Abstract
CPEB is a sequence-specific RNA binding protein that promotes polyadenylation-induced translation in early development, during cell cycle progression and cellular senescence, and following neuronal synapse stimulation. It controls polyadenylation and translation through other interacting molecules, most notably the poly(A) polymerase Gld2, the deadenylating enzyme PARN, and the eIF4E-binding protein Maskin. Here, we report that CPEB shuttles between the nucleus and cytoplasm and that its export occurs via the CRM1-dependent pathway. In the nucleus of Xenopus oocytes, CPEB associates with lampbrush chromosomes and several proteins involved in nuclear RNA processing. CPEB also interacts with Maskin in the nucleus as well as with CPE-containing mRNAs. Although the CPE does not regulate mRNA export, it influences the degree to which mRNAs are translationally repressed in the cytoplasm. Moreover, CPEB directly or indirectly mediates the alternative splicing of at least one pre-mRNA in mouse embryo fibroblasts as well as certain mouse tissues. We propose that CPEB, together with Maskin, binds mRNA in the nucleus to ensure tight translational repression upon export to the cytoplasm. In addition, we propose that nuclear CPEB regulates specific pre-mRNA alternative splicing.
Keywords: CPEB, polyadenylation, translation
INTRODUCTION
In early embryonic development prior to the onset of robust transcription, most protein production is directed predominantly by maternally inherited mRNAs. In Xenopus, these maternal mRNAs are dormant in oocytes arrested at the end of prophase, which resembles G2 of the mitotic cell cycle. Upon stimulation of oocyte maturation by progesterone, the cells re-enter the meiotic divisions and arrest again at metaphase II. During this time, a number of mRNAs that have relatively short poly(A) tails (∼20–40 nucleotides [nt]) undergo poly(A) elongation (to ∼150 nt), which is coincident with translational activation. Two regulatory elements in mRNA 3′ untranslated regions (UTR), the cytoplasmic polyadenylation element (CPE) and the polyadenylation hexanucleotide AAUAAA, are necessary for polyadenylation (Mendez and Richter 2001). The CPE is recognized by the RNA binding protein CPEB (Hake and Richter 1994) while AAUAAA is bound by the multi-subunit complex CPSF (Dickson et al. 1999). In oocytes, prior to the onset of polyadenylation, CPEB is associated with both the poly(A) polymerase Gld2 (Barnard et al. 2004) and the poly(A)-specific ribonuclease PARN. These two enzymes are both active in oocytes, but because PARN activity is more robust, the poly(A) tail is kept short (Kim and Richter 2006). In addition to these factors, CPEB also interacts with Maskin (Stebbins-Boaz et al. 1999), a eukaryotic initiation factor (eIF) 4E binding protein that prevents eIF4G from joining the cap binding complex (eIF4F) and thus inhibits translation initiation. When the oocytes are stimulated by progesterone, CPEB is phosphorylated on S174 (Mendez et al. 2000a), which in turn causes CPEB to strongly associate with CPSF (Mendez et al. 2000b) and expels PARN from the polyadenylation complex. Thus, poly(A) tail growth occurs by default, because PARN is no longer present to remove Gld2-catalyzed polyadenylation (Kim and Richter 2006).
Symplekin (Keon et al. 1996; Takagaki and Manley 2000; Barnard et al. 2004), which may act as a scaffold upon which multiple factors are assembled, and ePAB, an embryonic-type poly(A) binding protein (Voeltz et al. 2001; Kim and Richter 2007), are two additional proteins in the polyadenylation complex. ePAB dissociates from the polyadenylation complex when CPEB undergoes a subsequent round of cdk1-catalyzed phosphorylations and binds the newly elongated poly(A) tail. Here, ePAB not only protects the tail from degradation, but also binds the initiation factor eIF4G. This interaction helps eIF4G displace Maskin from and itself bind to eIF4E (Cao and Richter 2002; Kim and Richter 2007), resulting in initiation. Thus, through interactions with multiple proteins, CPEB acts as a translational switch during the meiotic divisions.
Several studies suggest that cytoplasmic regulation of mRNA begins with the association of nuclear-cytoplasmic shuttling factors. For example, nuclear binding of hnRNP I to Vg1 mRNA remodels the RNP complex so that Vg1RBP/vera can subsequently bind and direct vegetal localization of the mRNA in Xenopus oocytes (Kress et al. 2004; Lewis et al. 2008). Moreover, the nuclear interaction between the yeast ASH1 mRNA and the She2 protein is important for recruiting the translation repressor Puf6p and Loc1p; such factors are responsible for asymmetric translation repression after cytokinesis (Gu et al. 2004; Du et al. 2008; Shen et al. 2009). These and several other reports (e.g., Hachet and Ephrussi 2004; Huynh et al. 2004; Shibuya et al. 2004; Yano et al. 2004; Huttelmaier et al. 2005) suggest that the nuclear history of mRNA can affect their cytoplasmic fate, possibly due to factors deposited on the RNAs during biogenesis and/or transport.
Recently, CPEB has been shown to shuttle between nucleus and cytoplasm (Rouget et al. 2006; Ernoult-Lange et al. 2009), although the significance of this phenomenon is unclear. We have also found that CPEB shuttles between nucleus and cytoplasm, and have investigated the functional importance of CPEB in the nucleus. In Xenopus oocytes, nuclear CPEB associates with transcriptionally active lampbrush chromosomes in an RNase-sensitive manner. CPEB co-immunoprecipitation experiments show that it binds nuclear CPE-containing RNA as well as several RNA processing factors. In the nucleus, Maskin, but not Gld2 or PARN, are components of the CPEB-containing RNP complex. Experiments involving the injection of plasmid DNA or RNA into the nucleus and RNA into the cytoplasm indicate that the CPE, and by extension CPEB and probably Maskin, bind RNA in the nucleus to ensure that it is tightly repressed following export to the cytoplasm. Thus, the nuclear experience of CPEB helps dictate the cytoplasmic fate of mRNA. In addition, because of this CPEB nuclear experience, we also investigated the possibility that it could mediate specific alternative exon usage; indeed, CPEB KO mouse embryonic fibroblasts (MEFs) and tissues derived from CPEB KO mice differentially splice at least one pre-mRNA that encodes collagen 9a1. Thus, CPEB mediates both cytoplasmic and nuclear RNA processing.
RESULTS
CPEB is a nuclear-cytoplasmic shuttling protein
In stage VI Xenopus oocytes, CPEB is almost exclusively cytoplasmic (Hake and Richter 1994), although a small amount is nuclear in stage I/II oocytes (Fig. 1A, the left panel shows the quality of the antibody; the right panel shows CPEB during oocyte maturation). However, when oocytes were treated with leptomycin B (LMB), which blocks protein nuclear export via the chromosome region maintenance 1 (CRM1)-dependent pathway, a substantial amount of CPEB was detected in the nucleus (germinal vesicle) (Fig. 1B). Moreover, while CPEB was cytoplasmic in MEFs transfected with DNA encoding mouse CPEB fused to the HA epitope, it became mostly nuclear when the cells were incubated with LMB (Fig. 1C). These results, which were also observed in HeLa cells and 293T cells (data not shown), suggest that CPEB shuttles between the nucleus and cytoplasm.
FIGURE 1.
CPEB is a nuclear-cytoplasmic shuttling protein. (A) Left: Western blot of Xenopus oocyte lysate demonstrated the specificity of CPEB antibody used in this study. Right: Nuclei and cytoplasms from oocytes of different stages were manually separated and probed on Western blots for CPEB and tubulin. (B) Stage VI oocytes were treated with 200 nM leptomycin B overnight; nuclei and cytoplasms were then manually separated and probed for CPEB, tubulin as a cytoplasmic marker, and histone H4 as a nuclear marker. (C) MEFs were transfected with CPEB-HA, some of which were then treated with 10 nM LMB for 5 h. The HA epitope was located by indirect immunofluorescence. (D) Lampbrush chromosomes were prepared and immunostained for symplekin and CPEB. Some preparations were treated with RNase before immunolocalization for CPEB. The chromosomes were also stained with DAPI.
To determine whether CPEB is associated with nascent transcripts in the oocyte nucleus, lampbrush chromosomes, structures of intense transcription that can readily be detected by light or fluorescence microscopy (Smillie and Sommerville 2002), were prepared. Figure 1D shows that both symplekin and CPEB were associated with the lampbrush chromosomes, but that at least in the case of CPEB, the association was RNase A-sensitive. These data indicate that CPEB interacts with nascent chromosome-associated transcripts in the nucleus.
Complex control of CPEB nuclear import
To identify the regions of CPEB that mediate its nuclear import and export, 3T3 cells were infected with retroviruses harboring HA-tagged wild type (WT) and deletion mutant constructs (Fig. 2A). Some of the cells were subsequently treated with LMB; CPEB was then localized by indirect immunofluorescence for the HA epitope (Fig. 2B). In the absence of a nuclear localization signal (NLS), CPEB would be expected to be cytoplasmic even when cells were incubated in the presence of LMB. Indeed, deletion of CPEB residues 206–510 in different constructs caused CPEB to remain cytoplasmic when cells were incubated with LMB (Fig. 2B). To quantify the amount of CPEB that was nuclear or cytoplasmic, we determined the percent of cells with localization characteristics similar to those shown in Figure 2C. The data compiled in Figure 2D demonstrate that, in the absence of LMB, all the CPEB proteins were predominantly cytoplasmic. In the presence of LMB, however, only deletion mutants 206–510 showed significant cytoplasmic staining. It should be noted that, in cells treated with LMB, CPEB was predominantly, but not entirely nuclear, perhaps indicating that either the CPEB NLS is not as strong as the NLSs of other proteins or that there is some specific cytoplasmic retention.
FIGURE 2.
CPEB nuclear localization domain. (A) Diagram of deletion mutant constructs of CPEB. PEST refers to a domain rich in proline, glutamic acid, serine, and threonine that is thought to be involved in protein destruction; RRM refers to RNA recognition motif, and ZF refers to zinc finger. (B) Immunocytochemistry of 3T3 cells expressing CPEB-HA full-length or deletions illustrated in panel A. (C) The nucleus-cytoplasm localization was quantified using an arbitrary score; this scoring system was used to analyze the relative localization of the CPEB proteins shown in panel B. Histograms of these data are presented in D; the numbers atop the bars refer to the total number of cells examined.
To further define the CPEB NLS, deletion mutations spanning residues 206 to 309 were generated, transfected into 3T3 cells that were then treated with LMB, and probed for HA as described above. Figure 3A demonstrates that, with the first set of deletions, CPEB Δ206–257 was evenly distributed in cells treated with LMB while CPEB Δ258–309 was strongly cytoplasmic. Consequently, we constructed the next set of proteins: CPEB Δ258–283 was uniformly distributed in cells treated with LMB while Δ284–309 was strongly cytoplasmic. In the next set of proteins expressed in cells treated with LMB, CPEB Δ284–296 was distributed throughout the cells while Δ297–309 and Δ297–307 were strongly cytoplasmic.
FIGURE 3.
Requirement for CPEB nuclear localization and RNA binding. (A) CPEB deletion mutants lacking regions between residues 206 and 309 were HA tagged, transfected into NIH 3T3 cells, incubated in the presence of LMB, and analyzed as in Fig. 2. Residues 297–307 were necessary for nuclear localization; each of the 11 residues in this region was changed to alanine and the nuclear localization examined as above. In each case, single alanine substitutions had no effect on nuclear localization. (B) HEK 293T cells were infected with HA-tagged CPEB or CPEB Δ297–307. An extract was then prepared, supplemented with the CPE-containing cyclin B1 3′ UTR, UV irradiated, and subjected to HA immunoprecipitation. The proteins were then analyzed by Western blot for HA (upper panel, two bands are evident; the higher one was likely generated from an upstream cryptic transcription start site of the C-pOZ vector.) and by autoradiography for proteins made radioactive by label transfer (lower panel).
To investigate further how residues 297–307 affected nuclear localization, each residue within peptide 297–307 was changed to alanine in separate constructs. When these were transfected into cells that were subsequently treated with LMB, all the ectopically expressed CPEBs became nuclear (Fig. 3A; Supplemental Fig. 1). Thus, the deletion mutation but not the point mutation affected CPEB nuclear localization.
CPEB residues 297–307 lie just upstream of RNA recognition motifs (RRMs) (residues 310–510), so while RRM prediction algorithms do not indicate that they are a part of the RNA binding region, we nonetheless suspected that they might be involved in RNA binding. To assess this, 293T cells were infected with a retrovirus harboring HA-tagged WT or Δ297–307 CPEB (the 293T cells were employed because they express high amounts of exogenous CPEB compared to MEFs or other somatic cells); extracts derived from these cells were supplemented with radiolabeled RNA (mouse cyclin B1 3′ UTR) containing or lacking CPEs followed by UV cross-linking, CPEB immunoprecipitation with antibody against HA, and resolution by SDS-PAGE. On Western blots, two bands derived from the plasmid were evident; the lower band was the predicted size of CPEB (∼68 kDa) and the upper band could be the protein product from an upstream transcription start site (Fig. 3B, upper panel). CPEB WT cross-linked to the CPE containing but not CPE lacking RNA, whereas Δ297–307 did not cross-link to either of the RNAs (Fig. 3B, lower panel). Thus, Δ297–307 was defective in both RNA binding and nuclear import, and this might imply that RNA binding is required for CPEB nuclear import. However, a zinc finger deletion mutant CPEB, which does not bind the CPE-RNA (Hake et al. 1998), entered the nucleus similar to WT (Fig. 2B,D). Thus, we conclude from the NLS screening that, while the proper folding of RRMs is important for both RNA binding and protein nuclear import, CPE-RNA binding is not essential for nuclear import.
The complex nature of the CPEB NLS was also suggested by the observation that residues 206–309, when fused to luciferase, were unable to promote nuclear entry (data now shown), implying that the sequence information was not sufficient for nuclear import. Finally, although we have serially deleted CPEB in its entirety, we were unable to identify a nuclear export signal (NES). However, Ernoult-Lange et al. (2009) have recently identified two redundant NESs in CPEB. When leucine and isoleucine residues in the NESs (NES95–104 LCLGLQSLSL and NES197–206 LSDLISSLRI) were replaced by alanine, CPEB accumulated in the nucleus independently of LMB treatment (Fig. 4A). These two NESs are conserved among vertebrate species, and the critical leucines are conserved in Drosophila (Fig. 4B).
FIGURE 4.
CPEB contains two redundant NESs in the N-terminal half of the protein. (A) When both of the NESs were mutated, CPEB accumulated in the nucleus independently of LMB treatment. (B) An alignment shows these two NESs are conserved among species. The critical leucine/isoleucine residues are also conserved in Drosophila orb1, but not other CPEB family proteins of any species (not shown).
CPEB associates with the nuclear RNA processing machinery
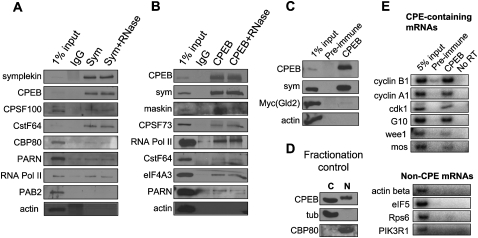
To begin to determine the function of CPEB in the nucleus, we conducted a series of co-immunoprecipitation experiments. First, because symplekin appears to act as a scaffold protein upon which the CPEB-containing cytoplasmic machinery is assembled (Barnard et al. 2004; Kim and Richter 2006), this protein was immunoprecipitated from the nucleus, where it is known to associate with the nuclear RNA processing machinery (Vethantham et al. 2007). Symplekin was immunoprecipitated from LMB-treated hand-isolated Xenopus oocyte stage VI nuclei (germinal vesicles) in the absence or presence of RNase A; the proteins that were co-precipitated were then identified by Western blotting (Fig. 5A). CPEB was strongly co-precipitated with symplekin from the nucleus, as were CPSF 100-kDa subunit and cleavage stimulatory factor 64 (CstF64). RNA polymerase II was also co-precipitated, but cap binding protein 80 (CBP80), PARN, PAB2, or actin either was not co-precipitated or were co-precipitated just barely above background, which could be due to nonspecific adsorption.
FIGURE 5.
CPEB is a component of the nuclear RNA processing machinery. (A) Symplekin was immunoprecipitated in the absence or presence of RNase A from ∼250 LMB-treated hand-isolated oocyte nuclei. A similar number was mock precipitated with nonspecific IgG. The precipitates were probed on Western blots for the proteins noted in the figure. Actin served as a negative control; 1% of the extract was also applied directly to the gel without immunoprecipitation. (B) Similar to panel A except that CPEB was immunoprecipitated from the nuclear extracts. (C) Oocytes were injected with mRNA encoding myc-tagged Gld2; following overnight culture, the nuclei were isolated and subjected to CPEB immunoprecipitation as in panel B and probed for the proteins noted in the figure. (D) Fractionation control from oocytes used in panels A–C and E; tubulin, a cytoplasmic protein, is entirely cytoplasmic, while CBP80, a nuclear protein, is entirely nuclear. (E) CPEB was immunoprecipitated from oocyte nuclei as before; the RNA was extracted from the precipitates and subjected to RT-PCR for the RNAs noted in the figure.
Other oocyte nuclei were used for CPEB co-immunoprecipitation experiments. In this case, symplekin, Maskin, CPSF73, RNA polymerase II, and eIF4A3 were all co-precipitated irrespective of the presence of RNase A. CstF64, PARN, and actin were not co-precipitated significantly above background (Fig. 5B). Moreover, CPEB was not co-precipitated with Gld2 (in this case, a heterologous myc-Gld2 fusion protein synthesized from injected mRNAs) (Fig. 5C). Finally, Figure 5D demonstrates that the hand isolation of oocyte nuclei was devoid of cytoplasmic contamination; tubulin, a cytoplasmic protein, was detected only in the cytoplasmic fraction, while CBP80, a nuclear protein, was detected only in the nuclear fraction. These results indicate that CPEB resides in a complex (or complexes) with specific components of the nuclear polyadenylation and RNA export machinery; they also suggest that its association with Maskin could be important for repressing translation once RNA is exported to the cytoplasm.
Although CPEB is a sequence-specific RNA binding protein, it associates with some of the general RNA processing machinery in the nucleus, suggesting it might be deposited on mRNA without sequence specificity. To investigate this possibility, CPEB was immunoprecipitated from hand-isolated nuclei followed by RNA extraction and RT-PCR for specific CPE-containing and CPE-lacking RNAs. Figure 5E shows that the CPE-containing RNAs cyclin B1, cyclin A1, cdk1, G10, wee1, and mos were all co-immunoprecipitated with CPEB. Conversely, none of the CPE-lacking RNAs, actin, eIF5, Rsp6, PIK3R1, was co-precipitated. Thus, although CPEB associates with general RNA processing machinery, it binds only to specific RNAs in the nucleus.
The nuclear experience of RNA and poly(A) metabolism
We considered a number of possible functions for nuclear CPEB including involvement in RNA processing (3′ end formation and splicing) and export. We also thought that nuclear CPEB might influence cytoplasmic polyadenylation and/or translation once the CPE-containing RNA was exported to the cytoplasm. To begin to examine these parameters, we injected plasmid DNA encoding luciferase fused to the cyclin B1 3′ UTR containing or lacking CPEs into oocyte nuclei (Fig. 6A). RNA derived from the injected DNA was exported to the cytoplasm with similar kinetics irrespective of the presence of CPEs (Fig. 6B). Thus, the CPE confers neither an advantage nor a disadvantage with respect to RNA biosynthesis or export.
FIGURE 6.
The nuclear experience of CPE does not mediate mRNA nuclear export or cytoplasmic deadenylation. (A) Diagram of experiment procedure for comparison of RNA export between CPE-containing and CPE-lacking luciferase mRNA. (B) Cytoplasmic luciferase mRNA levels following plasmid injection as determined by radioactive semiquantitative RT-PCR (upper panel). The relative mRNA levels are graphed in the lower panel. (C) Diagram of experiment procedure for comparison of deadenylation between cytoplasm-injected and nucleus-injected cyclin B1 mRNA. (D) Deadenylation assay. A radiolabled and polyadenylated partial cyclin B1 mRNA was injected into the nucleus or cytoplasm of oocytes; after overnight incubation, the cytoplasmic fraction was collected for RNA extraction and analysis on a denaturing polyacrylamide gel. (E) Deadenylation assay of CPE-containing RNA. Oocytes were injected with in vitro transcribed RNA or plasmid DNA; RNA collected over several hours was analyzed by ligation-mediated PAT assay (see Materials and Methods). Lower panels are ethidium bromide stained agarose gels showing RT-PCR products of cyclin B1 RNA; cyclin B1 mRNA started to accumulate ∼3 h after injection in the nucleus and ∼6 h in the cytoplasm. Note that because the RT-PCR does not distinguish endogenous from ectopic cyclin B1 3′ UTR, a band is present in the cytoplasmic fraction of noninjected oocytes.
When injected into the cytoplasm, CPE-containing RNA is deadenylated while CPE-lacking RNA is not (Kim and Richter 2006, 2007). To determine whether the nuclear experience of RNA has an effect on deadenylation, RNA containing or lacking the CPE, polyadenylated in vitro with 100–120 adenylate residues, was injected into the nucleus or cytoplasm of oocytes (Fig. 6C). The RNA was then analyzed by gel electrophoresis 16 h after injection. The RNA injected into the cytoplasm had a similar stability irrespective of the presence or absence of CPEs (Fig. 6D). Moreover, as reported previously (Kim and Richter 2006), the CPE-containing, but not CPE-lacking RNA was deadenylated when injected directly into the cytoplasm [Fig. 6D, cf. frog A, frog B (lanes C) and probe p(A)]. Surprisingly, however, when either RNA was injected into the nucleus, very strong deadenylation was evident [Fig. 6D, cf. frog A, frog B (lanes N) and probe p(A)]. A short time course demonstrated that this deadenylation occurred very rapidly (within 20 min, independently of the CPE) and took place in the cytoplasm following nuclear export (data not shown). In contrast to these results, cytoplasmic CPE-containing RNA derived from injected plasmid DNA was deadenylated in the cytoplasm with similar kinetics compared to when CPE-containing polyadenylated RNA was injected directly into the cytoplasm (Fig. 6E. Note that the PCR-based PAT assay to detect RNA derived from the injected plasmid also detects the endogenous RNA; hence, an RT-PCR signal is detected in the noninjected [NI] lane. The lower two panels of cytoplasmic and nuclear mRNA detect overall levels, both endogenous and ectopically expressed. The accumulation of cytoplasmic RNA is evident at 6 h, the same time when polyadenylation of the RNA is observed). Thus, while these data do not show a difference in deadenylation between CPE-containing and CPE-lacking RNA, they do demonstrate that injected RNA is rapidly deadenylated when it is injected directly into the nucleus but not when it is derived from de novo transcription. We interpret these results to mean that a factor(s) is deposited on the poly(A) tail of nascent RNA that protects it from rapid removal and that such a factor(s) is not present on the poly(A) tail of RNA injected directly into the nucleus (see Discussion).
The nuclear experience of CPEB and translational control
To determine whether the nuclear experience of CPEB could influence translation, we used the luciferase-cyclin B1 reporter system described in Figure 7A. Plasmid DNA encoding this construct, containing or lacking 3′ UTR CPEs, was injected into oocyte nuclei; 0–12 h later, extracts were prepared and divided into two portions, one for luciferase assays and one for RNA measurement. A calculation of the translational efficiency of each construct (luciferase activity/mass amount of RNA), shows that CPE-lacking mRNA was much more efficiently translated than CPE-containing mRNA; by 12 h, it was translated with ∼10-fold greater efficiency (Fig. 7B). Similarly, the CPE-lacking mRNA [with ∼100 base poly(A) tail] injected into the cytoplasm was translated more efficiently than the CPE-containing mRNA; by 12 h, it was translated with approximately threefold greater efficiency (Fig. 7B). In both nuclear and cytoplasmic injection, no difference in the timing of when CPE-dependent translation repression took place was observed (both started 3–6 h after injection). We next collected oocytes injected as in panel A that were incubated for 12–16 h and calculated the relative translational efficiencies of CPE-lacking to CPE-containing mRNA derived from nuclear or cytoplasmic injection (Fig. 7C). In this case, CPE-containing (WT) mRNA was more translationally repressed in a statistically significant manner compared to CPE-lacking mRNA (mt) when it experienced the nuclear milieu (translation efficiency mt/WT, approximately threefold when injected in the cytoplasm and approximately sixfold when injected in the nucleus). We infer from these results that the binding of CPEB (and probably Maskin) to CPE-containing mRNA in the nucleus imposes a tight translational regulation in the cytoplasm.
FIGURE 7.
The nuclear experience of CPE-containing mRNAs mediates tight translational repression. (A) Diagram of experimental procedure for comparing CPE-dependent translational repression with or without the nucleus experience. (B) Time course of translational efficiency of reporters (luciferase activity/RNA) containing or lacking CPEs derived from plasmid DNA-injected oocytes (top). Time course of translational efficiency of the constructs noted above that were synthesized in vitro and then injected into the cytoplasms of oocytes (bottom). (C) Comparison of the translational efficiencies from panel B of plasmid-injected nuclei versus RNA-injected cytoplasm (dark gray bars). Also shown is a comparison of the translational efficiencies between RNA-injected nuclei versus cytoplasm (light gray bars). The RNA was collected 12 to 16 h after injection, and the translation efficiency was determined as in B. The bars represent the fold difference of translational efficiency of RNA lacking the CPE (mt) divided by that of RNA containing the CPE (WT). Statistical analysis was by a one-tailed paired t-test.
CPEB and alternative splicing
Because CPEB shuttles to the nucleus where it associates with nascent transcripts (Fig. 1D), we considered the possibility that it might regulate alterative splicing as well as influence mRNA polyadenylation and translation. To investigate this possibility, we employed MEFs derived from WT and CPEB KO mice. RNA from four pairs of WT and KO MEFs were screened on an Affymetrix exon array platform (GeneChip Mouse Exon 1.0 ST Arrays). While bioinformatic analysis indicated a number of pre-mRNAs with skipped exons, we could validate only the one encoding collagen 9a1 (Col9a1). Figure 8A shows three sets of WT and CPEB KO MEFs where exon 34 was skipped. The examination of tissue from WT and CPEB KO mice demonstrates exon 34 skipping in brain and ear, which has high collagen content (Fig. 8B). However, the effect of CPEB on Col9a1 splicing was not the same in all tissues. For example, in testis, exon 35, instead of exon 34, was preferentially skipped in the absence of CPEB, whereas in heart, exon 35 was skipped independent of CPEB. These results demonstrate that CPEB mediates alternative pre-mRNA splicing, although they do not indicate whether this is a direct effect.
FIGURE 8.
CPEB mediates alternative pre-mRNA splicing. (A) RT-PCR (dCTP-[α-P32] incorporation) analysis of exons 33–36 of the collagen 9a1 mRNA from three different WT and CPEB KO MEF lines. (B) RT-PCR analysis of collagen 9a1 mRNA exons 33–36 from different tissues of WT and CPEB KO mice.
DISCUSSION
In this study, we demonstrate that CPEB shuttles between the nucleus and cytoplasm, that it has a complex NLS, that it along with RNA processing machinery associates with RNA in the nucleus, and that its nuclear experience is important for repressing translation in the cytoplasm. As depicted in Figure 9, we envisage that CPEB forms RNPs with nascent RNAs, and these RNPs include, among other factors, eIF4A3, CPSF, and Maskin. These factors may also be associated with RNA polymerase II. The RNP is exported to the cytoplasm where it reforms with components of the cytoplasmic polyadenylation machinery including Gld2, PARN, symplekin, and CPSF.
FIGURE 9.
Model of CPEB-mediated translational control. A CPE-containing RNA is recognized by a CPEB and Maskin-containing protein complex in the nucleus either co-transcriptionally or soon after transcription is complete. After export from the nucleus, a cytoplasmic RNP complex is assembled that includes the poly(A) polymerase Gld2 and the deadenylating enzyme PARN. PARN is expelled from the complex upon progesterone-induced and aurora A-catalyzed CPEB phosphorylation; Gld2 then elongates the poly(A) tail by default. Maskin is phosphorylated at this time. These events, as well as its association with an embryonic poly(A) binding protein (not shown), lead to the replacement of Maskin for eIF4G on eIF4E. As a consequence, translation is activated. The association of eIF4A3 with the cytoplasmic complex is conjectural.
We propose that the association of CPEB and Maskin with RNA in the nucleus is important for translational silencing upon export to the cytoplasm. The RNP resides in this translationally dormant state until progesterone signaling induces activation of the kinase Aurora A, which phosphorylates CPEB, leading to the expulsion of PARN from the RNP complex. This event results in Gld2-catalyzed polyadenylation and subsequent translation.
We also used MEFs derived from CPEB knockout mice to demonstrate that CPEB influences specific alternative splicing. While we do not know if CPEB acts directly or indirectly to regulate nuclear RNA processing, these data demonstrate that CPEB modulates gene expression at multiple levels and does not solely control cytoplasmic polyadenylation and translation. Therefore, the biological consequences of the loss of CPEB could in part be due to defective splicing as well as defective translation.
RNA nuclear and cytoplasmic regulation
Over the past decade, several lines of evidence have emerged showing that nuclear RNA processing events influence the cytoplasmic fates of mRNAs. It was originally suggested by Braddock et al. (1990) that a factor that blocks translation could be deposited on mRNA prior to export. In a similar vein, Bouvet and Wolffe (1994) showed that transcription is involved in relaying a negative translation factor to nascent RNA in the nucleus. More recently, it was reported that the reporter plasmid DNAs used as templates for the synthesis of mRNA affected the mechanism by which miRNAs repress translation (Kong et al. 2008), presumably due to different factors associated with newly transcribed RNAs that contain sites complementary to miRNA. Moreover, RNA splicing seems to be particularly important for regulating mRNA localization and translation. For example, insertion of a 5′ intron into an intronless gene enhances translation of the derived mRNA by 10-fold (Matsumoto et al. 1998). Molecules deposited during splicing, such as the EJC components eIF4A3, Y14, and Mago, are responsible for mRNA localization and following translational control (Hachet and Ephrussi 2004; Palacios et al. 2004). The EJC is also involved in nonsense-mediated mRNA decay (NMD), a cytoplasmic process. In this case, EJC components including UPF2 and UPF3 reside 20–24 bases 5′ of exon–exon junctions. They recruit and/or activate ribosome-associated UPF1, which ultimately induces NMD. Thus, NMD would be activated during an initial translational event (i.e., the “pioneer” round) (Maquat 2004; Brogna and Wen 2009). In addition to NMD components, shuttling hnRNP proteins must also experience a nuclear milieu to affect, at least the case of hnRNP D, cytoplasmic RNA stability (Chen et al. 2004).
CPEB is also deposited on nuclear RNA, but only those that contain a CPE (Fig. 5E). This would seem to be paradoxical given our other observations that show CPEB to be a component of the general RNA processing machinery such as RNA polymerase II, CPSF, and eIF4A3. This implies that the so-called general machinery may be associated with specific components. Such specific components may be transiently associated with the general machinery that could lend it diversity. Notably, there appears to be considerable remodeling of RNPs in the nucleus of cells, or when the RNPs enter the cytoplasm. For example, RNA localization in the vegetal cortex of the Xenopus oocyte cytoplasm is initiated by interactions with RNA binding proteins in the nucleus (Kress et al. 2004), at least with the protein Vg1RBP/vera. Moreover, PTB/hnRNP I plays a critical role in this process by coordinating and remodeling the association of Vg1RBP/Vera with the Vg1 target mRNA (Lewis et al. 2008). While we do not propose a remodeling activity for CPEB, these examples make clear that dynamic changes among RNA binding proteins occur as RNAs undergo nuclear export.
We suspected that the interaction of CPEB with nuclear RNA might facilitate the CPE-dependent cytoplasmic deadenylation. Therefore, we compared the efficiencies of the deadenylation process between nuclear and cytoplasm-injected RNA (Fig. 6). However, the in vitro transcribed RNA injected into the nucleus was rapidly cleaved and/or degraded while the little amount that was exported to the cytoplasm was rapidly deadenylated irrespective of a CPE. We hypothesize that the injected RNA would not be associated with a factor, for example, nuclear poly(A) binding protein, that would be deposited on nascent RNA during transcription or soon thereafter. In the absence of this factor, surveillance may be triggered to destroy the RNA. Such a surveillance mechanism would apply only to the nucleus since, in the cytoplasm, CPE-lacking RNAs contain stable poly(A) tails following cytoplasmic injection.
An additional function of CPEB: Alternative splicing
While nuclear CPEB (and Maskin) binds RNA in the nucleus to tightly regulate translation in the cytoplasm (Fig. 7C), our data suggest that CPEB is also involved in splicing regulation (Fig. 8). Because the sequences near the intron–exon boundary of an alternative exon (exon 34 of Col9a1) contain CPEs, we surmised that CPEB might directly control alternative processing. Consequently, we generated a minigene containing exon 34 surrounded by ∼500 bases of intron sequence; however, when co-transfected with heterologous CPEB, we could detect no change in its splicing pattern. Moreover, overexpression of CPEB in KO MEFs also did not rescue the splicing pattern of endogenous Col9a1 pre-mRNA (data not shown). We are unsure whether the sequence information of the minigene was sufficient to direct splicing or why overexpression of CPEB failed to induce the alternative splicing. We would expect the expression of CPEB to rescue the WT splicing pattern, even if CPEB was acting indirectly to induce exon skipping.
The complex nature of nuclear localization of CPEB
Recently, Ernoult-Lange et al. (2009) have also shown that CPEB traffics to the nucleus, in this case, in HeLa cells. Those investigators also found CPEB to be associated with nuclear foci that they referred to as Crm1 nucleolar bodies. Using HA-tagged CPEB, we have been unable to confirm these findings; we did not observe CPEB in any discreet region in the nucleus. However, because those investigators employed GFP-CPEB for this identification, we thought that perhaps the tag used to identify heterologus CPEB could influence its subcellular localization. Consequently, we fused GFP to CPEB and repeated our experiments; again we could not identify discreet subnuclear regions where CPEB accumulated. We do not know why these localization results differ from those of Ernoult-Lange et al. (2009).
We have found that the RRMs of CPEB are involved in nuclear import. While we did not identify a canonical NLS in this region, we nonetheless replaced several positively charged residues with alanine; none elicited a defect in nuclear import (data not shown). We hypothesize that the proper folding of RRMs, and hence CPEB's interaction with other factors, is necessary for efficient nuclear import. Irrespective of the precise nature and sequence of the CPEB NLSs, we identify two new functions for CPEB: it associates with mRNP in the nucleus to reinforce cytoplasmic translational repression, and it regulates alternative splicing of a specific pre-mRNA.
MATERIALS AND METHODS
Antibodies
Antibodies used in this study include: rabbit anti-CPEB (Hake and Richter 1994), rabbit anti-HA (Covance), mouse anti-α-tubulin (Sigma-Aldrich), rabbit anti-histone H4 (Upstate), mouse anti-symplekin (BD Transduction Laboratories), mouse anti-RNA polymerase II 8WG16 (Upstate), rabbit anti-CPSF73 (a gift from D. Bentley, University of Colorado), rabbit anti-CPSF100 (a gift from J. Manley, Columbia University) (Takagaki and Manley 2000), rabbit anti-CstF64 (a gift from C. Milcarek, University of Pittsburgh) (Shell et al. 2005), rabbit anti-CBP80 (a gift from E. Izaurralde, Max Planck Institute) (Izaurralde et al. 1994), rabbit anti-PARN (a gift from M. Wormington, University of Virginia), rabbit anti-PAB2 (a gift from E. Wahle, University of Halle) (Krause et al. 1994), mouse anti-actin, rabbit anti-Maskin (Stebbins-Boaz et al. 1999), mouse anti-eIF4A3 (a gift from A. Krainer, Cold Spring Harbor Laboratory), and mouse anti-myc 9E10 (Hake and Richter 1994).
Immunocytochemistry
3T3 cells were seeded to ∼50% confluency on the coverslip the day before infection. Mouse CPEB-containing virus made from the C-pOZ retroviral system (Groisman et al. 2006) was filtered and applied to the 3T3 cells. After overnight incubation to allow protein expression, the cells were washed twice with PBS and fixed with 4% paraformadehyde in cytoskeleton buffer (10 mM PIPES at pH 6.8, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, and 1 mM EGTA) 10 min at room temperature. The cells were washed with PBS and then permeabilized with 0.5% Triton X-100 in cytoskeleton buffer 5 min at room temperature. After washing twice, the cells were blocked in TBS-1 (10 mM Tris-HCl at pH 7.7, 150 mM NaCl, 3 mM KCl, 1.5 mM MgCl2. 0.05% Tween 20, 0.1% bovine serum albumin [BSA], 0.2% glycine) supplemented with 10% goat serum for at least 30 min. After blocking, the cells were incubated with HA antibody (1:1000) in PBS-1 with goat serum for 1 h, followed by five washes in PBS. The cells were then incubated in fluorescence-conjugated secondary antibody for 1 h. Following three washes, the cells were stained with DAPI to visualize the nucleus; they were mounted in Prolong Gold antifade reagent (Invitrogen).
Lampbrush chromosomes
The nucleus was dissected from stage VI oocytes in 5:1 isolation buffer (83 mM KCl, 17 mM NaCl, 6.5 mM Na2HPO4, 3.5 mM KH2PO4, 1 mM MgCl2, and 1 mM DTT) and transferred to coverslips with wax wells containing dispersal buffer (20.7 mM KCl and 4.3 mM NaCl). After removing the nuclear membrane to liberate the nuclear matrix, the chromosomes were allowed to spread for 30 min. The chromosomes were centrifuged onto glass slides at 2000g for 10 min and then fixed in cold methanol for 20 min. Following several washes, the chromosome preparation was blocked in 2% BSA in PBS for 30 min at room temperature, followed by immunocytochemistry.
Immunoprecipitation
CPEB and symplekin antibodies as well as control IgG were conjugated to protein A-sepharose 4B (Invitrogen) or anti-mouse dynabeads (Invitrogen) overnight at 4°C, and washed to remove free antibodies; 500 to 1000 nuclei from stage VI oocytes were homogenized in IP buffer (150 mM NaCl, 25 mM HEPES-KOH at pH 7.5, 10% glycerol, 1 mM MgCl2, 2 mM sodium orthovanadate, 2 mM β-glycerophosphate, 1 mM phenylmethylsulphonylfluoride [PMSF], 1 mM DTT, 2 mM EDTA, 0.5% Triton X-100, and protease inhibitor cocktail [Roche]). The oocyte lysate was precleared and incubated with antibody-conjugated beads overnight at 4°C with or without 50 μg/mL RNase A (Sigma-Aldrich) as indicated. The collected beads were then washed five times before boiling in SDS-sample buffer.
RNP-IP
Two thousand to 3000 LMB-treated hand-isolated nuclei were homogenized in RNP-IP buffer (150 mM NaCl, 25 mM Tris-HCl at pH 7.5, 1 mM DTT, 1 mM PMSF, 2 mM MgCl2, 10% glycerol, 0.5% Nonidit P-40, protease inhibitor cocktail, and 100 units/mL RNaseOUT [Invitrogen]) and precleared with IgG-conjugated beads for 30 min before incubated with antibody-conjugated beads for 3 h. The beads were washed four times, treated with five units of DNase I for 15 min at 30°C, and then the RNA on the beads was extracted by Trizol (Invitrogen). The purified RNA was subject to RT-PCR with the following primers:
For cyclin B1: 5′-GCATATGGCCAAGAACATCATCAAGG-3′ and 5′-CATGTTAAAATGAGCTTTATTAAAACCAG-3′;
For cyclin A1: 5′-CACCAATTCTGTCTTGGTGC-3′ and 5′-CAGTTGAGGGGAAGTATTGA-3′;
For cdk1: 5′-CCAAGTGGATCCGACAAGAC-3′ and 5′-CAGCGCTACTTTAGCAGAAAT-3′;
For G10: 5′-CAACTTTGGAACCAACTGTATT-3′ and 5′-CCAGAAGTCAGTTAGAATTGC-3′;
For wee1: 5′-CTCCAGAAACAGCTCAATGT-3′ and 5′-AACACTCGTCCTTCCCAGAA-3′;
For mos: 5′-CCATGGGGCAATTCATACCA-3′ and 5′-GGCCCATTCACACTTCTGAT-3′;
For actin-β: 5′-GAATGCAGAAAGAAATAACTGC-3′ and 5′-TGGAGCCACCAATCCAGAC-3′;
For eIF5: 5′-GCAAAGAGAAAGAAAATGGTTC-3′ and 5′-GCGTCTCTGAGCCTCTGC-3′;
For Rsp6: 5′-GAAGCAGCGTACTCAAAAGAA-3′ and 5′-AGCCTGCGCCTCTTCGC-3′; and
For PIK3R1: 5′-TCCTTGTGCGAGAGAGCAG-3′ and 5′-GAACCCAAAACCAGTATGCG-3′.
UV cross-linking and immunoprecipitation
HEK 293T cells were infected with retrovirus carrying CPEB-HA or Δ297–307 CPEB-HA and incubated overnight to allow protein expression. The cells were homogenized in IP buffer (see above) and incubated with 2 × 106 cpm of mouse cyclin B1 3′ UTR (containing CPEs; in some cases, the CPEs were mutated) in 2× gel retardation buffer (20 mM HEPES at pH 7.6, 2 mM MgCl2, 0.2 mM ZnCl2, 100 mM KCl, 20% glycerol, and 2 mM DTT) supplemented with 2.5 mg/mL heparin, 50 μg/mL tRNA, 0.5 mM DTT, and 0.6 unit/μL RNaseOUT, 10 min on ice and 10 min at room temperature. The protein–RNA mixture (20 μL) was applied per well on a Nunclon Δ Surface plate (Nunc) and UV-irradiated with 440 mJ (Stratalinker UV Crosslinker, Strategene) on iced water. Following 2 μg of RNase A treatment at 37°C for 30 min, the mixture was subject to IP with anti-HA antibody followed by boiling in SDS sample buffer and analysis by Western blotting and autoradiography.
Deadenylation assay
In vitro transcription using mMESSAGE mMACHINE T7 Ultra (Ambion) from linearized pBSSK-xCCNB1C WT or CPE mutant plasmid was performed with 20 μCi UTP-[α-P32]. The mRNA was polyadenylated with Escherichia coli poly(A) polymerase (Ambion) followed by LiCl precipitation. About 103 cpm of polyadenylated mRNA was injected into the nucleus or the cytoplasm of stage VI oocytes. After incubation, the mRNA was recovered in PAS buffer (0.1 M Tris at pH 7.6, 1% SDS, 6% p-Aminosalicylic Acid [PAS] and 1 mM EDTA at pH 8) and extracted with phenol/chloroform. The purified mRNA was analyzed on 4% denaturing polyacrylamide gel (SequaGel Sequencing System, National Diagnostics) and visualized using a PhosphoImager.
For analysis of in vivo transcribed RNA, a ligation-mediated PAT assay was performed (Rassa et al. 2000). RNA extracted from injected oocytes was ligated to 0.5 μg of P1′ primers (5′-P-GGTCACCTTGATCTGAAGC-NH2-3′) using T4 RNA ligase at 37°C for 30 min and inactivated at 95°C for 5 min. The RNA was subjected to an RT reaction with P1-Anchor primer (5′-GCTTCAGATCAAGGTGACCTTTTT-3′), followed by RNase H digestion. The cDNA was used for PCR with P1-Anchor primer and xCCNB1-f(PAT) primer (5′-GTGGCATTCCAATTGTGTATTGTT-3′), supplemented with dATP-[α-P32]. The PCR product was resolved on a 4% polyacrylamide gel.
Exon array analysis
Four pairs of WT and CPEB KO male MEFs from the same litter were collected at passages 3–5. The RNA was purified using Qiagen RNeasy Mini Kit and its integrity was examined by gel electrophoresis. The microarray hybridization (Affymetrix GeneChip Mouse Exon 1.0 ST Arrays) was carried out at the Protein and Nucleic Acid Facility of Stanford School of Medicine. Statistical analysis was performed according to published procedures (Xing et al. 2006, 2008). The inclusion of exon 34 of Col9a1 was validated by RT-PCR with primers: 5′-GGAGATATGGGACAACCTGG-3′ and 5′-GCTGGCTGCCATTTCCGC-3′.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank D. Bentley (University of Colorado), J. Manley (Columbia University), C. Milcarek (University of Pittsburgh), E. Izaurralde (Max Planck Institute), M. Wormington (University of Virginia), E. Wahle (University of Halle), A. Krainer (Cold Spring Harbor Laboratory), and C. Peterson (University of Massachusetts) for gifts of antibodies. We are also grateful to all of the current and previous laboratory members for discussions and technical support, especially J.H. Kim (Research Institute, National Cancer Center of South Korea) and M.-C. Kan (University of Massachusetts). V.E. was supported by NRSA F32 CA12496. This work is supported by NIH grants GM46779 and HD37267 (J.D.R.) and NIH grant HG004634 (Y.X.). Additional core support from the Diabetes and Endocrinology Research Center Program Project (DK32520) is gratefully acknowledged.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1779810.
REFERENCES
- Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Bouvet P, Wolffe AP. A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell. 1994;77:931–941. doi: 10.1016/0092-8674(94)90141-4. [DOI] [PubMed] [Google Scholar]
- Braddock M, Thorburn AM, Chambers A, Elliott GD, Anderson GJ, Kingsman AJ, Kingsman SM. A nuclear translational block imposed by the HIV-1 U3 region is relieved by the Tat-TAR interaction. Cell. 1990;62:1123–1133. doi: 10.1016/0092-8674(90)90389-v. [DOI] [PubMed] [Google Scholar]
- Brogna S, Wen J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat Struct Mol Biol. 2009;16:107–113. doi: 10.1038/nsmb.1550. [DOI] [PubMed] [Google Scholar]
- Cao Q, Richter JD. Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 2002;21:3852–3862. doi: 10.1093/emboj/cdf353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Xu N, Zhu W, Shyu AB. Functional dissection of hnRNP D suggests that nuclear import is required before hnRNP D can modulate mRNA turnover in the cytoplasm. RNA. 2004;10:669–680. doi: 10.1261/rna.5269304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson KS, Bilger A, Ballantyne S, Wickens MP. The cleavage and polyadenylation specificity factor in Xenopus laevis oocytes is a cytoplasmic factor involved in regulated polyadenylation. Mol Cell Biol. 1999;19:5707–5717. doi: 10.1128/mcb.19.8.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du TG, Jellbauer S, Muller M, Schmid M, Niessing D, Jansen RP. Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA. EMBO Rep. 2008;9:781–787. doi: 10.1038/embor.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernoult-Lange M, Wilczynska A, Harper M, Aigueperse C, Dautry F, Kress M, Weil D. Nucleocytoplasmic traffic of CPEB1 and accumulation in Crm1 nucleolar bodies. Mol Biol Cell. 2009;20:176–187. doi: 10.1091/mbc.E08-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman I, Ivshina M, Marin V, Kennedy NJ, Davis RJ, Richter JD. Control of cellular senescence by CPEB. Genes & Dev. 2006;20:2701–2712. doi: 10.1101/gad.1438906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Deng Y, Zenklusen D, Singer RH. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes & Dev. 2004;18:1452–1465. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- Hake LE, Richter JD. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- Hake LE, Mendez R, Richter JD. Specificity of RNA binding by CPEB: Requirement for RNA recognition motifs and a novel zinc finger. Mol Cell Biol. 1998;18:685–693. doi: 10.1128/mcb.18.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Huynh JR, Munro TP, Smith-Litiere K, Lepesant JA, St Johnston D. The Drosophila hnRNPA/B homolog, Hrp48, is specifically required for a distinct step in osk mRNA localization. Dev Cell. 2004;6:625–635. doi: 10.1016/s1534-5807(04)00130-3. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- Keon BH, Schäfer S, Kuhn C, Grund C, Franke WW. Symplekin, a novel type of tight junction plaque protein. J Cell Biol. 1996;134:1003–1018. doi: 10.1083/jcb.134.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Richter JD. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Kim JH, Richter JD. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes & Dev. 2007;21:2571–2579. doi: 10.1101/gad.1593007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong YW, Cannell IG, de Moor CH, Hill K, Garside PG, Hamilton TL, Meijer HA, Dobbyn HC, Stoneley M, Spriggs KA, et al. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci. 2008;105:8866–8871. doi: 10.1073/pnas.0800650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause S, Fakan S, Weis K, Wahle E. Immunodetection of poly(A) binding protein II in the cell nucleus. Exp Cell Res. 1994;214:75–82. doi: 10.1006/excr.1994.1235. [DOI] [PubMed] [Google Scholar]
- Kress TL, Yoon YJ, Mowry KL. Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J Cell Biol. 2004;165:203–211. doi: 10.1083/jcb.200309145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RA, Gagnon JA, Mowry KL. PTB/hnRNP I is required for RNP remodeling during RNA localization in Xenopus oocytes. Mol Cell Biol. 2008;28:678–686. doi: 10.1128/MCB.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. Nonsense-mediated mRNA decay: Splicing, translation, and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Wassarman KM, Wolffe AP. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 1998;17:2107–2121. doi: 10.1093/emboj/17.7.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Richter JD. Translational control by CPEB: A means to the end. Nat Rev Mol Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature. 2000a;404:302–307. doi: 10.1038/35005126. [DOI] [PubMed] [Google Scholar]
- Mendez R, Murthy KG, Ryan K, Manley JL, Richter JD. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol Cell. 2000b;6:1253–1259. doi: 10.1016/s1097-2765(00)00121-0. [DOI] [PubMed] [Google Scholar]
- Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- Rassa JC, Wilson GM, Brewer GA, Parks GD. Spacing constraints on reinitiation of paramyxovirus transcription: The gene end U tract acts as a spacer to separate gene end from gene start sites. Virology. 2000;274:438–449. doi: 10.1006/viro.2000.0494. [DOI] [PubMed] [Google Scholar]
- Rouget C, Papin C, Mandart E. Cytoplasmic CstF-77 protein belongs to a masking complex with cytoplasmic polyadenylation element-binding protein in Xenopus oocytes. J Biol Chem. 2006;281:28687–28698. doi: 10.1074/jbc.M601116200. [DOI] [PubMed] [Google Scholar]
- Shell SA, Hesse C, Morris SM, Jr, Milcarek C. Elevated levels of the 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection. J Biol Chem. 2005;280:39950–39961. doi: 10.1074/jbc.M508848200. [DOI] [PubMed] [Google Scholar]
- Shen Z, Paquin N, Forget A, Chartrand P. Nuclear shuttling of She2p couples ASH1 mRNA localization to its translational repression by recruiting Loc1p and Puf6p. Mol Biol Cell. 2009;20:2265–2275. doi: 10.1091/mbc.E08-11-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya T, Tange TØ, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat Struct Mol Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- Smillie DA, Sommerville J. RNA helicase p54 (DDX6) is a shuttling protein involved in nuclear assembly of stored mRNP particles. J Cell Sci. 2002;115:395–407. doi: 10.1242/jcs.115.2.395. [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JD. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol Cell. 1999;4:1017–1027. doi: 10.1016/s1097-2765(00)80230-0. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vethantham V, Rao N, Manley JL. Sumoylation modulates the assembly and activity of the pre-mRNA 3′ processing complex. Mol Cell Biol. 2007;27:8848–8858. doi: 10.1128/MCB.01186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Ongkasuwan J, Standart N, Steitz JA. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes & Dev. 2001;15:774–788. doi: 10.1101/gad.872201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Kapur K, Wong WH. Probe selection and expression index computation of Affymetrix exon arrays. PLoS One. 2006;1:e88. doi: 10.1371/journal.pone.0000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Stoilov P, Kapur K, Han A, Jiang H, Shen S, Black DL, Wong WH. MADS: A new and improved method for analysis of differential alternative splicing by exon-tiling microarrays. RNA. 2008;14:1470–1479. doi: 10.1261/rna.1070208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, López de Quinto S, Matsui Y, Shevchenko A, Shevchenko A, Ephrussi A. Hrp48, a Drosophila hnRNPA/B homolog, binds and regulates translation of oskar mRNA. Dev Cell. 2004;6:637–648. doi: 10.1016/s1534-5807(04)00132-7. [DOI] [PubMed] [Google Scholar]