L, Sutter M, Felden B, Ban N. Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature. 2003;424:699–703. doi: 10.1038/nature01831. [DOI] [PubMed] [Google Scholar]
- Hallier M, Ivanova N, Rametti A, Pavlov M, Ehrenberg M, Felden B. Pre-binding of small protein B to a stalled ribosome triggers trans-translation. J Biol Chem. 2004;279:25978–25985. doi: 10.1074/jbc.M314086200. [DOI] [PubMed] [Google Scholar]
- Hanawa-Suetsugu K, Takagi M, Inokuchi H, Himeno H, Muto A. SmpB functions in various steps of trans-translation. Nucleic Acids Res. 2002;30:1620–1629. doi: 10.1093/nar/30.7.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno H, Sato M, Tadaki T, Fukushima M, Ushida C, Muto A. In vitro trans translation mediated by alanine-charged 10Sa RNA. J Mol Biol. 1997;268:803–808. doi: 10.1006/jmbi.1997.1011. [DOI] [PubMed] [Google Scholar]
- Karzai AW, Susskind MM, Sauer RT. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Gillet R, Li W, Gursky R, Frank J. Cryo-EM visualization of transfer messenger RNA with two SmpBs in a stalled ribosome. Proc Natl Acad Sci. 2006;103:16484–16489. doi: 10.1073/pnas.0607438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC. Biology of trans-translation. Annu Rev Microbiol. 2008;62:133–151. doi: 10.1146/annurev.micro.62.081307.162948. [DOI] [PubMed] [Google Scholar]
- Klaholz BP, Myasnikov AG, Van Heel M. Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature. 2004;427:862–865. doi: 10.1038/nature02332. [DOI] [PubMed] [Google Scholar]
- Kurita D, Sasaki R, Muto A, Himeno H. Interaction of SmpB with ribosome from directed hydroxyl radical probing. Nucleic Acids Res. 2007;35:7248–7255. doi: 10.1093/nar/gkm677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Metzinger L, Hallier M, Felden B. The highest affinity binding site of small protein B on transfer messenger RNA is outside the tRNA domain. RNA. 2008;14:1761–1772. doi: 10.1261/rna.1185808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonin-Lecomte S, Germain-Amiot N, Gillet R, Hallier M, Ponchon L, Dardel F, Felden B. Ribosome hijacking: A role for small protein B during trans-translation. EMBO Rep. 2009;10:160–165. doi: 10.1038/embor.2008.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- Penczek PA, Frank J, Spahn CM. A method of focused classification, based on the bootstrap 3D variance analysis, and its application to EF-G–dependent translocation. J Struct Biol. 2006;154:184–194. doi: 10.1016/j.jsb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rudinger-Thirion J, Giege R, Felden B. Aminoacylated tmRNA from Escherichia coli interacts with prokaryotic elongation factor Tu. RNA. 1999;5:989–992. doi: 10.1017/s135583829999101x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuette JC, Murphy FVt, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Semenkov YP, Rodnina MV, Wintermeyer W. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat Struct Biol. 2000;7:1027–1031. doi: 10.1038/80938. [DOI] [PubMed] [Google Scholar]
- Shimizu Y, Ueda T. The role of SmpB protein in trans-translation. FEBS Lett. 2002;514:74–77. doi: 10.1016/s0014-5793(02)02333-5. [DOI] [PubMed] [Google Scholar]
- Shoji S, Walker SE, Fredrick K. Ribosomal translocation: One step closer to the molecular mechanism. ACS Chem Biol. 2009;4:93–107. doi: 10.1021/cb8002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya T, Nameki N, Hosoi H, Suzuki S, Hatanaka H, Fujii M, Terada T, Shirouzu M, Inoue Y, Shibata T, et al. Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus. FEBS Lett. 2003;535:94–100. doi: 10.1016/s0014-5793(02)03880-2. [DOI] [PubMed] [Google Scholar]
- Stagg SM, Frazer-Abel AA, Hagerman PJ, Harvey SC. Structural studies of the tRNA domain of tmRNA. J Mol Biol. 2001;309:727–735. doi: 10.1006/jmbi.2001.4632. [DOI] [PubMed] [Google Scholar]
- Stark H, Rodnina MV, Rinke-Appel J, Brimacombe R, Wintermeyer W, van Heel M. Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature. 1997;389:403–406. doi: 10.1038/38770. [DOI] [PubMed] [Google Scholar]
- Valle M, Gillet R, Kaur S, Henne A, Ramakrishnan V, Frank J. Visualizing tmRNA entry into a stalled ribosome. Science. 2003;300:127–130. doi: 10.1126/science.1081798. [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- van Heel M, Gowen B, Matadeen R, Orlova EV, Finn R, Pape T, Cohen D, Stark H, Schmidt R, Schatz M, et al. Single-particle electron cryo-microscopy: Towards atomic resolution. Q Rev Biophys. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- Villa E, Sengupta J, Trabuco LG, LeBarron J, Baxter WT, Shaikh TR, Grassucci RA, Nissen P, Ehrenberg M, Schulten K, et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci. 2009;106:1063–1068. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- Zwieb C, Wower I, Wower J. Comparative sequence analysis of tmRNA. Nucleic Acids Res. 1999;27:2063–2071. doi: 10.1093/nar/27.10.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]