Abstract
The genetic networks controlling stem cell identity are the focus of intense interest, due to their obvious therapeutic potential as well as exceptional relevance to models of early development. Genome-wide mapping of transcriptional networks in mouse embryonic stem cells (mESCs) reveals that many endogenous noncoding RNA molecules, including long noncoding RNAs (lncRNAs), may play a role in controlling the pluripotent state. We performed a genome-wide screen that combined full-length mESC transcriptome genomic mapping data with chromatin immunoprecipitation genomic location maps of the key mESC transcription factors Oct4 and Nanog. We henceforth identified four mESC-expressed, conserved lncRNA-encoding genes residing proximally to active genomic binding sites of Oct4 and Nanog. Accordingly, these four genes have potential roles in pluripotency. We show that two of these lncRNAs, AK028326 (Oct4-activated) and AK141205 (Nanog-repressed), are direct targets of Oct4 and Nanog. Most importantly, we demonstrate that these lncRNAs are not merely controlled by mESC transcription factors, but that they themselves regulate developmental state: knockdown and overexpression of these transcripts lead to robust changes in Oct4 and Nanog mRNA levels, in addition to alterations in cellular lineage-specific gene expression and in the pluripotency of mESCs. We further characterize AK028326 as a co-activator of Oct4 in a regulatory feedback loop. These results for the first time implicate lncRNAs in the modulation of mESC pluripotency and expand the established mESC regulatory network model to include functional lncRNAs directly controlled by key mESC transcription factors.
Keywords: ncRNA, mouse embryonic stem cells/mESCs, differentiation, Gomafu, RNCR2, C18ORF22
INTRODUCTION
The study of pluripotent mouse embryonic stem cells (mESCs) has yielded important insights into pre-implantation mouse development and genomic reprogramming (Smith 2001). Derived from the pluripotent epiblast of the mouse blastocyst, mESCs can self-renew while retaining a normal karyotype, in response to recombinant leukemia inhibitory factor (LIF) and fetal bovine serum (FBS) (Williams et al. 1988). Withdrawal of essential pluripotency maintenance factors allows mESCs to differentiate into cell types of the three principal embryonic germ layers: mesoderm, endoderm, and ectoderm (Keller 1995). In particular, withdrawal of LIF and application of retinoic acid (RA) result in loss of pluripotency and in predominantly ectodermal differentiation of mESCs (Bibel et al. 2004).
mESC pluripotency requires the expression of the PIT/OCT/UNC (POU) homeodomain Class 5 Transcription Factor 1 (Pouf51) or Oct4 (Nichols et al. 1998) and the homeobox transcription factor Nanog (Chambers et al. 2003; Mitsui et al. 2003). Depletion of Oct4 in both pre-implantation embryos and mESCs leads to trophectoderm lineage differentiation of the inner cell mass (Niwa et al. 2000). Overexpression of Nanog confers LIF-independent self-renewal of ES cells, whereas its knockdown results in parietal and visceral endoderm differentiation (Chambers et al. 2003; Mitsui et al. 2003).
Oct4 and Nanog are part of a core transcriptional regulatory network that is required for mESC gene expression regulation (Loh et al. 2006). Nanog is partly dependent on Oct4 by virtue of its participation in the well-characterized Oct4/Sox2/Nanog regulatory triumvirate (Rodda et al. 2005). Furthermore, induced expression of Oct4 (Takahashi and Yamanaka 2006) and Nanog (Yu et al. 2007) along with other key mESC-enriched regulatory proteins can promote an mESC-like phenotype in nominally lineage-restricted somatic cell populations. Continued study of Oct4- and Nanog-regulated genes is likely to provide further insight into the maintenance of mESC pluripotency.
Accordingly, our emerging focus area in mESC genomics concerns Oct4- and Nanog-regulated RNA transcripts that do not encode protein but may modulate mESC pluripotency or differentiation at the RNA level. Generally, RNAs are categorized into two distinct classes: messenger RNAs (mRNAs), which are translated into proteins, and the non-protein-coding RNAs (ncRNAs), which function directly as structural, catalytic, or regulatory RNAs without ever being translated into protein (Huttenhofer et al. 2005). Although considerable attention has been devoted to short regulatory RNAs in recent years (Sen and Roy 2007), there is another noteworthy class of potential regulatory RNAs in the transcriptome. We refer to this class as long ncRNAs (lncRNAs). Up to two orders of magnitude more numerous than miRNAs (Carninci and Hayashizaki 2007), these lncRNAs possess mRNA-like features, as they generally undergo splicing, are transcribed by RNA polymerase II, and are polyadenylated (Sone et al. 2007). However, they do not possess protein-coding open reading frames; the few reading frames that may be gleaned in some of these transcripts are generally not conserved between closely related species, and would encode very short peptides without any protein database hits. The small number of lncRNAs that have been characterized to date suggests that they have crucial diverse biological roles including those in early developmental and in postnatal, organ-specific contexts (Blackshaw et al. 2004; Sonkoly et al. 2005; Young et al. 2005; Feng et al. 2006; Ginger et al. 2006; Rinn et al. 2007; Sone et al. 2007), and that protein-coding potential is not necessary for fulfillment of these roles. A notable property of some mammalian lncRNAs is their ability to participate in regulatory ribonucleoprotein complexes and to exert a mechanistically heterogeneous repertoire of regulatory activities that include direct co-regulation (co-activation or co-repression) of key transcription factors in development and disease (Lanz et al. 1999; Willingham et al. 2005; Feng et al. 2006; Zhou et al. 2007). Still other mammalian lncRNAs regulate transcription factors indirectly, for example through chromatin remodeling (Rinn et al. 2007). These regulatory properties firmly establish those known lncRNAs as high-level control elements in gene regulatory networks. There is substantial evidence, for an increasing number of lncRNAs, that lncRNAs have diverse cellular functions; this evidence is not limited to differential expression (Blackshaw et al. 2004; Sonkoly et al. 2005; Young et al. 2005; Ginger et al. 2006; Sone et al. 2007; Johnson et al. 2009).
In this study, we asked whether conserved, Oct4- and Nanog-targeted lncRNAs have functional roles in mESCs. Starting from Oct4 and Nanog chromatin immunoprecipitation followed by paired-end tag sequencing (chromatin-immunoprecipitated [ChIP-PET]) experimental results in mESCs, we performed a genome-wide computational analysis of positional relationships between the genomic binding sites of the two transcription factors and GenBank cDNA and EST alignments to nearby genomic sequences. We thus inferred four evolutionarily conserved lncRNAs that are genomically encoded at loci that also contain Oct4 and Nanog binding sites, are expressed in mESCs, and exhibit specific expression responses to RA-induced mESC differentiation. Knockdown of two lncRNAs (RNCR2/AK028326/Gomafu/Miat and AK141205) altered Oct4 and/or Nanog transcript levels and modulated mESC differentiation toward specific lineages in the presence of LIF. Overexpression of both the lncRNAs, in separate experiments, enhanced mesodermal, endodermal, and ectodermal differentiation in the presence of LIF. Our data suggest that these conserved lncRNAs, which are directly controlled by two known mESC transcription factors, contribute to the regulation of mESC pluripotency and differentiation.
RESULTS
Identification of putative Oct4- and Nanog-targeted conserved lncRNAs
To identify ncRNAs modulated by pluripotency-associated transcription factors, we searched for candidate lncRNA genes throughout the entire catalog of genomic proximal target genes. Proximal targets were defined as genes that, based on their genomic position, mapped in close proximity to Oct4 and Nanog high-confidence binding sites that had been inferred by paired end-tag sequencing of ChIP-PET DNA (Loh et al. 2006). By manual annotation, we determined that 105 (10%) of the 1083 Oct4 binding sites had lncRNA genes as their proximal genomic targets (<10 kb genomic distance from a gene to the binding site) (see Materials and Methods), and, similarly, that 335 (11%) of the 3006 Nanog binding sites had lncRNA genes as their proximal targets. Hence, 10% of transcription factor binding sites in this study resided nearer to lncRNA genes than to any protein-coding genes. Intriguingly, these findings suggested that lncRNA genes represent a significant fraction of proximal transcription factor targets detectable in genome-wide studies of Oct4 and Nanog transcription factor binding sites discovered by ChIP-PET (Loh et al. 2006), and led us to investigate whether these lncRNAs impact mESC pluripotency.
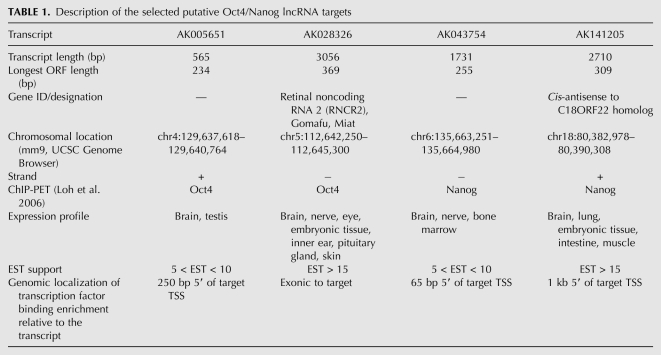
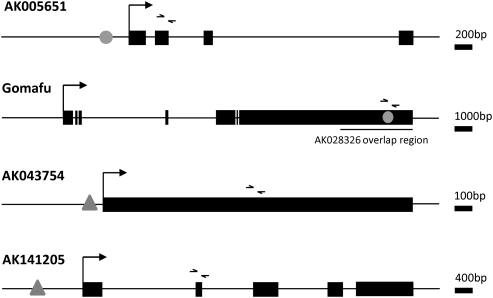
Of 105 putative Oct4 noncoding target genes, 29 possessed substantial interspecies conservation of their exonic sequence visible in the UCSC Genome Browser MultiZ track. One hundred forty-two of the 335 putative Nanog noncoding targets were similarly conserved. Because conservation generally indicates sequence evolution constraints due to the preservation of a function, we limited further analysis to four lncRNAs with strong genomic conservation of exonic sequence (Supplemental Fig. 2), substantial mammalian cDNA/EST support, and ChIP-PET evidence of Oct4 or Nanog binding specifically at the 5′ end of, or internal to, the lncRNA-encoding gene in mESCs (Table 1). Only two Oct4 and two Nanog targets fit these stringent criteria, namely AK005651, AK028326 (Gomafu; synonym: Miat), AK043754, and AK141205 (Fig. 1), whose genomic properties are described in Table 1. None of these four lncRNA-encoding loci matched any genomically encoded microRNAs in their exons or introns, according to the “MicroRNAs from miRBase” track of the UCSC Genome Database (mm9). Therefore, these lncRNAs are not likely to function as precursors of smaller RNAs.
TABLE 1.
Description of the selected putative Oct4/Nanog lncRNA targets
FIGURE 1.
Graphical representation of the lncRNA genomic loci and approximate sites of transcription factor interaction. (Black boxes) Exonic sequences, commencing at the transcriptional start site (right-angle arrow); (gray ovals) putative Oct4 binding sites supported by Oct4 ChIP-PET data; (gray triangles) putative Nanog binding sites supported by Nanog ChIP-PET data; (small arrowheads) forward and reverse primer binding sites for real-time PCR primers. Gene length is indicated by the scale bar provided.
Distinguishing coding from noncoding RNAs remains a nontrivial problem, although a combination of methods involving open reading frame (ORF) discovery, sequence homology (Dinger et al. 2008a), and the coding potential calculator (CPC), a support vector machine method (Kong et al. 2007), can address this problem to a significant extent. The functional annotation of the mammalian genome (FANTOM) mammalian transcriptome project established a 100-amino acid cutoff as the minimal ORF length for an RNA to be considered protein-coding, a distinction supported by multiple lines of evidence (Dinger et al. 2008a). We used the NCBI ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) to search for positive-strand ORFs of the four mouse lncRNAs under study, and found that only AK141205 had a positive-strand ORF longer than 100 amino acids. Pollard et al. (2006) suggest that excessive nonsynonymous substitutions in ORFs, along with differences in start/stop codon positions between closely related species (e.g., human/chimpanzee), are reasons to doubt the protein-coding capacity of an RNA. We performed BLAST (Altschul et al. 1990) and BLAT (Kent 2002) searches of AK141205’s 102-amino acid ORF at both the nucleotide and protein levels and did not detect protein homologies in any species, or nucleotide homologies (by BLAT) even in the closely related sequenced species relative to mouse, the rat. This lack of ORF conservation argues against any protein-coding capacity of mouse AK141205. CPC scores for the complete FASTA sequences of all four lncRNA candidates were negative, meaning that the RNAs were CPC-classified as noncoding. The sub-100-amino acid ORFs of three RNAs, AK141205 ORF nonconservation, and negative CPC scores together indicate that these four RNAs are not likely to function by encoding proteins.
Oct4 and Nanog lncRNA targets are differentially expressed during mESC differentiation
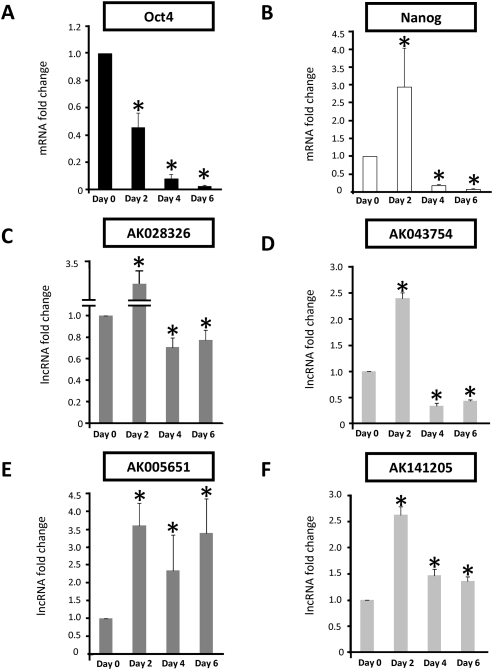
To test the four lncRNAs for a role in pluripotency, we assayed their response to RA-induced differentiation over a 6-d time course (Fig. 2). RA treatment promoted mESC differentiation, with concomitant down-regulation of Oct4 and Nanog mRNA levels as expected, showing that the differentiation stimulus was successful as pluripotency was robustly ablated. (Fig. 2A,B). Transcripts for all four lncRNAs were observed in undifferentiated mESCs, albeit at low levels (threshold cycle analysis showed that exponential quantitative PCR products derived from equal amounts of cDNA template were observed at 31–32 cycles on average). The transcription of all four changed rapidly in response to mESC differentiation. Relative to undifferentiated mESCs, Oct4 target AK028326 (Miat) and Nanog target AK043754 expression was significantly decreased (P < 0.05, N = 6 replicates) by day 6 (Fig. 2C,D). AK005651 transcription was induced by RA treatment but remained significantly elevated (P < 0.05, N = 6 replicates relative to undifferentiated mESCs) between days 2 and 6 (Fig. 2E). Induction of AK141205 by RA (Fig. 2F) was observed by day 2 and remained elevated (P < 0.05, N = 6 replicates relative to undifferentiated mESCs) at day 4 and day 6.
FIGURE 2.
Retinoic acid (RA) treatment simultaneously induces mESC differentiation and impacts lncRNA transcription. (A) Oct4 expression was highest in undifferentiated mESCs and rapidly decreased over the 6-d time course. (B) Nanog expression was initially elevated on RA treatment but was rapidly down-regulated by days 4 and 6. The expression of putative Oct4-regulated lncRNAs (C,E: dark gray bars) and putative Nanog-regulated lncRNAs (D,F: light gray bars) was examined in differentiating mESCs. Transcription of the lncRNAs AK028326 (C) and AK043754 (D) was elevated by day 2 and down-regulated relative to undifferentiated mESCs by days 4 and 6. In contrast, transcription of AK005651 (E) and, to a lesser extent, AK141205 (F) remained elevated relative to undifferentiated mESCs at all time points examined. (*) Significant difference relative to undifferentiated mESCs (P < 0.05, N = 6 replicates).
Oct4 and Nanog RNAi modulates lncRNA transcription
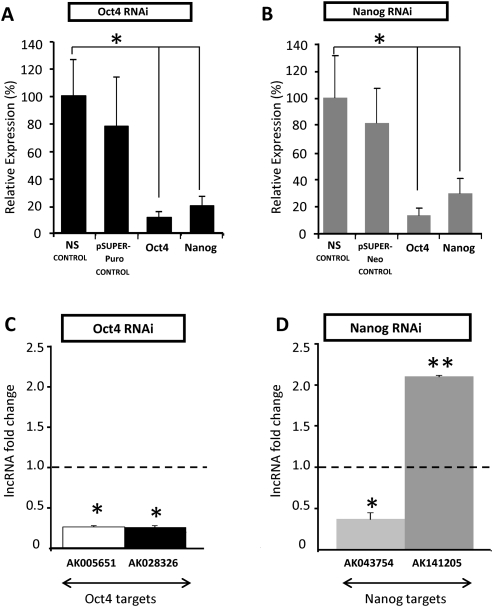
Oct4 and Nanog binding site localization near the transcription start sites of the lncRNA genes suggested direct regulation of the lncRNAs by Oct4 and Nanog. To test the hypothesis that expression levels of these lncRNAs are directly Oct4- and Nanog-dependent, as opposed to being merely responsive to RA treatment for some other reason, we evaluated whether RNAi-mediated down-regulation of Oct4 and Nanog in the absence of exogenous RA treatment could similarly alter lncRNA levels. ShRNA transfection directed against Oct4 and Nanog robustly reduced mRNA levels of both Oct4 and Nanog concurrently, 3 d post-transfection (Fig. 3A,B). In response to Oct4 RNAi (Fig. 3C), the putative Oct4 lncRNA targets AK005651 and AK028326 (Miat) were transcriptionally down-regulated (P < 0.05). Similarly, Nanog RNAi (Fig. 3D) induced down-regulation of the putative Nanog lncRNA target AK043754 (P < 0.05). In contrast, the putative Nanog lncRNA target AK141205 was significantly up-regulated (P < 0.01). These post-RNAi lncRNA expression level changes, given the Oct4 binding site at the AK028326 locus and the Nanog binding site at the AK043754 locus, are consistent with a role for endogenous Oct4 and Nanog in activating transcription of the lncRNAs AK028326 and AK043754, respectively. These data also suggest that Nanog may directly repress transcription of lncRNA AK141205, which would be consistent with the elevated transcription of that lncRNA in RA-treated mESCs (Fig. 2F). However, since reduced Oct4 and Nanog levels themselves induce differentiation, the post-RNAi lncRNA expression level changes may have also been caused indirectly, rather than solely through reduced Oct4 and Nanog binding to lncRNA promoters. Our observations on lncRNA expression mirror those on the expression of protein-coding genes directly regulated by these two transcription factors: In addition to activated targets of Oct4 and Nanog (Loh et al. 2006), there are well-characterized examples of transcriptional repression by these transcription factors, including the repression of Cdx2 by Oct4 (Hay et al. 2004) and the PRC-dependent Oct4-mediated suppression of other targets (Mikkelsen et al. 2008).
FIGURE 3.
Differential expression of lncRNA upon robust Oct4 and Nanog RNAi knockdown. (A) shRNA-directed knockdown resulted in greater than or ∼80% reduction of Oct4 mRNA and Nanog mRNA, respectively, compared with the non-silencing control (NS control) and the pSUPER-PURO vector-only control (pSUPER-PURO control), 3 d post-transfection. (B) shRNA-directed knockdown resulted in comparable levels of Nanog mRNA reduction and Oct4 mRNA compared with the NS control and the pSUPER-NEO vector-only control (pSUPER-NEO control), 3 d post-transfection. (C) Oct4 RNAi resulted in down-regulation of the two putative Oct4 targets, AK005651 and AK028326. (D) Nanog RNAi resulted in down-regulation of the putative Nanog target AK043754 and up-regulation of AK141205. (Asterisks) Significant difference from control samples (*, P < 0.05; **, P < 0.01) relative to the NS control (dotted black line).
We noted that the significant up-regulation of the putative Oct4 target AK005651 upon RA-induced differentiation between days 2 and 6 (which suggests an association of this lncRNA with differentiation) is inconsistent with its down-regulation upon Oct4 RNAi (which suggests that the same lncRNA is positively associated with Oct4 and by proxy with pluripotency). This discordance might imply that this lncRNA is located at a convergence of multiple regulatory pathways where its expression is determined by system inputs other than merely RA exposure and Oct4/Nanog levels. The reproducibility and consistent directionality of the expression response argue against a nonspecific variable response to cellular stressors inherent in the experiments. Therefore, AK005651 was eliminated from further study. Consequently, we chose to study AK141205, as it was the only lncRNA that was up-regulated during RA-induced differentiation and putatively repressed by Nanog. AK028326 (Gomafu/Miat) was selected as the other candidate lncRNA for functional studies, due to its down-regulation during differentiation and its putative activation by Oct4. We declined to further pursue AK043754, as it would have been a second lncRNA putatively activated by an ES transcription factor in our analysis.
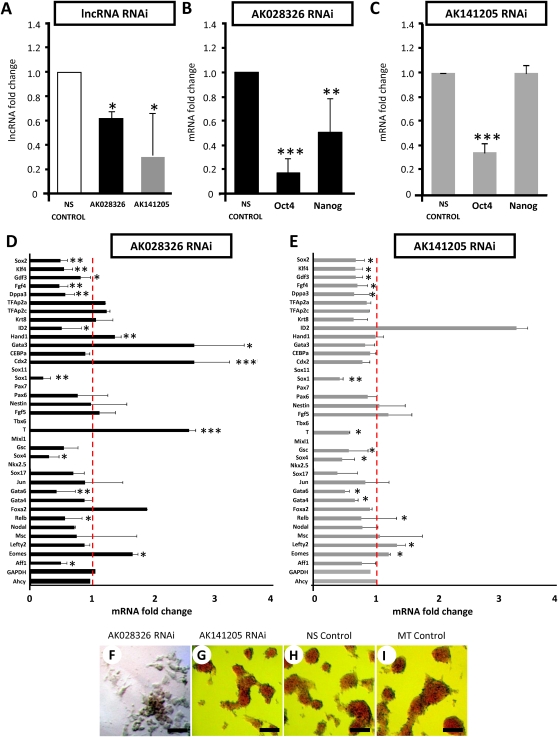
Directed knockdown of the lncRNA AK028326 (Gomafu/Miat) promotes loss of pluripotency and co-activates Oct4
To check whether lncRNA transcript level reduction was correlated with alteration of pluripotency and whether the lncRNAs might participate in a feedback loop affecting the levels of their own regulators, we examined whether RNAi against the lncRNAs AK028326 (Gomafu/Miat) and AK141205 could change Oct4 and Nanog mRNA levels in self-renewing mESCs. Three sequential rounds of siRNA transfection significantly depleted endogenous levels of AK028326 and AK141205 relative to nonsilencing control siRNA-transfected mESCs (Fig. 4A), confirming that these lncRNAs are susceptible to Dicer-mediated suppression. RNAi of AK028326 (Fig. 4B) resulted in reduced mRNA levels of Oct4 and Nanog. In conjunction with Oct4-mediated activation of AK028326 (Fig. 3C,D), the result suggests the potential for an autofeedback loop in which AK028326 in turn maintains or increases Oct4 mRNA levels; this hypothetical AK028326-dependent stabilization or up-regulation of Oct4 would be abolished upon AK028326 RNAi, as observed. The lesser significance of the effect of AK028326 RNAi on Nanog is consistent with the fact that AK028326 is a genomic Oct4 target but not a genomic Nanog target.
FIGURE 4.
Expression of lncRNAs, pluripotent markers, trophectoderm markers, and epiblast lineage markers in response to lncRNA silencing (RNAi). (A) AK028326 and AK141205 are significantly down-regulated in response to siRNA treatment. (B) AK028326 RNAi was associated with significant down-regulation of both Oct4 and Nanog. (C) AK141205 RNAi was associated with significant down-regulation of Oct4, but Nanog levels remained unchanged. (D) In AK028326 siRNA-treated mESCs, there was a significant enhancement of the mesodermal marker T/Brachyury and the trophectoderm markers Cdx2, Hand1, Gata3, and Eomes; down-regulation of pluripotent markers; and unchanged levels of the housekeeping genes Gapdh and Ahcy. (E) In AK141205 siRNA-treated mESCs, there was general down-regulation of the pluripotent and epiblast lineage markers, up-regulation of the Oct4-repressed marker Id2, and unchanged levels of Gapdh and Ahcy. (F–I) Alkaline phosphatase staining of mESCs treated with siRNAs against AK028326 (F), AK141205 (G), NS control (H), or mock transfection control (MT control) (I). Scale bar, 100 μm. (Asterisks) Significant difference from control samples (*, P < 0.05; **, P < 0.01; ***, P < 0.001) relative to NS control (dotted red line).
In addition to Oct4 and Nanog, we evaluated the mRNA levels of other well-described pluripotent markers positively regulated by Oct4: Sox2, Klf4, Gdf3, Fgf4, and Dppa3/Stella. All five markers were down-regulated when AK028326 (Gomafu/Miat) was suppressed by RNAi (Fig. 4D), further supporting the possibility that AK028326 is a pluripotency-promoting direct target of Oct4 as well as a co-activator of Oct4 in a potential synergistic feedback mechanism.
In light of the initial evidence that lncRNA RNAi can reduce Oct4 and Nanog levels (Fig. 4B,C), we examined mRNA levels of definitive lineage-associated genes to determine whether lncRNA RNAi also promoted mESC differentiation (Fig. 4D). The significant reduction of Oct4 mRNA by AK028326 RNAi resulted to elevated levels of the mesodermal marker T/Brachyury (P < 0.001 relative to nonsilencing control-transfected cells, N = 6 replicates). AK028326 RNAi also reduced the mRNA level of Sox1 (P < 0.001 relative to nonsilencing control-transfected cells, N = 6 replicates), a neural-specific transcription factor. However, other endodermal (Gata6) and mesodermal (MyoD) markers were down-regulated (P < 0.01 relative to nonsilencing control-transfected cells, N = 6 replicates). Nevertheless, the partial differentiation observed upon AK028326 RNAi is broadly consistent with the paradigm that AK028326, as an Oct4-activated gene, may have a pluripotency-promoting function.
In order to examine if Oct4 mRNA reduction as a consequence of AK028326 (Gomafu/Miat) RNAi was concordant with differentiation toward the trophoblast lineage, we assayed AK028326-knockdown cells for trophoblast markers (Fig. 4D) and found significant upregulation of Cdx2 (Ralston and Rossant 2008) (P < 0.001), Hand1 (P < 0.01), Eomes (Russ et al. 2000) (P < 0.05), and Gata3 (P < 0.05) (Fig. 4D). Since Cdx2 is endogenously Oct4-repressed, its activation upon AK028326 RNAi is consistent with AK028326-dependent positive regulation of Oct4, and with Cdx2 up-regulation upon knockdown of Oct4 itself (Niwa et al. 2000). Therefore, expression level changes expected upon Oct4 reduction are actually occurring as a result of lncRNA-dependent decrease of Oct4 mRNA levels. This may suggest a potential role of AK028326 in inhibiting differentiation along the trophoblast lineage.
Directed knockdown of the lncRNA AK141205 promotes loss of pluripotency and indirectly modulates Oct4
RNAi against AK141205 (Fig. 4C) also resulted in significantly lower levels of Oct4. However, the Nanog mRNA level was unchanged. Considering the Nanog binding site at the AK141205 transcriptional start region, this may indicate that AK141205 helps Nanog to modulate Oct4 activity. This also suggests that AK141205 and Nanog do not form an autofeedback loop.
In contrast to AK028326 RNAi, AK141205 RNAi (Fig. 4E) did not significantly elevate mRNA levels of differentiation-associated definitive lineage markers, and also resulted in general down-regulation of meso- and endodermal markers, in addition to Sox1 (P < 0.01). Expression of the housekeeping genes Gapdh and Ahcy remained unchanged. Consistent with the down-regulation of Oct4 upon AK141205 RNAi, Id2, a gene negatively regulated by Oct4 (Loh et al. 2006), was strongly up-regulated as a result of AK141205 RNAi.
In summary, we have established that RNAi directed against these lncRNAs significantly altered the mRNA level of Oct4, a key transcription factor that binds the genomic locus encoding one of the lncRNAs and is necessary for mESC pluripotency. We have also shown that RNAi of these lncRNAs leads to highly specific effects on downstream lineage-specific differentiation factors.
Directed knockdown of the lncRNAs AK028326 (Miat/Gomafu) and AK141205 affects cell proliferation and morphology
Loss of AK028326 had stronger effects on DNA synthesis as a proxy for cell proliferation, which inversely correlates with differentiation, than did loss of AK141205. AK028326-RNAi mESCs incorporated markedly less BrdU (Supplemental Fig. 3A), whereas BrdU incorporation by AK141205-RNAi cells was not significantly different from that of controls. No significant difference in cell apoptosis as examined by Annexin-V and Propidium Iodide (PI) flow cytometry was observed between control-transfected and either lncRNA RNAi-transfected culture, either with or without LIF (data not shown).
Analysis of alkaline phosphatase (AP) activity, a marker of the pluripotent cell, and cell morphology showed that AK028326-RNAi transfected mESCs were morphologically more differentiated, with flat squamous cells surrounding residual dark red colonies in the presence of LIF (Fig. 4F). In contrast, AK141205-RNAi transfected mESCs (Fig. 4G) and control-transfected mESCs (Fig. 4H,I) exhibited morphologically similar, homogenously AP-positive cell colonies. Thus, loss of both AK141205 and AK028326 reduced Oct4 mRNA, but only loss of AK028326 reduced both cell proliferation and morphological differentiation. This post-AK028326-RNAi phenotype is consistent with a potential role of AK028326 as a pluripotency-maintaining direct target of Oct4.
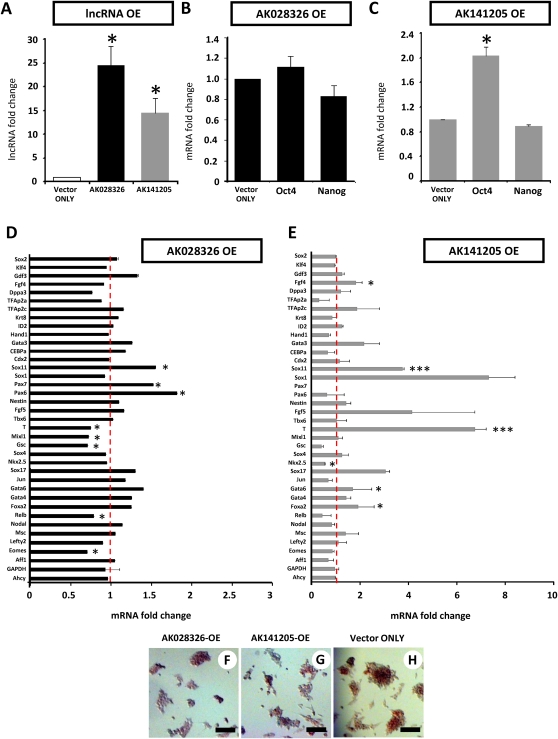
Overexpression of the lncRNA gene AK028326 (Gomafu/Miat) promotes mESC lineage-specific differentiation
We next asked whether overexpression of AK028326 could modulate pluripotency-associated transcript levels or mESC differentiation. LncRNA AK028326 was cloned into the pcDNA3.1 (+) vector (Invitrogen) downstream from the CMV promoter and upstream of the Bgh (A) polyadenylation sequence, to ensure expression of just the lncRNA, without any lengthy or ORF-containing vector sequences. Overexpression (OE) of AK028326 (Fig. 5A), a fragment of the larger (∼9.0-kb) Gomafu/Miat lncRNA transcript (Sone et al. 2007) marginally altered Oct4 mRNA level (Fig. 5B). In light of Oct4 binding at the AK028326 locus and of the RNAi-based evidence for Oct4/AK028326 mutual positive regulation, the OE result may imply that an increase in AK028326 RNA level beyond a certain threshold does not result in elevating Oct4 mRNA level, or that the Oct4/AK028326 autofeedback loop is self-dampening in a manner that precludes escalation of endogenous AK028326 RNA levels. However, lineage-specific marker gene analysis was suggestive of enhanced differentiation upon AK028326-OE (Fig. 5D), as evidenced by increased transcription of the ectodermal markers Pax6, Pax7, and Sox11 (P < 0.001 relative to vector only-control, N = 6 replicates). Mesodermal marker T/Brachyury, as well as associated primitive streak genes such as Mixl1 (Ng et al. 2005) and Gsc, were down-regulated upon AK028326-OE (P < 0.001 relative to vector only-control, N = 6 replicates). These data indicated that overexpression of the AK028326 lncRNA, despite the fact that it may comprise merely a partial fragment of the Gomafu/Miat transcriptional unit, was sufficient to alter cell differentiation and promote acquisition of ectodermal lineage transcription. Sox2 and Gdf3, pluripotent markers positively regulated by Oct4, were mildly up-regulated in AK028326-OE, consistent with the idea of an Oct4/AK028326 pluripotency-associated synergy.
FIGURE 5.
Expression patterns of lncRNAs, pluripotent markers, trophectoderm markers, and epiblast lineage markers in response to lncRNA overexpression (OE). (A) LncRNAs AK028326 and AK141205 are significantly overexpressed relative to the vector-only control. (B) AK028326 overexpression marginally altered Oct4 and Nanog transcription. (C) AK141205 overexpression significantly up-regulated Oct4 but not Nanog transcription relative to vector-only control. (D) In response to AK028326 overexpression, there was significant enhancement of mesodermal marker T/Brachyury and Sox4, the epiblast marker Fgf5, the ectodermal markers Sox11, Pax6, and Pax7, in addition to unchanged levels of the housekeeping genes Gapdh and Ahcy. (E) In response to AK141205 overexpression, there was significant enhancement of mesodermal marker T/Brachyury, the epiblast marker Fgf5, the ectodermal markers Sox1 and Sox11, and endoderm markers, in addition to unchanged levels of Gapdh and Ahcy. (F–H) AP staining of AK028326-OE (F), AK141205-OE (G), and vector-only control (H). Scale bar, 100 μm. (Asterisks) Significant difference from control samples (*, P < 0.05; ***, P < 0.001) relative to the pCAG control (dotted red line).
Overexpression of the lncRNA gene AK141205 also promotes mESC lineage-specific differentiation
We then asked whether overexpression of AK141205 could modulate pluripotency-associated transcript levels or mESC differentiation. The lncRNA AK141205, which possessed a bona fide internal consensus AATAAA polyadenylation signal sequence immediately upstream of its 3′ end, was cloned into the pCAG-IRES-EGFP vector downstream from the CAG promoter and upstream of the IRES-EGFP. By 3′ RACE experiments (data not shown), we demonstrated the absence of the downstream IRES-EGFP vector sequence in the overexpressed transcripts, and expression of just the lncRNA with termination shortly after the endogenous poly(A) signal. Mirroring, and reciprocally to, the AK141205 RNAi outcome, overexpressed AK141205 RNA levels (Fig. 5A) saw significant elevations of endogenous levels of Oct4 mRNA, whereas Nanog mRNA levels were not significantly perturbed (Fig. 5C).
Similarly to AK028326-OE, in AK141205-OE mESCs (Fig. 5E), elevated levels of differentiation lineage-associated transcripts were observed including the mesodermal marker T/Brachyury, columnar epiblast marker Fgf5, and ectodermal markers Sox1 and Sox11 (P < 0.001 relative to vector-only control, N = 6 replicates), suggestive of enhanced meso- and ectodermal differentiation in self-renewing mESCs. Mixl1 and Gsc were down-regulated and so were most other definitive lineage markers such as Aff1, Relb, Eomes, and Nkx2.5 (P < 0.05 relative to vector-only control, N = 6 replicates). The significant up-regulation of Oct4 mRNA upon AK141205 overexpression was also correlated with the up-regulation of endoderm markers, Sox17 (P < 0.001 relative to vector-only control, N = 6 replicates), Gata6, and Foxa2 (P < 0.05 relative to vector-only control, N = 6 replicates). Only one pluripotency marker was up-regulated (Fgf4 [P < 0.05]), while others remained unchanged (Klf4 and Sox2). Overall, AK141205 OE results are consistent with the idea that AK141205 is a Nanog-repressed, differentiation-promoting lncRNA, since they indicate up-regulation of specific, but not all, classes of differentiation markers as a consequence of the OE. Trophoblast markers were not significantly up-regulated. The housekeeping genes Gadph and Ahcy remained unchanged in both AK028326 and AK141205-OE (Fig. 5D,E).
To evaluate whether cell proliferation, apoptosis, or morphology were altered in lncRNA OE, cultures were trypsinized and plated in mESC media in the presence of LIF. Consistent with transcriptional evidence of enhanced cell differentiation, AK141205-OE mESCs exhibited reduced BrdU incorporation in response to LIF (data not shown). Similarly to lncRNA-RNAi cultures, apoptosis post-lncRNA-OE was not significantly different from control-transfected cultures, either in the presence or absence of LIF (data not shown). Therefore, apoptosis-inducing major cellular stress, a potential nonspecific cause of the diverse marker expression profile changes we observed, was not a contributing factor to the experiments.
Both AK028326-OE mESCs (Fig. 5F) and (to a lesser extent) AK141205-OE (Fig. 5G) exhibited flattened, smaller colonies with reduced AP staining relative to pCAG control cultures (Fig. 5H), suggestive of enhanced cell differentiation. These data further indicate that overexpression of lncRNA transcripts may significantly perturb cell proliferation.
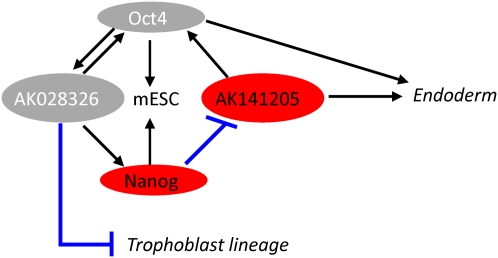
Together, our findings suggest the existence of a simple dampening autofeedback regulatory loop in the case of AK028326, and a Nanog-mediated effect on Oct4 through the known Oct4/Nanog transcription factor network in the case of AK141205. Consequently, we have formulated a model that incorporates the two lncRNAs into the known Oct4/Nanog regulatory network, both as signaling intermediaries between Oct4 and Nanog and as determinants of specific lineage decisions (Fig. 6).
FIGURE 6.
A model for AK028326 and AK141205 lncRNA placement into the Oct4 and Nanog regulatory network. (Black lines) Positive regulation; (blue lines) repression. Oct4 and Nanog are required for self-renewal, and Oct4 activates AK028326, while Nanog represses AK141205. Reduction of AK028326 exhibited a correlated reduction in Oct4 mRNA levels with a concordant up-regulation of Trophoblast lineage transcripts, though the mechanism is unclear. Overexpression of AK141205 led to up-regulation of Oct4 with a corresponding up-regulation of endodermal markers.
DISCUSSION
LncRNAs are abundantly encoded in mammalian genomes, numbering in the tens of thousands, in contrast to microRNA-encoding genes, which are an order of magnitude less numerous. However, functionalizing the rich repertoire of long non-protein-coding transcripts remains a challenge. Recently, the role of lncRNAs in pluripotency was examined in another study by interpreting the genomic context of the ncRNAs relative to nearby protein-coding genes and expression upon embryoid body (EB) differentiation. Out of the 945 ncRNAs expressed during EB differentiation, 174 were differentially expressed, many correlating with pluripotency and cell fate decisions (Dinger et al. 2008b). AK028326 (synonyms: Gomafu, Miat) was among the 174 differentially expressed lncRNAs, but it was not singled out for in-depth regulatory or functional analysis in that study. The other three lncRNAs that we considered were not mentioned there. In contrast to our work, Dinger et al. (2008b) did not enterprise to integrate ChIP-PET and lncRNA expression data to derive conclusions about direct regulation of lncRNAs by specific transcription factors. Their timely observations, nevertheless, emphasize the potentially widespread functional inputs of numerous lncRNAs toward mESC pluripotency and differentiation.
Our approach is unique in that the four candidate lncRNA genes were selected based on their proximity to experimentally (ChIP) supported Oct4 and Nanog binding sites. Furthermore, the Oct4 and Nanog binding sites of Gomafu/Miat and AK141205, respectively, were supported by five and seven, respectively, distinct overlapping PET sequences, a high extent of support that is generally consistent with successful chIP-qRTPCR validation of the binding sites inferred by chIP-PET (Loh et al. 2006, see their Supplemental Table 1 and Supplemental Figs. 1, 3, and 4). This integration of experimental evidence for transcription factor binding with lncRNA annotation at the bound loci contrasts with other studies to date, which generally either consider lncRNA expression in isolation from the transcriptional control thereof, or rely on computational predictions of transcription factor binding. The interspecies conservation of the four lncRNAs was suggestive of functional importance, despite their evident lack of protein-coding capacity. Our study has integrated sequence analysis, expression data for three classes of genes (transcription factors, their lncRNA direct targets, and panels of cellular lineage markers), ChIP transcription factor binding data, and system perturbations (RNAi and overexpression) to show that four conserved lncRNAs respond to an ES differentiation stimulus and are under direct Oct4/Nanog control, based both on ChIP-PET (Oct4 and Nanog genomic recruitment) data and Oct4/Nanog RNAi. It would be of interest to further evaluate how lncRNAs might integrate into the core transcriptional regulatory networks of mESCs. We propose that two of these lncRNAs are capable of altering the transcription of two key transcription factors, Oct4 and Nanog, and therefore directly contribute to differentiation and dedifferentiation of perturbed mESCs.
LncRNA AK028326 has been previously described as retinal noncoding RNA 2 (RNCR2), strongly expressed in the developing retina (Blackshaw et al. 2004). Subsequent studies have demonstrated that RNCR2/AK028326 is a 3′ terminal fragment of a ∼9-kb lncRNA, Gomafu/Miat, widely expressed in central nervous system neurons (Sone et al. 2007). Our study provides the first evidence that RNCR2/AK028326 is also expressed in mESCs and may be regulated by ES-associated transcription factors, expanding our understanding of this particular lncRNA by implicating it in regulatory networks outside of the central nervous system.
In our analysis of the AK028326 locus, we relied on reagents based solely on the AK028326 cDNA sequence. Our original annotations described AK028326 as the nearest target to the Oct4 ChIP-PET-supported binding site in the region; in addition, during the early stages of our work, EST evidence was lacking to conclusively link AK028326 to the Gomafu/Miat transcriptional unit (Sone et al. 2007). Both AK028326 and the Gomafu/Miat cDNAs were derived from a 5′ cap-trapped, dT-primed cDNA library, suggesting that multiple transcriptional initiation and termination events, generating a variety of mature transcripts, including possibly the original AK028326 isoform, may take place at this locus. Although our results explore the synergy of Oct4 and AK028326 expression, they portray an apparently contradictory increase in mRNA levels of several differentiation markers when AK028326 is overexpressed. While this particular finding may be due to overexpression far beyond maximum endogenous levels that saturates the regulatory network and leads to unanticipated cell differentiation triggers, our results also do not exclude functional outcomes associated with the longer transcripts from this locus, whose expression we did not investigate.
Of potential relevance to our AK028326 work is the finding that MIAT, an lncRNA encoded at the human chromosome 22 locus orthologous to AK028326/Gomafu, is genetically associated with heart disease (Ishii et al. 2006) and, similarly to AK028326/Gomafu, exhibits multiple alternative transcription initiation and termination sites, which are supported by cDNA evidence and are accompanied in EST data by different expression specificities of the different transcripts encoded at the locus. It is therefore remarkable that RNAi and overexpression of even a partial fragment of this particular lncRNA are sufficient to promote mESC differentiation under self-renewing conditions, and to promote meso- and ectodermal gene transcription, respectively.
We provide evidence that AK141205, a novel and potentially Nanog-repressed lncRNA, positively regulates Oct4. LncRNAs may prove to be important intermediaries in the Oct4/Sox2/Nanog network. The observation that AK141205 is repressed by Nanog contrasts with the fact that the vast majority of Nanog targets are activated by Nanog (Loh et al. 2006). When overexpressed, AK141205 led to an increase in Oct4 mRNA and to a corresponding up-regulation of endodermal markers, in addition to initiating meso- and ectodermal differentiation. AK141205 is a member of a genomically encoded sense–antisense gene pair, and its 3′ terminal exon overlaps the terminal exons of the mouse homolog of the human C18ORF22 gene that is encoded by the opposite strand of the genome. Although cis-antisense transcription of lncRNAs may regulate their partner protein-coding genes (Carninci and Hayashizaki 2007), C18ORF22 is homologous to mitochondrial ribosome-binding factor A and hence seems to lack a stem-cell-specific function. Although all four lncRNAs contain sequence segments predicted by multiple methods (Gruber et al. 2008) to encode intramolecular helices punctuated by hairpin loops (Supplemental Fig. 4), the detection of such RNA secondary structures by structure prediction software is not unique to lncRNAs and occurs in mRNAs and miRNA precursors as well, hindering the interpretation of these structure results specifically from an lncRNA function viewpoint. The mechanisms by which AK028326 and AK141205 exert their impact remain unknown but worthy of future studies.
Given the recent evidence that lncRNAs such as NRON, Evf-2, and MEG3 may directly co-regulate transcription factors, testing for direct interactions between Oct4, Nanog, and these two lncRNAs in the nucleus is a promising potential avenue for future investigation, especially given that nuclear localization of several other functional lncRNAs, including a regulatory lncRNA that forms a ribonucleoprotein complex with transcription factors in vivo (Bond et al. 2009), has been highlighted in recent studies (Hutchinson et al. 2007; Clemson et al. 2009). In view of these precedents, testing for interactions of the lncRNAs Gomafu (Miat) and AK141205 with the Oct4 and Nanog proteins may be promising and can be accomplished by RIP-ChIP experiments in which the mRNA and lncRNA populations of these proteins are identified through hybridization to a custom array representing lncRNAs, or by placing lncRNAs on affinity columns to trap their interacting proteins.
A number of well-studied ncRNAs are poorly conserved and yet have important regulatory roles (Avner and Heard 2001). Given this, the extensive conservation of the four lncRNAs in our study is intriguing and only helps bolster our argument that these genes are functional. In fact, discovery of the nucleocytoplasmic transport function of the lncRNA NRON was made possible by prioritizing lncRNAs from a large pool of candidates on the basis of interspecies conservation (Willingham et al. 2005). Our findings should help elucidate mechanisms by which lncRNAs can modulate pluripotency. Our findings should stimulate further research in functional genomics of both conserved and nonconserved ncRNAs as targets of known transcription factors from the increasing number of genome-scale ChIP-sequencing experiments. Such future studies of mESC transcription factors and their lncRNA targets should expand our grasp of stem cell genomic regulatory networks and potentially facilitate the derivation of rational therapeutic interventions exploiting the regulatory potential of mESC lncRNAs.
MATERIALS AND METHODS
Computational evaluation of lncRNAs
To identify ncRNAs modulated by pluripotency-associated transcription factors, we evaluated all genomic proximal target genes of Oct4 and Nanog. High-confidence genomic Oct4 and Nanog binding sites were inferred by paired end-tag sequencing of chromatin-immunoprecipitated DNA. High confidence was defined as a minimum of three unique ChIP-PET sequences sharing overlaps (“moPET3+”) (Loh et al. 2006). Starting from Supplemental Table 1 of that publication, we defined proximal target genes as genes containing binding sites of either of the two mESC transcription factors in their 5′ proximal (<10 kb upstream of the transcription start site) and intragenic regions (Loh et al. 2006). Within this subset of the Nanog and Oct4 target genes, we searched for candidate lncRNA genes, which we operationally defined as having longest positive-strand ORFs of <100 amino acids devoid of both BLASTP high-complexity and Conserved Domain Database (CDD) homologs. We then focused on Oct4 and Nanog lncRNA proximal targets exhibiting expressed sequence tag (EST) and/or cDNA support, as well as evidence of genomic (MultiZ), including transcribed (xenoRNA), multispecies sequence conservation in the UCSC Genome Browser (Kent et al. 2002).
Cell culture
All cell culture reagents were supplied by Invitrogen unless otherwise specificed. Mouse E14 (American Type Culture Collection: CRL-1821) mESCs were cultured in 5% CO2/O2 in feeder-free mESC Medium, Dulbecco's modified Eagle's medium (DMEM) supplemented with 15% heat-inactivated ES-standard fetal bovine serum, 100 μM nonessential amino acids, 2 mM L-glutamine, 55 nM β-mercaptoethanol, 100 U/mL penicillin/100μg/mL streptomycin, and mouse Leukemia Inhibitory Factor (mLIF; 103 U/mL, Chemicon) on 0.1% gelatin-coated plates (Nunc GmbH). For differentiation, mESCs were replated at medium density (0.2 × 106 cells/well) to 0.1% gelatin-coated 12-well plates in mESC media for 12–16 h, washed once in Dulbecco's phosphate buffered saline (DPBS), and replaced with mESC medium without LIF and supplemented with all-trans RA (RA, 100 nM; Sigma-Aldrich).
RNA interference (RNAi)
Short-hairpin RNA interference (shRNA)
Short-hairpin RNA interference (shRNA)-encoding constructs directed against Oct4 and Nanog were cloned into the RNA pol-III driven pSUPER-PURO and pSUPER-NEO-EGFP vectors, as previously described (Loh et al. 2006). A scrambled nonsilencing short-hairpin RNA-encoding construct (pSUPER-NEO-EGFP) and the original vectors (pSUPER-PURO, p-SUPER-NEO) were used as positive controls. Transfection of constructs (2.0 μg/well, diluted in Opti-MEM-I reduced serum medium; Invitrogen) was performed using Lipofectamine 2000 (Invitrogen) in serum-free mESC medium followed by replacement of serum post-transfection. Twenty-four hours post-transfection, mESC media was replaced and supplemented with selection antibiotics (Puromycin, 500 ng/mL; Neomycin [G418], 300 ng/mL). mESC media with selection was continued for 3 d with media changes every day before RNA isolation.
Small interfering RNAi (siRNA) experiments
Dharmacon On-Target SMARTpool double-stranded oligonucleotides were directed against mouse AK028326 and AK141205 (Dharmacon). Dharmacon si-CONTROL nontargeting siRNA pool and mock-transfected (MT) mESCs were used as negative controls. mESCs were transfected with the Dharmafect-2 Reagent according to the manufacturer's protocol. mESCs were seeded to 12-well plates at a density of 0.2 × 106 cells/well and transfected in serum-free mESC medium. Following transfection, 100% of the medium was replaced with mESC medium and the mESCs were fed daily. Passaging and re-transfection of siRNAs was performed at days 2 and 4 after the initial siRNA transfection as previously described. RNA was extracted from mESCs 6 d after initial plating (5 d post-transfection).
LncRNA overexpression experiments
AK028326
Mouse cDNA encoding AK028326 was obtained from I.M.A.G.E (Invitrogen). Primers that incorporated 5′ BamHI and 3′ NotI restriction sites were used to amplify the AK028326 transcript. To make overexpression (OE) constructs, PCR products were digested with BamHI and NotI (New England Biolabs), purified, and subcloned into the RNA pol-II driven pCDNA3.1 (+) vector immediately after the CMV promoter and upstream of the Bgh polyadenylation site (Invitrogen).
AK141205
RIKEN mouse cDNA encoding AK141205 was obtained from K. K. DNAFORM. Primers that incorporated 5′ SalI and 3′ XmaI restriction sites were used to amplify the AK141205 transcript. To make overexpression (OE) constructs, PCR products were digested with SalI and XmaI (New England Biolabs), purified, and subcloned into the RNA pol-II driven pCAG-IRES-EGFP-NEO vector immediately downstream the CAG promoter and upstream of the IRES-EGFP (Clontech).
Primer sequences are provided in Supplemental Table 1. OE constructs were sequence-characterized prior to transfection. AK028326-OE and AK141205-OE constructs were transfected into mESCs and Neomycin-selected for 3 d without passaging as previously described (Zhang et al. 2006).
RNA extraction, reverse transcription, and quantitative real-time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen), purified with the RNeasy Minikit (Qiagen), and DNaseI treated (Ambion). Oligo-d(T)-primed reverse transcription total RNA was performed using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions. To examine mRNA levels of lncRNAs, customized FAM-TAMRA probes were designed against the lncRNAs AK005651, AK028326, AK043754, and AK142105. Primer–probe information is provided in Supplemental Table 2. PCR was performed using mESC E14 RNA, and a “no reverse transcriptase” control (no RT control) was included to preclude the possibility of genomic contamination. Confirmatory agarose gels were run for the amplicons generated by the Taqman primer–probe sets (Supplemental Fig. 1).
Taqman Gene Expression Assays (Applied Biosystems) were used to evaluate pluripotent, trophectoderm, and epiblast lineage marker gene expression (Supplemental Table 3). Quantitative real-time PCR was performed using Taqman Universal PCR Master Mix (Applied Biosystems) in 384-well optical plates on the ABI 7900HT FAST Real-time PCR System (Applied Biosystems). Amplicons were analyzed using the Sequence Detection System 2.2 software (Applied Biosystems).
BrdU analysis
Cell proliferation was evaluated following a 12-h pulse of 5-Bromo-2-Deoxyuridine (BrdU). Cells were trypsinized, and BrdU incorporation was evaluated by ELISA according to the manufacturer's instructions (BrdU cell proliferation assay kit; Millipore).
Alkaline phosphatase (AP) detection
X-Phos/NBT detection of AP activity in mESCs was performed as previously described (Fields-Berry et al. 1992). mESCs were briefly fixed in 4% paraformaldehyde v/v DPBS, washed, and incubated for 30 min at 65°C. DPBS was replaced by room temperature detection buffer (100 mM Tris-HCl, 100 mM NaCl, 50 mM MgCl2) for 15–30 min before application of reaction buffer (0.1 mg/mL 5-bromo-4-chloro-3-indolyl-phosphate [BCIP] and 1 mg/mL nitro blue tetrazolium [NBT] v/v detection buffer, 0.5 mM Levamisole hydrochloride) and incubation for 48 h at 4°C. mESCs were rinsed in DPBS, and representative cells were photographed using a Lumar V12 inverted microscope (Carl Zeiss).
Statistical analysis
Two independent biological and three technical replicates for each experiment were conducted in parallel unless otherwise stated. The statistical significance (P-values) in mean values of two-sample comparison was determined with unpaired Student's t-test.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Dr. David Rodda, Dr. Rory Johnson, Dr. Andrew Hutchins, Dr. Leah Vardy, Mr. Galih Kunarso, Dr. Manjiri Bakre, Dr. Ralf Jauch, and Dr. Michael Rossbach for assistance with protocol development, as well as for fruitful discussions and critical comments of the manuscript. We are indebted to Nurul Ain binte Hashim and Kennt Boey for their genome-wide manual annotation to Nanog ChIP-PET targets. We thank Edwin Lian-Chong Ng and Rick Smith for technical assistance. We thank A*STAR (Agency of Science, Technology, and Research; Republic of Singapore) for GIS (Genome Institute of Singapore) competitive funding (intramural grant, budget number GIS/06-114101-OOE, to L.L.), which made this project possible.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1441510.
REFERENCES
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Avner P, Heard E. X-chromosome inactivation: Counting, choice, and initiation. Nat Rev Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- Bibel M, Richter J, Schrenk K, Tucker KL, Staiger V, Korte M, Goetz M, Barde YA. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat Neurosci. 2004;7:1003–1009. doi: 10.1038/nn1301. [DOI] [PubMed] [Google Scholar]
- Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AM, Vangompel MJ, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Hayashizaki Y. Noncoding RNA transcription beyond annotated genes. Curr Opin Genet Dev. 2007;17:139–144. doi: 10.1016/j.gde.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput Biol. 2008a;4:e1000176. doi: 10.1371/journal.pcbi.1000176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008b;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes & Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields-Berry SC, Halliday AL, Cepko CL. A recombinant retrovirus encoding alkaline phosphatase confirms clonal boundary assignment in lineage analysis of murine retina. Proc Natl Acad Sci. 1992;89:693–697. doi: 10.1073/pnas.89.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginger MR, Shore AN, Contreras A, Rijnkels M, Miller J, Gonzalez-Rimbau MF, Rosen JM. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci. 2006;103:5781–5786. doi: 10.1073/pnas.0600745103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay DC, Sutherland L, Clark J, Burdon T. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells. 2004;22:225–235. doi: 10.1634/stemcells.22-2-225. [DOI] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-1-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhofer A, Schattner P, Polacek N. Noncoding RNAs: Hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al. Identification of a novel noncoding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet. 2006;51:1087–1099. doi: 10.1007/s10038-006-0070-9. [DOI] [PubMed] [Google Scholar]
- Johnson R, Teh CH, Jia H, Vanisri RR, Pandey T, Lu ZH, Buckley NJ, Stanton LW, Lipovich L. Regulation of neural macroRNAs by the transcriptional repressor REST. RNA. 2009;15:85–96. doi: 10.1261/rna.1127009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller GM. In vitro differentiation of embryonic stem cells. Curr Opin Cell Biol. 1995;7:862–869. doi: 10.1016/0955-0674(95)80071-9. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O'Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Ng ES, Azzola L, Sourris K, Robb L, Stanley EG, Elefanty AG. The primitive streak gene Mixl1 is required for efficient haematopoiesis and BMP4-induced ventral mesoderm patterning in differentiating ES cells. Development. 2005;132:873–884. doi: 10.1242/dev.01657. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Ralston A, Rossant J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev Biol. 2008;313:614–629. doi: 10.1016/j.ydbio.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- Sen CK, Roy S. miRNA: Licensed to kill the messenger. DNA Cell Biol. 2007;26:193–194. doi: 10.1089/dna.2006.0567. [DOI] [PubMed] [Google Scholar]
- Smith AG. Embryo-derived stem cells: Of mice and men. Annu Rev Cell Dev Biol. 2001;17:435–462. doi: 10.1146/annurev.cellbio.17.1.435. [DOI] [PubMed] [Google Scholar]
- Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. 2007;120:2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- Sonkoly E, Bata-Csorgo Z, Pivarcsi A, Polyanka H, Kenderessy-Szabo A, Molnar G, Szentpali K, Bari L, Megyeri K, Mandi Y, et al. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J Biol Chem. 2005;280:24159–24167. doi: 10.1074/jbc.M501704200. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15:501–512. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, Lou Y, Yang J, Ma Y, Chai L, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8:1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A. Activation of p53 by MEG3 noncoding RNA. J Biol Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]