Abstract
Hen1 is an RNA ribose 2′-O-methyltransferase that modifies the 3′ terminal nucleoside of eukaryal small regulatory RNAs. Here, we report that Hen1 homologs are present in bacterial proteomes from eight different phyla. Bacterial Hen1 is encoded by the proximal ORF of a two-gene operon that also encodes polynucleotide kinase-phosphatase (Pnkp), an RNA repair enzyme. Purified recombinant Clostridium thermocellum Hen1 is a homodimer of a 465-amino acid polypeptide. CthHen1 catalyzes methyl transfer from AdoMet to the 3′ terminal nucleoside of an RNA oligonucleotide, but is unreactive with a synonymous DNA oligonucleotide or an RNA with a single 3′-terminal deoxyribose sugar. CthHen1 is optimally active at alkaline pH and dependent on manganese. Activity is inhibited by AdoHcy and abolished by mutations D291A and D316A in the putative AdoMet-binding pocket. The C-terminal fragment, Hen1-(259–465), comprises an autonomous monomeric methyltransferase domain.
Keywords: AdoMet, transmethylation, RNA end-healing
INTRODUCTION
Endoribonuclease toxins play important roles in defending bacteria against nonself species and viruses. Colicin D and colicin E5 are secreted ribotoxins that kill nonimmune Escherichia coli cells by breaking the anticodon loops of essential tRNAs, thereby inhibiting cellular protein synthesis (Ogawa et al. 1999; Tomita et al. 2000; Yajima et al. 2006). Colicin efficacy relies on the inability of E. coli to repair the damaged tRNAs. Other ribotoxins are maintained intracellularly in a latent state until activated by stress or virus infection. For example, E. coli responds to bacteriophage T4 infection by turning on the tRNA endonuclease PrrC, which incises tRNALys at a single phosphodiester in the anticodon loop, thereby shutting off protein synthesis before the phage can replicate (Amitsur et al. 2003; Blanga-Kanfi et al. 2006). The virus evades the host response by encoding a two-component RNA repair system, consisting of T4 polynucleotide kinase-phosphatase (Pnkp) and T4 RNA ligase 1 (Rnl1), which heals the broken 2′,3′ cyclic phosphate and 5′-OH tRNA ends (by hydrolysis of the 2′,3′ cyclic phosphate to form a 3′-OH and phosphorylation of the 5′-OH to form a 5′-PO4), and then seals them to restore the pool of functional tRNA (Amitsur et al. 1987).
tRNA- and mRNA-damaging endoribonuclease toxins are widely distributed among bacterial taxa (Zhang et al. 2005; Zhu et al. 2006; Davidoff and Kaufmann 2008; Nariya and Inouye 2008; Yamaguchi et al. 2009). The prevalence of ribotoxins raises the question of whether RNA repair systems might also exist in bacterial proteomes as a means of evading programmed RNA breakage. The identification of RNA healing and sealing enzymes in several bacterial species suggested such an RNA repair capacity (Martins and Shuman 2004, 2005; Raymond and Shuman 2007).
Clostridium thermocellum Pnkp (CthPnkp) is the exemplary bacterial end-healing enzyme. CthPnkp is a homodimer of an 870-amino acid polypeptide protein composed of N-terminal kinase, central metallophosphoesterase, and C-terminal ligase-like domains (Fig. 1A; Martins and Shuman 2005). The CthPnkp kinase module catalyzes phosphoryl transfer from ATP to the 5′-OH terminus of RNA polynucleotides. The metallophosphoesterase domain releases Pi from 2′-PO4, or 3′-PO4, or 2′,3′ cyclic phosphate ribonucleotide ends (Keppetipola and Shuman 2006a,b, 2007). Packaging of the kinase and phosphatase activities within a single polypeptide implied an RNA repair function for CthPnkp, analogous to that of T4 Pnkp. Indeed, CthPnkp was able to heal a broken tRNA with 2′,3′ cyclic phosphate and 5′-OH termini, yielding 3′-OH and 5′-PO4 ends that were substrates for sealing by T4 Rnl1 (Keppetipola et al. 2007). The case was fortified by the structural homology of the CthPnkp C-terminal domain to RNA ligases and the demonstration that CthPnkp reacts with ATP to form a covalent enzyme-AMP adduct, just as RNA ligases do during strand joining (Martins and Shuman 2005).
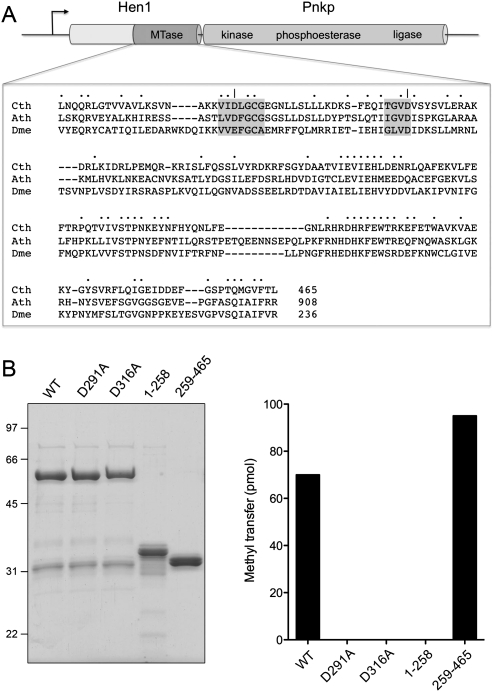
FIGURE 1.
Clostridium thermocellum Hen1. (A) The adjacent co-oriented ORFs encoding the Hen1 (465-amino acid) and Pnkp (870-amino acid) polypeptides comprise a putative two-gene operon. Pnkp is a trifunctional RNA repair enzyme composed of 5′ OH polynucleotide kinase, 2′,3′ phosphoesterase, and ligase-like adenylyltransferase domains. Hen1 is composed of an N-terminal domain unique to the bacterial Hen1 clade and a C-terminal methyltransferase domain (MTase) that is homologous to the methyltransferase domains of eukaryal Hen1 RNA methyltransferase enzymes. The amino acid sequences of the Clostridium thermocellum (Cth), Arabidopsis thaliana (Ath), and Drosophila melanogaster (Dme) Hen1 methyltransferase domains are aligned. (•) Positions of side-chain identity/similarity in all three proteins. (–) Gaps in the alignment. The conserved AdoMet-binding motifs are shaded in gray. (|) The Asp291 and Asp316 residues that were mutated to alanine. (B) CthHen1 purification and methyltransferase activity. (Left) Aliquots (5 μg) of recombinant full-length wild-type (WT) CthHen1, mutants D291A and D316A, and the N-terminal (1–258) and C-terminal (259–465) polypeptide fragments were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (kilodalton) of marker polypeptides are indicated on the left. (Right) Reaction mixtures (10 μL) containing 25 mM Tris-HCl (pH 8.5), 0.5 mM MnCl2, 20 μM [3H-CH3]AdoMet, 10 μM 24-mer RNA, and 4 μM WT or mutant CthHen1 as specified were incubated for 30 min at 45°C. The extents of 3H-methyl transfer to RNA are plotted.
At the time of its discovery and initial characterization, CthPnkp homologs with the same trifunctional domain organization were detected in only a few other bacteria (Martins and Shuman 2005). Presently, we find CthPnkp homologs in 40 bacterial species, representing eight different phyla (Supplemental Table S1). The instructive point of the phylogenetic comparison was the realization that the 465-amino acid protein encoded by the Cthe_2767 ORF located immediately upstream of the C. thermocellum pnkp gene is also conserved in the same 40 bacteria (Supplemental Table S1). The C-terminal half of Cthe_2767 polypeptide is homologous to the ribose 2′-O-methyltransferase module of eukaryal metazoan and plant Hen1 proteins, which modify the 3′-terminal ribonucleotide of small regulatory RNAs (Fig. 1A; Yu et al. 2005; Yang et al. 2006; Horwich et al. 2007; Kirino and Mourelatos 2007; Saito et al. 2007). The N-terminal half of Cthe_2767 is conserved among the bacterial Hen1 homologs only. Henceforth, we shall refer to the Cthe_2767 polypeptide as CthHen1 and to the suite of its bacterial homologs as a distinctive bacterial Hen1 clade, which is more widely distributed among bacteria taxa than was appreciated initially (Tkaczuk et al. 2006).
The C. thermocellum hen1 and pnkp ORFs are co-oriented and packed tightly so that the Hen1 translation stop codon overlaps (with a −1 frame shift) the Pnkp translation start codon. In contrast, the Cthe_2769 ORF downstream from pnkp is transcribed in the opposite orientation; moreover, there is a gap of 126 nucleotides between the hen1 translation start codon and the stop codon of the preceding Cthe_2766 ORF. This arrangement is highly suggestive of a hen1 pnkp operon. Indeed, the hen1 and pnkp ORFs are arrayed as a two-component cassette in the other bacteria listed in Supplemental Table S1, with a few exceptions (e.g., Herpetosiphon aurantiacus and Maricaulis marinus) in which the hen1 and pnkp genes are separated by a co-oriented ORF to form a multigene cassette.
The conserved clustering of Pnkp with a putative methyltransferase (Hen1) suggests that Hen1 might have a role in bacterial RNA end modification and, perhaps, in RNA repair. Here, we report the physical and functional characterization of recombinant CthHen1.
RESULTS AND DISCUSSION
Recombinant CthHen1 is an RNA methyltransferase
Previous studies of eukaryal Hen1 were performed with recombinant proteins produced in bacteria as GST-fusions and isolated by glutathione-affinity chromatography (Yu et al. 2005; Horwich et al. 2007; Kirino and Mourelatos 2007; Saito et al. 2007). Here, we produced a His6-tagged version of CthHen1 in order to avoid the issue of forced dimerization by the GST-domain. Isolation of His6CthHen1 from a soluble bacterial extract by Ni-affinity chromatography yielded a predominant ∼58 kDa polypeptide (Fig. 1B).
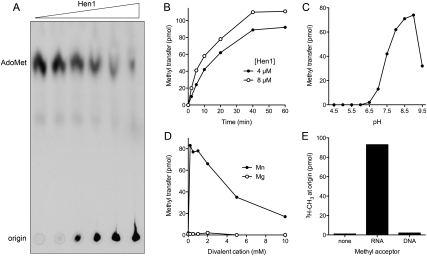
The methyltransferase activity of CthHen1 was demonstrated by incubating the protein with 20 μM [3H-CH3]-AdoMet and 10 μM of a 24-mer RNA oligonucleotide substrate, which resulted in label transfer from AdoMet to the RNA to form a polyanionic methylated product that remained at the origin during PEI-cellulose TLC and was well separated from the cationic AdoMet substrate (Fig. 2A). The extent of methylation was proportional to input CthHen1 (Fig. 2A). The methylated product accumulated steadily with time, attaining a plateau value corresponding to ∼1 pmol of 3H-CH3 transfer per pmol of input RNA oligonucleotide (Fig. 2B). The initial rate of product formation was proportional to the CthHen1 concentration (Fig. 2B); we estimated a turnover number of 0.11 min−1. The rate decreased with time as the AdoMet donor was depleted and the available acceptor sites on the RNA were methylated. Activity was optimal at pH 8.5–9.0 in Tris-HCl buffer, decreased to 18% of the optimum at pH 7.0 (Tris-HCl), and was virtually abolished at pH <6.5 (Tris-acetate) (Fig. 2C). The optimal temperature for CthHen1 methyltransferase activity was 45°–50°C (data not shown); activity was 70%, 79%, and 48% of the peak value at 55°C, 37°C, and 25°C, respectively (data not shown).
FIGURE 2.
Characterization of the CthHen1 RNA methyltransferase activity. (A) Methyl transfer to RNA. Reaction mixtures (10 μL) containing 25 mM Tris-HCl (pH 8.0), 5 mM DTT, 0.5 mM MnCl2, 20 μM [3H-CH3]AdoMet, 10 μM 24-mer RNA, and increasing concentrations of CthHen1 (0.06, 0.6, 1.25, 2.5, 5, and 9.8 μM, proceeding from left to right) were incubated for 60 min at 45°C and then quenched with EDTA. Aliquots of the mixtures were analyzed by PEI-cellulose TLC in 0.2 M ammonium sulfate. The 3H-labeled material was visualized by autoradiography. (B) Time course. Reaction mixtures containing 25 mM Tris-HCl (pH 8.5), 0.5 mM MnCl2, 20 μM [3H-CH3]AdoMet, 10 μM 24-mer RNA, and 4 or 8 μM CthHen1 were incubated at 45°C. Aliquots (10 μL) were withdrawn at the times specified and quenched immediately with EDTA. The extents of 3H-methyl transfer to RNA are plotted as a function of reaction time. (C) pH dependence. Reaction mixtures (10 μL) containing 25 mM Tris buffer (either Tris-acetate at pH 4.5–6.5 or Tris-HCl at pH 7.0–9.5), 0.5 mM MnCl2, 20 μM [3H-CH3]AdoMet, and 4 μM CthHen1 as specified were incubated for 30 min at 45°C. The extents of 3H-methyl transfer to RNA are plotted as a function of pH. (The pH values represent that of a 1M solution of Tris buffer at 22°C.) (D) Divalent cation dependence. Reaction mixtures (10 μL) containing 25 mM Tris-HCl (pH 8.0), either 0, 0.2, 0.5, 1, 2, 5, or 10 mM MnCl2 or MgCl2 as specified, 20 μM [3H-CH3]AdoMet, 10 μM 24-mer RNA, and 5 μM CthHen1 were incubated for 30 min at 45°C. The extents of 3H-methyl transfer to RNA are plotted as a function of divalent cation concentration. (E) Methyl acceptor specificity. Reaction mixtures (10 μL) containing 25 mM Tris-HCl (pH 8.5), 0.5 mM MnCl2, 20 μM [3H-CH3]AdoMet, 8 μM CthHen1, and either 10 μM 24-mer RNA, 10 μM 24-mer DNA, or no added nucleic acid (none) were incubated for 30 min at 45°C. The extents of 3H-methyl transfer to the TLC origin are plotted.
CthHen1 methyltransferase activity depended on inclusion of a divalent cation cofactor. The metal requirement was satisfied by manganese, optimally at 0.2–1 mM MnCl2 (Fig. 2D). In contrast, magnesium was ineffective in the range of from 0.2 to 10 mM MgCl2 (Fig. 2D). CthHen1 failed to promote methyl transfer to a synonymous 24-mer DNA oligonucleotide under the same conditions that resulted in methylation of a 24-mer RNA (Fig. 2E). A simple interpretation of this strict RNA specificity is that CthHen1, like its eukaryal homologs, is a ribose O2′-methyltransferase.
CthHen1 is a homodimer
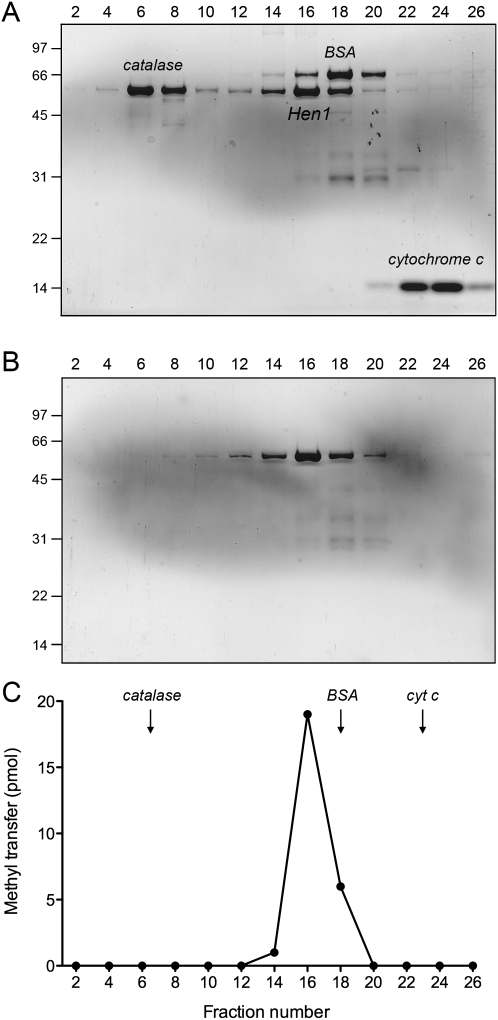
The recombinant full-length CthHen1 protein was subjected to zonal velocity sedimentation in a 15%–30% glycerol gradient, either alone or with marker proteins catalase (native size 248 kDa), BSA (66 kDa), and cytochrome c (12 kDa) included as internal standards. His6CthHen1 (calculated to be a 56 kDa polypeptide) sedimented as a discrete peak (fraction 16) on the heavy side of BSA (Fig. 3A). The protein alone sedimented identically in a parallel gradient (Fig. 3B). The RNA methyltransferase activity profile paralleled the abundance of the CthHen1 polypeptide and peaked at fraction 16 (Fig. 3C). We surmise from these results that the methyltransferase activity is intrinsic to CthHen1, and that the enzyme is a dimer in solution (perhaps with an asymmetric shape).
FIGURE 3.
Glycerol gradient sedimentation. Zonal velocity sedimentation was performed as described in the Materials and Methods. Aliquots (15 μL) of even-numbered glycerol gradient fractions of CthHen1 cosedimented with marker proteins (A), or by itself (B) were analyzed by SDS-PAGE. The Coomassie-blue stained gels are shown. The CthHen1, catalase, BSA, and cytochrome c polypeptides are indicated in A. Molecular weight calibrations for the PAGE analysis (kilodalton) are indicated on the left. The methyltransferase activity profile for the glycerol gradient in B is shown in C. Reaction mixtures (10 μL) containing 25 mM Tris-HCl (pH 8.5), 1 mM MnCl2, 20 μM [3H-CH3]AdoMet, 10 μM 24-mer RNA, and 2 μL of the indicated gradient fraction were incubated for 60 min at 45°C. The extents of 3H-methyl transfer to RNA are plotted.
Essential AdoMet-binding aspartates and delineation of an autonomous methyltransferase domain
A primary structure alignment of the bacterial, plant, and metazoan Hen1 proteins highlights the presence of a canonical AdoMet binding site composed of two peptide motifs: 289VIDLGCG295 and 313TGVD316 in CthHen1 (Fig. 1A). The Asp316 equivalent typically engages in a bidentate hydrogen-bonding interaction with the AdoMet ribose hydroxyls. The Asp291 equivalent coordinates the AdoMet methionine amine. To query whether these aspartates are functionally relevant in CthHen1, we mutated them individually to alanine and purified the recombinant D291A and D316A proteins in parallel with wild-type CthHen1 (Fig. 1B, left panel). The D291A and D316A mutations abolished RNA methyltransferase activity (Fig. 1B, right panel), implying that bacterial Hen1 relies on a classical AdoMet binding site. Previous studies of Arabidopsis and Drosophila Hen1 had shown that RNA methylation was abolished by more drastic quadruple mutations (DFGCG to NAVAV in Arabidopsis; EFGCA to QAVAV in Drosophila) in the proximal AdoMet-binding motif (Yu et al. 2005; Saito et al. 2007).
In order to evaluate the role of the distinctive N-terminal domain of bacterial Hen1 proteins and to test whether the C-terminal Hen1-like segment containing the methyltransferase motifs suffices for activity, we produced and purified recombinant versions of the N-domain (amino acids 1–258) and the C-domain (amino acids 259–465, initiating just 33 residues upstream of the essential Asp291) (Fig. 1B). As expected, the isolated N-domain had no methyltransferase activity; the instructive finding was that the isolated C-domain was at least as active in RNA methylation as full-length wild-type CthHen1 (Fig. 1B). Additional experiments indicated that the rate of methylation by the isolated C-domain (4 μM) was about 1.7 times that of an equivalent concentration of full-length CthHen1 (data not shown). We conclude that the C-terminal module of bacterial Hen1 is an autonomous catalytic unit.
The native sizes of the isolated N- and C- domains were gauged by glycerol gradient sedimentation with internal standards (Supplemental Fig. S1). The CthHen1-C protein (calculated to be a 27 kDa polypeptide) sedimented between cytochrome c and BSA, albeit closer to the cytochrome c peak (Supplemental Fig. S1B). We surmise that Hen1-C is a monomer. The methyltransferase activity profile was coincident with the abundance of the Hen1-C polypeptide (Supplemental Fig. S2). The Hen1-N protein (a 32 kDa polypeptide) sedimented as a heavier species, overlapping the light side of the BSA peak (Supplemental Fig. S1A). We surmise that Hen1-N is a dimer and that the dimeric quaternary structure of full-length CthHen1 is conferred by an N-terminal dimerization module specific to the bacterial Hen1 clade.
Analysis of the reaction product
For the purpose of product analysis, the RNA methylation reaction was performed with the 24-mer RNA methyl acceptor (at 40 μM) in twofold excess over [3H-CH3]-AdoMet (20 μM) in order to maximize the extent of label transfer to the RNA. As shown in Figure 4, the majority of the label was transferred to RNA to form a polyanionic species retained at the origin during PEI-cellulose TLC. Appearance of this labeled material depended on the 24-mer RNA and either full-length CthHen1 (Fig. 4A) or CthHen1-(259–465) (Fig. 4B). Aliquots of the reaction mixtures were subjected to alkaline hydrolysis with 0.3 M KOH for 15 h at 37°C, then neutralized with 0.3 M HCl and analyzed by TLC in parallel with control samples that were incubated for 15 h at 37°C in 0.3 M KCl (Fig. 4). The treatments of control reaction mixtures lacking Hen1 highlighted the chemical lability of [3H-CH3]-AdoMet (Parks and Schlenk 1958). Incubation in strong alkali transformed the AdoMet virtually quantitatively into a single 3H-labeled species that migrated faster than the parent compound; this species comigrated with 35S-methionine (data not shown). In contrast, incubation in KCl resulted in incomplete AdoMet depletion and formation of two 3H-labeled decay products, one of which comigrated with the alkaline hydrolysis product 3H-methionine. The other more abundant decay product migrated behind AdoMet at the same position as the minor labeled species present in the original sample of [3H-CH3]-AdoMet after the 30 min incubation in the absence of CthHen1. This species (indicated by the asterisk in Fig. 4) is either methylthioadenosine or S-ribosylmethionine (Parks and Schlenk 1958).
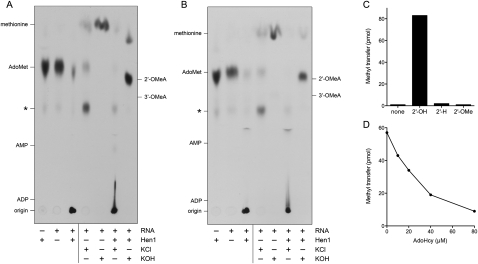
FIGURE 4.
Product analysis and substrate specificity. (A,B) Reaction mixtures (30 μL) containing 25 mM Tris-HCl (pH 8.5), 0.5 mM MnCl2, 20 μM [3H-CH3]AdoMet, 8 μM CthHen1 (A) or 8 μM CthHen1(259–465) (B), and 40 μM 24-mer RNA (where indicated by +) were incubated for 30 min at 45°C and then quenched with EDTA. The RNA-containing mixtures were split into three aliquots. One was stored at −20°C, while the other two were adjusted to either 0.3 M KOH or 0.3 M KCl and incubated for 15 h at 37°C. The KOH samples were then neutralized by adding HCl to 0.3 M final concentration. The samples were analyzed by PEI cellulose TLC in 50 mM ammonium sulfate. Unlabeled marker nucleotides analyzed in parallel were detected by UV autofluorescence. The 3H-labeled material was visualized by autoradiography. (C) Methyl acceptor specificity. Reaction mixtures (10 μL) containing 25 mM Tris-HCl (pH 8.5), 0.5 mM MnCl2, 20 μM [3H-CH3]AdoMet, 8 μM CthHen1, and either no added nucleic acid (none) or 10 μM 24-mer RNA with a 3′ terminal ribose (2′-OH), a 3′-terminal deoxyribose (2′-H), or a 3′-terminal 2′-O-methylribose (2′-OMe), were incubated for 30 min at 45°C. The extents of 3H-methyl transfer to the TLC origin are plotted. (D) Product inhibition. Reaction mixtures (10 μL) containing 25 mM Tris-HCl (pH 8.5), 0.5 mM MnCl2, 20 μM [3H-CH3]AdoMet, 10 μM 24-mer RNA, 4 μM CthHen1, and 0, 10, 20, 40 or 80 μM AdoHcy were incubated for 30 min at 45°C. The extents of 3H-methyl transfer to RNA are plotted as a function of AdoHcy concentration.
The terminal nucleoside of the 24-mer RNA acceptor is guanosine, and the expected alkaline hydrolysis product if CthHen1 is a 3′-terminal ribose methyltransferase would be either 2′-O-methylguanosine or 3′-O-methylguanosine. In contrast, ribose 2′-O-methylation at an internal nucleoside would yield a labeled HONmpNp dinucleotide product with a nominal net charge of –3. The instructive finding was that KOH treatment of the labeled RNA product of the CthHen1 and CthHen1-(259–465) reactions quantitatively released the 3H-label from the origin to form a predominant species that migrated two-thirds of the way up the chromatogram, at the same position as a cold 2′-O-methyladenosine standard, which was separated by this procedure from the 3′-O-methyladenosine standard (Fig. 4; note that the analogous pair of 2′-O-methylguanosine and 3′-O-methylguanosine nucleosides are not available commercially). There was no labeled product of alkaline hydrolysis migrating in the vicinity of nucleotide markers ADP (nominal charge –3) or AMP (nominal charge –2) (Fig. 4). Thus, we surmise that CthHen1 methylated the 3′-terminal ribose O2′ of the RNA acceptor.
Additional evidence for the terminal ribose O2′ as the target of CthHen1 methylation emerged from a comparison of 24-mer RNA substrates of identical nucleobase sequence that differed only with respect to the 3′ pentose sugar moiety. Whereas the RNA with a terminal ribose was efficiently methylated by CthHen1, we detected no methyl transfer to synonymous RNAs with either a single 3′-terminal deoxyribose sugar (2′-H) or a single 3′-terminal 2′-O-methyl ribose sugar (Fig. 4C).
Finally, we found that label transfer from 20 μM [3H-CH3]-AdoMet to the RNA 3′ end was inhibited in a concentration-dependent fashion by the reaction product AdoHcy (Fig. 4D). We estimated an IC50 of 25 μM AdoHcy, indicating that CthHen1 has similar affinity for the substrate AdoMet and the product AdoHcy.
Mechanistic implications
Notwithstanding the novelty of targeted methylation of RNA 3′ ends and the importance of eukaryal Hen1 enzymes in microRNA and piwiRNA biogenesis and stability (Park et al. 2002; Yu et al. 2005; Horwich et al. 2007; Kurth and Mochizuki 2009), there is relatively little known about the mechanism of the Hen1 methylation reaction. The present characterization of bacterial Hen1 as a metal-dependent RNA 3′-terminal ribose 2′-O-methyltransferase instantly distinguishes Hen1 from the metal-independent viral cap ribose 2′-O-methyltransferase enzyme family, exemplified by vaccinia virus VP39, that has been characterized in depth—structurally and mechanistically (Hodel et al. 1998; Li et al. 2004; Li and Gershon 2006). VP39 and CthHen1 catalyze the same chemical reaction at opposite ends of the RNA chain, yet only the latter enzyme requires a metal.
The preferential use of manganese by CthHen1 is intriguing, but not without precedent among metal-dependent O-methyltransferase enzymes (Kauss and Hassid 1967; Veser 1987; Kelm et al. 1998; Lukacin et al. 2004). The exemplar of the metal-dependent O-methyltransferase clade is catechol O-methyltransferase (COMT), which transfers the methyl group from AdoMet to one of two vicinal hydroxyl groups on the catechol ring. A view of the active site of rat liver COMT, from the crystal structure of the enzyme bound to AdoMet, Mg2+, and a catechol analog inhibitor (Vidgren et al. 1994), is shown in Supplemental Figure S3. The structure illustrates how the metal ion plays a critical role in organizing the active site and positioning the catechol hydroxyl for in-line attack on the AdoMet methyl carbon. The octahedral metal coordination complex is occupied by two aspartate carboxylate oxygens, one asparagine amide oxygen, one water, and both vicinal hydroxyls of the catechol ring. We noted that the metal-binding Asp141 of COMT, which also coordinates the AdoMet amine (Supplemental Fig. S3), is located within a peptide motif (139FLDHW) that is conserved in CthHen1 (367VIEHL) and its Arabidopsis (VIEHM) and Drososphila (LIEHV) homologs (Fig. 1A). COMT Asp141 is situated 52 residues downstream from the Glu90 side chain (in the AdoMet binding motif) that coordinates the AdoMet ribose hydroxyls (Vidgren et al. 1994). In CthHen1, the conserved Glu369 residue (which, we propose, corresponds to COMT Asp141) is spaced similarly 54 residues downstream from the essential Asp316 (equivalent to COMT Glu90). We further speculate that the metal-binding function of the Asp169–Asn170 dipeptide of COMT (Supplemental Fig. S3) might be fulfilled in CthHen1 by a Asp417–His418 dipeptide that is conserved in the fly and plant Hen1 homologs (Fig. 1A). Participation of a histidine in the metal-binding complex could explain the preference of CthHen1 for manganese versus magnesium.
By analogy to COMT (Vidgren et al. 1994), and in light of the utilization of substrates with vicinal hydroxyls by both COMT and CthHen1, we surmise that the likely mechanism of CthHen1 transmethylation entails: (1) metal coordination of the RNA 3′ terminal ribose 2′ and 3′ hydroxyls to position the ribose O2′ for attack on the AdoMet methyl carbon; and (2) metal-assisted lowering of the pKa of the ribose 2′ hydroxyl. This mechanism easily accounts for the specificity of Hen1 for methylation of the terminal RNA nucleoside and blockade of terminal ribose methylation by a 3′-phosphomonoester (Saito et al. 2007), insofar as metal coordination of vicinal ribose hydroxyls is not possible at internal positions of an RNA chain or at an RNA-3′-phosphate end. The proposed mechanism of dual hydroxyl coordination by metal also explains why a terminal 3′-deoxy modification (3′-H) suppresses ribose 2′-O-methylation by Arabidopsis Hen1 (Yu et al. 2005).
Biological implications
The genetic clustering of the bacterial RNA 3′-end-modifying enzyme Hen1 in an operon with the RNA end-healing enzyme Pnkp is tantalizing with respect to potential RNA repair pathways. Although Pnkp can heal broken RNA ends for sealing by RNA ligases (Keppetipola et al. 2007), and Pnkp itself has an RNA ligase-like adenylyltransferase domain, our initial inability to detect RNA sealing by CthPnkp in vitro (using substrates that are sealed by T4 RNA ligases) raised the prospects that either CthPnkp can seal only specific RNAs, or that its full RNA ligation activity depends on another bacterial protein (Martins and Shuman 2005). Hen1 is a plausible candidate for this role. Our preliminary experiments indicate that mixing the isolated homodimeric Hen1 N-terminal domain with recombinant Pnkp homodimer results in formation of a higher order complex that is detectable by glycerol gradient sedimentation. However, we have not been able to reconstitute a ligase activity by simply mixing full-length Hen1 with Pnkp. It is conceivable that the two proteins need to be coexpressed in vivo in order to form a ligation-competent RNA repair complex.
Packaging of a site-specific RNA methylase with an RNA repair enzyme is loosely reminiscent of DNA restriction-modification operons. An appealing scenario is that the Hen1–Pnkp system is an antidote to target-specific ribotoxins that generate 2′,3′ cyclic phosphate/5′-OH breaks. Methylation of the ribose O2′ by Hen1 after 3′ end healing would generate an NmpN′ repair junction after sealing, which should then be immune to recleavage by the toxin endoribonuclease. Any number of other functions can also be envisioned for Hen1–Pnkp in bacterial RNA transactions, e.g., in RNA recombination, genesis of regulatory RNAs, etc. The landscape is uncharted and free of preconceptions, given the scant knowledge of RNA biology for the various bacterial taxa that carry the Hen1–Pnkp cassette.
MATERIALS AND METHODS
Recombinant CthHen1
The Cthe_2767 ORF encoding CthHen1 was amplified by PCR from Clostridium thermocellum genomic DNA with sense and antisense primers designed to introduce a BamHI site preceding the ATG start codon and a SalI site downstream from the stop codon. The PCR product was restricted with endonucleases BamHI and SalI and inserted between the BamHI and SalI sites in pETduet-1 to generate a phage T7 RNA polymerase-driven expression vector encoding the 465-amino acid CthHen1 polypeptide fused to an N-terminal His6 tag. Missense mutations D291A and D316A were introduced into the hen1 gene by two-stage overlap extension PCR. A truncated ORF encoding the N-terminal domain (1–258) was generated by PCR amplification using an antisense primer that introduced a stop codon in lieu of the Lys259 codon and a SalI site downstream. A truncated ORF encoding the C-terminal domain (259–465) was generated by PCR amplification using a sense primer that introduced a start codon in lieu of the Leu258 codon and an upstream BamH1 site. The truncated ORFs were inserted into pETduet-1. The pET plasmid inserts were sequenced completely to verify the intended coding sequence and exclude acquisition of unwanted coding changes. The pET-His6CthHen1 plasmids were transformed into E. coli BL21(DE3). Cultures (1-L) amplified from single transformants were grown at 37°C in Luria-Bertani medium containing 0.1 mg/mL ampicillin until the A600 reached ∼0.6. The cultures were adjusted to 0.5 mM isopropyl-β-D-thiogalactopyranoside and 2% (v/v) ethanol and then incubated at 17°C for 16 h with constant shaking. Cells were harvested by centrifugation, and the pellets were stored at −80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 30 mL of buffer A (50 mM Tris-HCl at pH 7.5, 0.5 M NaCl, 10% sucrose). Cell lysis was achieved by the addition of lysozyme, PMSF, and Triton X-100 to final concentrations of 1 mg/mL, 0.2 mM, and 0.1%, respectively. The lysates were sonicated to reduce viscosity, and insoluble material was removed by centrifugation. The soluble extracts were applied to 5-mL columns of Ni-nitrilotriacetic acid-agarose (Qiagen) that had been equilibrated with buffer A. The columns were washed with buffer B (50 mM Tris-HCl at pH 7.5, 0.25 M NaCl, 0.05% Triton X-100, 10% glycerol) containing 25 mM imidazole and eluted stepwise with buffer B containing 50 and 100 mM imidazole. The polypeptide compositions of the column fractions were monitored by SDS-PAGE. The peak 100 mM imidazole eluate fractions containing the CthHen1 polypeptide were pooled and dialyzed overnight against buffer C (50 mM Tris-HCl at pH 7.5, 0.1 M NaCl, 2 mM DTT, 2 mM EDTA, 10% glycerol). The dialysates were applied to 2.5-mL columns of DEAE-cellulose equilibrated in buffer C; the full-length wild-type CthHen1, CthHen1-Ala mutants, and the C-terminal domain CthHen1(259–465) were recovered in the DEAE flow-through. The N-terminal domain CthHen1(1–258) adhered to the DEAE resin and was eluted with 200 mM NaCl. Protein concentrations were determined by using the Bio-Rad dye reagent with bovine serum albumin as the standard. The yield of His6CthHen1 was 10 mg from a 1-L bacterial culture.
Glycerol gradient sedimentation
Aliquots (2 nmol) of full-length CthHen1 or the isolated N- and C-terminal domains were mixed with catalase, BSA, and cytochrome c (50 μg each). These mixtures, or samples of CthHen1, CthHen1(1–258), and CthHen1(259–465) alone, were applied to 4.8-mL 15%–30% glycerol gradients containing 50 mM Tris-HCl (pH 7.5), 0.1 M NaCl, 2 mM DTT, 2 mM EDTA, 0.1% Triton X-100. The gradients were centrifuged at 50,000 rpm for 16 h in a Beckman SW55i rotor. Fractions (∼0.2 mL) were collected from the bottoms of the tubes.
Methyltransferase assay
Reaction mixtures containing 25 mM Tris-HCl (pH 8.5), 0.5 mM MnCl2, 20 μM [3H-CH3]AdoMet, 10 μM 24-mer synthetic RNA oligonucleotide (5′-CACUAUCGGAAUAAGGGCGACACG) (Dharmacon), and CthHen1 (plus 0.2 mM EDTA, 0.2 mM DTT, and 10 mM NaCl contributed by the enzyme buffer), were incubated at 45°C. The reactions were quenched by adding 1 μL of 55 mM EDTA. Aliquots (4 μL) were spotted on a polyethyleneimine cellulose TLC plate, which was developed with 0.2 M (NH4)2SO4. For product analyses, the dried TLC plate was sprayed with Enhance (Perkin Elmer) and radiolabeled material was visualized by autoradiography after exposing the plate to X-ray film for 1 d at −80°C. To quantify the methyltransferase activity, the 3H-AdoMet and 3H RNA-containing (origin) portions of the lanes were cut out and the radioactivity in each was determined by liquid scintillation counting.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This research was supported by NIH grant GM42498. S.S. is an American Cancer Society Research Professor.
NOTE ADDED IN PROOF
While this paper was under review, Raven Huang and colleagues reported that coexpression of Anabaena variabilis Pnkp and Hen1 in E. coli yielded a Pnkp–Hen1 complex with RNA methyltransferase and RNA ligase activity (Chan et al. 2009).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1926510.
REFERENCES
- Amitsur M, Levitz R, Kaufman G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase, and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitsur M, Benjamin S, Rosner R, Chapman-Shimshoni D, Meidler R, Blanga S, Kaufmann G. Bacteriophage T4-encoded Stp can be replaced as activator of anticodon nuclease by a normal host cell metabolite. Mol Microbiol. 2003;50:129–143. doi: 10.1046/j.1365-2958.2003.03691.x. [DOI] [PubMed] [Google Scholar]
- Blanga-Kanfi S, Amitsur M, Azem A, Kaufmann G. PrrC-anticodon nuclease: Functional organization of a prototypal bacterial restriction RNase. Nucleic Acids Res. 2006;34:3209–3219. doi: 10.1093/nar/gkl415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CM, Zhou C, Huang R. Reconstituting bacterial RNA repair and modification in vitro. Science. 2009;326:247. doi: 10.1126/science.1179480. [DOI] [PubMed] [Google Scholar]
- Davidoff E, Kaufmann G. RloC: A wobble nucleotide-excising and zinc-responsive bacterial tRNase. Mol Microbiol. 2008;69:1560–1574. doi: 10.1111/j.1365-2958.2008.06387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel AE, Gershon PD, Quiocho FA. Structural basis for sequence-nonspecific recognition of 5′-capped mRNA by a cap-modifying emzyme. Mol Cell. 1998;1:443–447. doi: 10.1016/s1097-2765(00)80044-1. [DOI] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matarnga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Kauss H, Hassid WZ. Biosynthesis of the 4-O-methyl-D-glycuronic acid unit of hemicellulose B by transmethylation from S-adenosyl-L-methionine. J Biol Chem. 1967;242:1680–1684. [PubMed] [Google Scholar]
- Kelm A, Shaw L, Schauer R, Reuter G. The biosynthesis of 8-O-methylated sialic acids in the starfish Asteria rubens: Isolation and characterization of S-adenosyl-L-methionine:sialate-8-O-methyltransferase. Eur J Biochem. 1998;251:874–884. doi: 10.1046/j.1432-1327.1998.2510874.x. [DOI] [PubMed] [Google Scholar]
- Keppetipola N, Shuman S. Mechanism of the phosphatase component of Clostridium thermocellum polynucleotide kinase-phosphatase. RNA. 2006a;12:73–82. doi: 10.1261/rna.2196406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppetipola N, Shuman S. Distinct enzymic functional groups are required for the phosphomonoesterase and phosphodiesterase activities of Clostridium thermocellum polynucleotide kinase-phosphatase. J Biol Chem. 2006b;281:19251–19259. doi: 10.1074/jbc.M602549200. [DOI] [PubMed] [Google Scholar]
- Keppetipola N, Shuman S. Characterization of the 2′,3′ cyclic phosphodiesterase activities of Clostridium thermocellum polynucleotide kinase-phosphatase and bacteriophage λ phosphatase. Nucleic Acids Res. 2007;35:7721–7732. doi: 10.1093/nar/gkm868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppetipola N, Nandakumar J, Shuman S. Reprogramming the tRNA splicing activity of a bacterial RNA repair enzyme. Nucleic Acids Res. 2007;35:3624–3630. doi: 10.1093/nar/gkm110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA. 2007;13:1397–1401. doi: 10.1261/rna.659307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth HM, Mochizuki K. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA. 2009;15:675–685. doi: 10.1261/rna.1455509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Gershon PD. pKa of the mRNA cap-sepcific 2′-O-metyltransferase catalytic lysine by HSQC NMR detection of a two-carbon probe. Biochemistry. 2006;45:907–917. doi: 10.1021/bi051736h. [DOI] [PubMed] [Google Scholar]
- Li C, Xia Y, Gao X, Gershon PD. Mechanism of RNA 2′-O-methylation: Evidence that the catalytic lysine acts to steer rather than deprotonate the target nucleophile. Biochemistry. 2004;43:5680–5687. doi: 10.1021/bi0359980. [DOI] [PubMed] [Google Scholar]
- Lukacin R, Matern U, Specker S, Vogt T. Cations modulate the substrate specificity of bifunctional class I O-methyltransferase from Ammi majus. FEBS Lett. 2004;577:367–370. doi: 10.1016/j.febslet.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Martins A, Shuman S. An RNA ligase from Deinococcus radiodurans. J Biol Chem. 2004;279:50654–50661. doi: 10.1074/jbc.M407657200. [DOI] [PubMed] [Google Scholar]
- Martins A, Shuman S. An end-healing enzyme from Clostridium thermocellum with 5′ kinase, 2′,3′ phosphatase, and adenylyltransferase activities. RNA. 2005;11:1271–1280. doi: 10.1261/rna.2690505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nariya H, Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Tomita K, Ueda T, Watanabe K, Uozumi T, Masaki H. A cytotoxic ribonuclease targeting specific tRNA anticodons. Science. 1999;283:2097–2100. doi: 10.1126/science.283.5410.2097. [DOI] [PubMed] [Google Scholar]
- Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks LW, Schlenk F. The stability and hydrolysis of S-adenosylmethionine: Isolation of S-ribosylmethionine. J Biol Chem. 1958;230:295–305. [PubMed] [Google Scholar]
- Raymond A, Shuman S. Deinococcus radiodurans RNA ligase exemplifies a novel ligase clade with a distinctive N-terminal module that is important for 5′-PO4 nick sealing and ligase adenylylation but dispensable for phosphodiester formation at an adenylylated nick. Nucleic Acids Res. 2007;35:839–849. doi: 10.1093/nar/gkl1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suziki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylatiom of Piwi-interacting RNA and their 3′ ends. Genes & Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkaczuk KL, Obarska A, Bujnicki JM. Molecular phylogenetics and comparative modeling of HEN1, a methyltransferase involved in plant microRNA biogenesis. BMC Evol Biol. 2006;6:6. doi: 10.1186/1471-2148-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Ogawa T, Uozumi T, Watanabe K, Masaki H. A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine tRNAs at their anticodon loops. Proc Natl Acad Sci. 2000;97:8278–8283. doi: 10.1073/pnas.140213797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veser J. Kinetics and inhibtion studies of catechol O-methyltransferase from the yeast Candida tropicalis. J Bacteriol. 1987;169:3696–3700. doi: 10.1128/jb.169.8.3696-3700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidgren J, Svensson LA, Liljas A. Crystal structure of catechol O-methyltransferase. Nature. 1994;368:354–358. doi: 10.1038/368354a0. [DOI] [PubMed] [Google Scholar]
- Yajima S, Inoue S, Ogawa T, Nonaka T, Ohsawa K, Masaki H. Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction. Nucleic Acids Res. 2006;34:6074–6082. doi: 10.1093/nar/gkl729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Park JH, Inouye M. MsqR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J Biol Chem. 2009;284:28746–28753. doi: 10.1074/jbc.M109.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ebright YW, Yu B, Chen X. HEN1 recognizes 21-24 nt small RNA duplexes and deposits a methyl group into the 2′OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006;34:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation is a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Hara H, Kato I, Inouye M. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J Biol Chem. 2005;280:3143–3150. doi: 10.1074/jbc.M411811200. [DOI] [PubMed] [Google Scholar]
- Zhu L, Zhang Y, Teh JS, Zhang J, Connell N, Rubin H, Inouye M. Characterization of mRNA interferases from Mycobacterium tuberculosis. J Biol Chem. 2006;281:18638–18643. doi: 10.1074/jbc.M512693200. [DOI] [PubMed] [Google Scholar]