Abstract
3′ Untranslated region (UTR)-dependent post-transcriptional regulation has emerged as a critical mechanism of controlling gene expression in various physiological contexts, including cellular differentiation events. Here, we examine the regulation of the 3′UTR of the die-1 transcription factor in a single neuron of the nematode C. elegans. This 3′UTR shows the intriguing feature of being differentially regulated across the animal's left/right axis. In the left gustatory neuron, ASEL, in which DIE-1 protein is normally expressed in adult animals, the 3′UTR confers no regulatory information, while in the right gustatory neuron, ASER, where DIE-1 is normally not expressed, this 3′UTR confers negative regulatory information. Here, we systematically analyze the cis-regulatory architecture of the die-1 3′UTR using a transgenic, in vivo assay system. Through extensive mutagenesis and sequence insertions into heterologous 3′UTR contexts, we describe three 25-base-pair (bp) sequence elements that are both required and sufficient to mediate the ASER-specific down-regulation of the die-1 3′UTR. These three 25-bp sequence elements operate in both a redundant and combinatorial manner. Moreover, there are not only redundant elements within the die-1 3′UTR regulating its left/right asymmetric activity but asymmetric 3′UTR regulation is itself redundant with other regulatory mechanisms to achieve asymmetric DIE-1 protein expression and function in ASEL versus ASER. The features of 3′UTR regulation we describe here may apply to some of the vast number of genes in animal genomes whose expression is predicted to be regulated through their 3′UTR.
Keywords: C. elegans, 3′UTR, miRNA, neuronal cell fate, cis-regulatory elements
INTRODUCTION
3′ Untranslated regions (3′UTRs) of mRNAs represent a rich playground for post-transcriptional gene regulatory events (Kuersten and Goodwin 2003; Wilkie et al. 2003; de Moor et al. 2005). These regulatory events are mediated by often poorly defined cis-regulatory elements embedded in 3′UTRs, and impact on various aspects of the activity of a mRNA, such as its translation or stability. Trans-acting factors that act through cis-regulatory elements in 3′UTRs have been relatively sparse until it was discovered, just a few years ago, that metazoans contain hundreds if not thousands of small regulatory RNAs of the miRNA family that bind to cis-regulatory elements in 3′UTR (and also elsewhere in mRNAs) (Ambros 2004; Ruvkun et al. 2004). Since miRNAs bind to their cis-regulatory sites in mRNAs with at least partial complementarity, computational algorithms have been developed to predict miRNA target interactions (Watanabe et al. 2007). Such algorithms predict that a substantial fraction (∼30%) of all 3′UTRs encoded in a genome contain cis-regulatory elements that may be targeted by miRNAs. However, relatively few studies have experimentally investigated whether a given 3′UTR (e.g., one that contains miRNA binding sites) indeed contains gene regulatory information, and even fewer studies have used unbiased experimental approaches to dissect the composition and underlying regulatory logic of cis-regulatory elements in 3′UTRs. The definition of cis-regulatory elements in 3′UTRs, therefore, significantly lags behind the definition of cis-regulatory elements hardwired into DNA and controlling transcription.
The experimental dissection of the regulatory properties and cis-regulatory elements of 3′UTRs has often led to unexpected results. For example, mutational analyses of three miRNA-regulated 3′UTRs in the nematode Caenorhabditis elegans, the lin-14 3′UTR (regulated by the lin-4 miRNA), the lin-41 3′UTR (regulated by the let-7 miRNA), and the cog-1 3′UTR (regulated by the lsy-6 miRNA) have revealed insights into the sequence features required for miRNA/mRNA target recognition that computational approaches fail to accurately predict (Ha et al. 1996; Vella et al. 2004; Didiano and Hobert 2006, 2008). These examples corroborate the importance of unbiased experimental analysis of 3′UTR regulation. These examples are also noteworthy because they demonstrate the power of using an appropriate in vivo system to analyze 3′UTR activity. Specifically, C. elegans allows one to easily generate transgenic animals that express a gfp-based reporter gene construct fused to a 3′UTR, thereby allowing us to assay 3′UTR activity not only in live animals, but also within their normal cellular context. This contrasts the heterologous cell culture contexts in which 3′UTR/miRNA interactions are often examined. In this article, we use the power of the C. elegans system to probe the regulatory capacity of another 3′UTR, that of the die-1 Zn finger transcription factor.
The die-1 gene displays an intriguing pattern of regulation. DIE-1 protein is expressed in a taste receptor neuron of the nematode C. elegans, called the left ASE (ASEL) (Chang et al. 2004). This taste receptor neuron is one of a pair of morphologically bilaterally symmetric neurons, the ASEL neuron and the right ASE (ASER) neuron. Even though scores of genes are expressed in both ASEL and ASER, DIE-1 protein expression is restricted to ASEL in adult animals, where it is required for appropriate execution of ASEL cell fate (Chang et al. 2004). The restriction of DIE-1 protein expression to ASEL is mimicked by its 3′UTR; that is, if the die-1 3′UTR is fused to a reporter gene that is transcribed in both ASEL and ASER, the reporter becomes down-regulated in ASER (Chang et al. 2004).
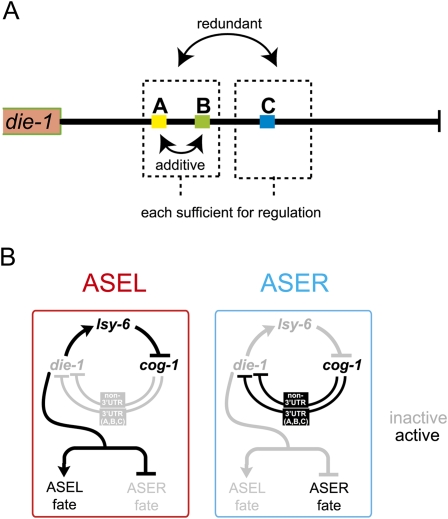
The differential regulation of the die-1 3′UTR in ASEL versus ASER is not the only 3′UTR that is differentially regulated in ASEL versus ASER. Another transcription factor, the COG-1 homeodomain protein, displays a reciprocal regulation to that of the DIE-1 transcription factor (Fig. 1A)—rather than being down-regulated in ASER, the COG-1 protein is down-regulated in ASEL (Chang et al. 2003). This down-regulation depends on the 3′UTR of the cog-1 gene and a cognate, ASEL-expressed miRNA, lsy-6 (Johnston and Hobert 2003). The 3′UTR regulation of cog-1 and die-1 depends on each other. The cog-1 gene, expressed in ASER, is genetically required for the down-regulation of the die-1 3′UTR in ASER, and the die-1 gene, expressed in ASEL, is genetically required for the down-regulation of the cog-1 3′UTR in ASEL (Johnston et al. 2005). These two factors are therefore arranged in the configuration of a double-negative bistable feedback loop (Fig. 1A).
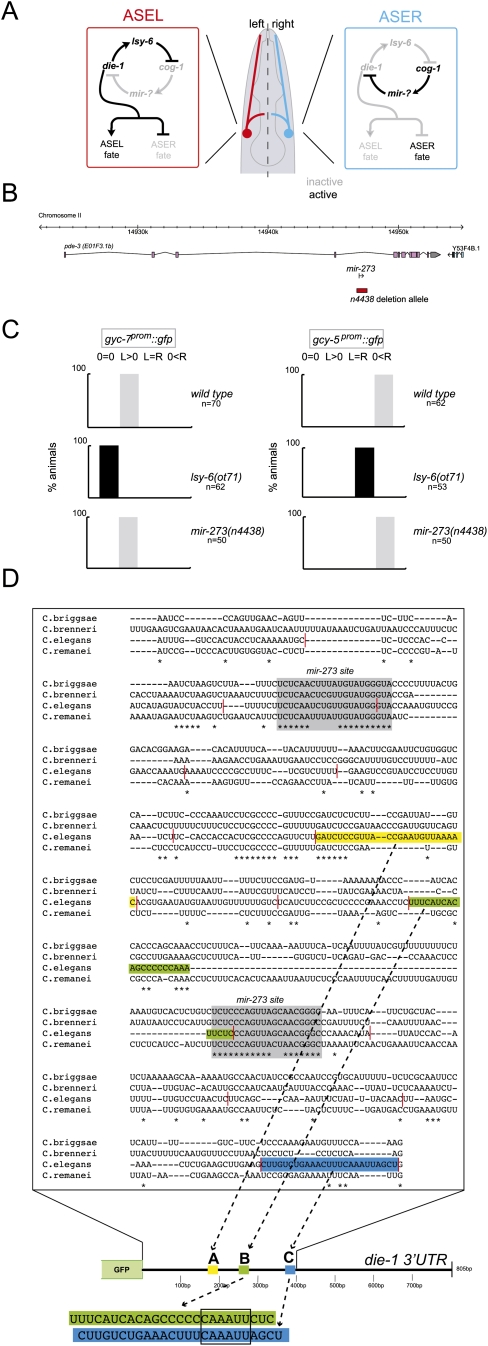
FIGURE 1.
Laterality of the ASE neurons. (A) Model of gene regulatory network responsible for the generation of ASE asymmetry (Hobert 2006). (B) mir-273 locus and deletion allele tested in this analysis. The deletion allele is from Miska et al. (2007). (C) mir-273 null mutant animals do not have an ASE asymmetry phenotype. lsy-6 loss-of-function and control data taken from Johnston and Hobert (2003). (D) Alignment of the C. elegans die-1 3′UTR with three other related nematode species. Multiple sequence alignments were generated with T-coffee (Notredame et al. 2000). Only the first 400 bp of the die-1 3′UTR is shown because it contains all regulatory information (see Fig. 3). Gray boxes indicate the most conserved, extended sequenced patches, both of which show matches to the mir-273 family of miRNAs (for miRNA/target pairing, see Supplemental Fig. 1). Sequence elements identified by mutational analysis motifs to be involved in 3′UTR regulation are colored. Red lines indicate the extent of the 25-bp scanning mutagenesis windows (Supplemental Figs. 2, 3).
The trans-acting factor that regulates the cog-1 3′UTR is well characterized. The die-1-regulated lsy-6 miRNA acts through two complementary sites in the cog-1 3′UTR to down-regulate cog-1. The impact of lsy-6 on cog-1 regulation has been documented by a variety of means, including mutant analysis of the cog-1 3′UTR, lsy-6 mutant alleles, and ectopic expression of lsy-6 (Johnston and Hobert 2003; Sarin et al. 2007; Didiano and Hobert 2008). The trans-acting factors that regulate the die-1 3′UTR are less well characterized. The die-1 3′UTR contains two phylogenetically conserved sites with complementarity to a family of seven related miRNAs (mir-273, mir-51, mir-52, mir-53, mir-54, mir-55, mir-56) (Chang 2004; Chang et al. 2004). As one would expect from down-regulation of the die-1 3′UTR in ASER, but not ASEL, several of these miRNAs are expressed in ASER, but not ASEL (Chang 2004; Chang et al. 2004). Moreover, ectopic expression of mir-273 (Chang et al. 2004) and other members of this family (this article) in ASEL results in down-regulation of die-1 activity in ASEL. However, no loss-of-function analysis of any of these miRNAs has been described to date. This is important as one can easily envision that misexpression/overexpression of miRNAs may reveal nonphysiological effects. In this article, we describe such genetic loss-of-function analysis, demonstrating that in spite of their appropriate expression and regulation, mir-273 family members do not appear to be genetically required regulators of die-1. Taking this result as a starting point, we then go on to undertake an unbiased mutational analysis of the die-1 3′UTR, leading to the identification of several cis-regulatory elements required for left/right asymmetric die-1 3′UTR regulation in the ASEL versus ASER neuron. These cis-regulatory elements act in a combinatorial and redundant manner to control 3′UTR activity. This regulatory logic is strikingly distinct from that of the cog-1 3′UTR, which is absolutely dependent on a single, nonredundant trans-acting factor. We also show that the 3′UTR regulation of cog-1 appears to be the dominant principle of left/right asymmetric regulation of cog-1 expression, while, in contrast, 3′UTR regulation does not appear to be required for restricting die-1 expression to ASEL versus ASER. The ASEL/ASER cell fate decision therefore embodies different principles of 3′UTR regulation.
RESULTS
Analysis of ASE laterality in miRNA mutants
A collection of reverse engineered miRNA knockout alleles (Fig. 1B; Miska et al. 2007) allowed us to test the prediction that mir-273 controls die-1 expression and, therefore, ASEL/R laterality. This prediction is based on (1) left/right asymmetric (“lateralized”) expression of mir-273 in ASER, where die-1 is down-regulated (Chang et al. 2004); (2) the ability of mir-273 to induce ASER fate upon ectopic misexpression (Chang et al. 2004); and (3) the phylogenetic conservation of the two predicted binding sites of mir-273 in the die-1 3′UTR (which is particularly notable as little else in the die-1 3′UTR is conserved) (Fig. 1). In spite of these predictions, we find that a null mutation of mir-273 has no effect on the left/right asymmetric adoption of ASEL and ASER fate, nor on the left/right asymmetric expression of a die-1 3′UTR sensor construct (Fig. 1C; Supplemental Table 1).
mir-273 is a member of a family of related miRNAs, the mir-51 family, of which there are seven members (mir-51 through mir-56 in addition to mir-273). Those miRNA also show potential pairing to the predicted mir-273 sites in the die-1 3′UTR (Supplemental Fig. 1). At least three additional members (mir-54, mir-55, and mir-56) are expressed in ASE with a strong bias to ASER, as would be expected from trans-acting factors down-regulating the die-1 3′UTR in ASER (Chang 2004). Moreover, as observed with mir-273, ectopic misexpression of several of these family members in ASEL can drive ASER fate in ASEL (Supplemental Table 2). However, recently generated knockout alleles that remove each of those family members (Miska et al. 2007) also do not affect ASEL/R laterality of 3′UTR sensor expression (Supplemental Table 1). As these miRNAs may act redundantly, we generated several multiple mutant combinations, including a septuple mutant strain in which all mir-51 family members are removed. Although these septuple mutants display embryonic arrest, they live long enough to develop ASE neurons that can be scored for laterality. We found that none of the analyzed mutant combinations affected ASE laterality in a manner expected from a derepression of die-1 expression in ASER, namely, a “2 ASEL phenotype” (Supplemental Table 1).
Mutational analysis of the die-1 3′UTR
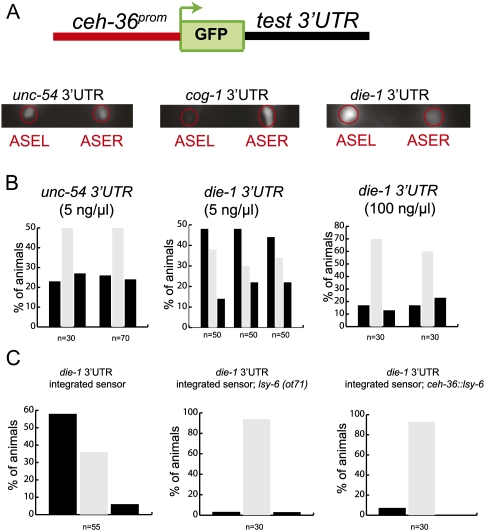
To investigate the mechanisms that control lateralized die-1 3′UTR regulation, we undertook an unbiased mutational analysis of the 3′UTR. We used a previously described sensor system to report on left/right asymmetric die-1 3′UTR regulation (Fig. 2A,B). As previously reported (Chang et al. 2004), this sensor contains the bilaterally (i.e., ASEL + ASER) expressed ceh-36 promoter, gfp, and the die-1 3′UTR. This sensor displays—in contrast to a sensor with a control 3′UTR—down-regulation of gfp expression in ASER versus ASEL (Fig. 2). As this sensor system is sensitive to dosage of the sensor transgene (Fig. 2B), we optimized the previously reported die-1 3′UTR sensor (as described by Chang et al. 2004) in various ways so as to minimize the risk of sensor overexpression artifacts (see Materials and Methods). Under these optimized conditions, left/right asymmetric expression is reliably reported by the die-1 3′UTR sensor while a control 3′UTR remains symmetrically expressed (Fig. 2). As expected from our previous analysis (Johnston et al. 2005), the asymmetry of the die-1 3′UTR reporter is regulated by the bistable feedback loop controlling functional ASE laterality (Fig. 2C). From here on we refer to the differential expression of the 3′UTR in ASEL and ASER as “asymmetric 3′UTR regulation.” The experimental power of this system lies in the facts (1) that it bears an internal control against which the extent of 3′UTR regulation can be compared (gfp fluorescence in ASEL versus ASER); (2) that it assays 3′UTR activity in the correct cellular context in which 3′UTR normally occurs, rather than in the heterologous cellular context that many 3′UTR studies utilize; and (3) that it does not average variable expression levels of heterologous cellular population, but rather is a single cell assay.
FIGURE 2.
Features of the die-1 3′UTR sensor system. (A) Schematic of the die-1 3′UTR sensor and gfp images that show expression of the sensor in transgenic adult worms (all scoring throughout this article is done in young adults). ASEL and ASER are labeled by red circles. AWC neurons that serve as internal control are out of the plane of view. The data on unc-54 and cog-1 have been described before and are taken from Didiano and Hobert (2006). (B) Quantification of 3′UTR sensor expression. Each set of three bars represents an independent transgenic line. Individual animals are scored and binned into one of three categories: gfp expression in ASEL > ASER (left black bar), ASEL = ASER (middle gray bar) or ASER > ASEL (right black bar). Multiple transgenic lines of the same construct are shown for unc-54 and die-1 3′UTRs. Constructs were injected at 5 ng/μL. Injection of the wild-type die-1 3′UTR sensor construct at high concentrations (100 ng/μL) results in a loss of the observed regulation. The unc-54 data were freshly rescored. (C) die-1 3′UTR regulation is dependent upon ASE asymmetry. An integrated die-1 3′UTR sensor construct (otIs260) displays slightly higher levels of regulation than the extrachromosomal lines shown in panel B. Regulation of the integrated die-1 3′UTR sensor construct is lost in 2-ASER (lsy-6) or 2-ASEL (ceh-36∷lsy-6) mutants. The lsy-6 allele used (ot71) has a 100% 2-ASER phenotype (Johnston and Hobert 2003). The ceh-36∷lsy-6 strain otIs204 expresses lsy-6 bilaterally in ASEL and ASER, causing a near 100% 2-ASEL phenotype, as previously described (Ortiz et al. 2009).
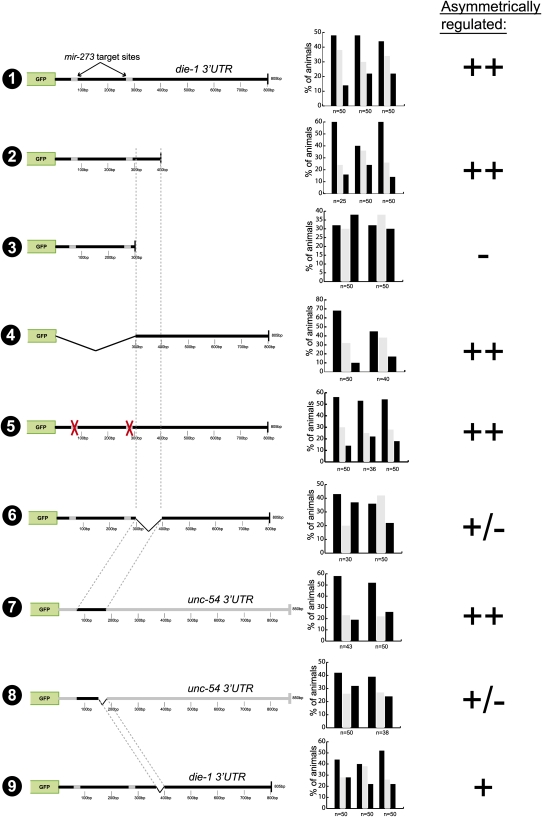
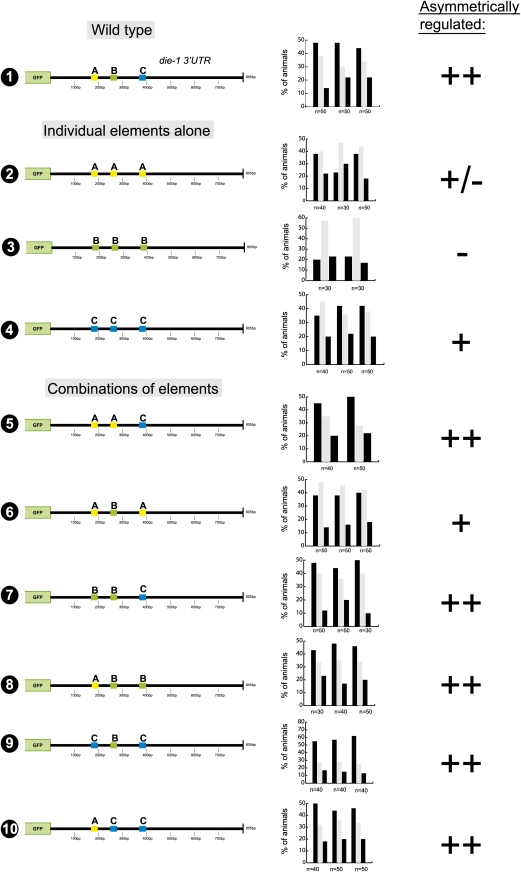
To define regulatory regions within the die-1 3′UTR, we deleted parts of the ∼800-bp 3′UTR and found that a 400-bp part of the 3′UTR is sufficient to confer left/right asymmetric regulation (Fig. 3, construct 2). These 400 bp are at best moderately conserved between several distinct C. elegans species with only two patches of extended conservation, corresponding to the sequences that are complementary to the mir-273 family (Fig. 1C; Supplemental Fig. 1). We generated a systematic collection of 3′UTR reporters in which consecutive 25-bp windows in the 400-bp 3′UTR are deleted. This mutational scanning analysis revealed that no single element is required for the asymmetric regulation of the 3′UTR (Supplemental Fig. 2). Note that these individual deletions removed either mir-273 site, as well as other small patches of conserved sequences.
FIGURE 3.
Mutational analysis of the die-1 3′UTR. Primary scoring data for multiple transgenic lines is shown, which is translated into the symbols ++, +, +/−, and −, which are used to indicate the levels of differential regulation of gfp expression in ASEL versus ASER (“asymmetric regulation”). These categories are based on asymmetry indices (A.I.s) calculated for each transgenic line, as described in the Materials and Methods.
Deletion of another 100 bp from this minimal 400-bp element completely abolished asymmetric 3′UTR regulation (Fig. 3, construct 3). Note that the remaining 300-bp fragment still contains the two mir-273 sites, yet is not asymmetrically regulated. This argues that these sites are not sufficient to confer regulation. In addition, mutation of both mir-273 sites simultaneously in the context of the whole die-1 3′UTR showed that the two sites are also not required for asymmetric 3′UTR regulation (Fig. 3, construct 5). Even though these findings differ from a previous mutational analysis of the mir-273 sites, in which different assay conditions were used (Chang et al. 2004; see Materials and Methods), they are consistent with the genetic analysis described above in which we noted no significant effect of the loss of mir-273 family members on ASE asymmetry (Fig. 1).
Defining three regulatory elements involved in asymmetric 3′UTR regulation
The definition of a 400-bp minimal element (sequence shown in Fig. 1C) and the importance of 100 bp at the 3′end of this minimal element provided us with a starting point for a further dissection of the 3′UTR. We found that the 100-bp element required for regulation of the 400-bp element is also required for regulation within the complete 3′UTR (Fig. 3, construct 6). Moreover, if they are transplanted into the heterologous and normally unregulated unc-54 3′UTR, these 100 bp are also sufficient to confer asymmetric 3′UTR regulation (Fig. 3, construct 7). The sufficiency of the 100-bp element to confer asymmetric regulation in the unc-54 3′UTR is largely impaired, if not abolished upon deletion of 25 bp at the 3′end of the 100-bp element (Fig. 3, in construct 8 removal of any other 25 bp from the first 75 bp did not result in a loss of regulation; data not shown). Surprisingly, when these 25 bp (called the C element) (Fig. 1D, indicated in color) are deleted in the context of the whole die-1 3′UTR, the sensor is still regulated (Fig. 3, construct 9). These findings argue that the 25-bp C element is required in an isolated context, but its loss can be compensated for by other redundantly working elements in the context of the complete die-1 3′UTR.
To identify these redundant elements we undertook another systematic scanning deletion analysis, deleting 25-bp windows at a time, but this time in the context of the deletion of the C element (Supplemental Fig. 3). This analysis revealed two additional elements (Fig. 1C, indicated in color), “A” (position 175–200 bp) and “B” (position 250–275 bp), which when deleted in the absence of the C element result in a loss of asymmetric 3′UTR regulation. Results with all deletion constructs are shown in Supplemental Figure 3, and the relevant constructs are shown in Figure 4. Note that the initial scanning analysis (Supplemental Fig. 2) had shown that neither deletion of the A nor the B element alone has any effect on 3′UTR regulation. Moreover, deletion of A and B without deleting C (Fig. 4, construct 5) does not abolish asymmetric 3′UTR regulation. Like the A + C and B + C double deletions, deleting all three sites together leads to a complete loss of regulation as well (Fig. 4, construct 6). Taken together, we have identified three 25-bp elements in the die-1 3′UTR that act in a redundant/combinatorial manner to post-transcriptionally down-regulate the expression of the die-1 3′UTR in ASER.
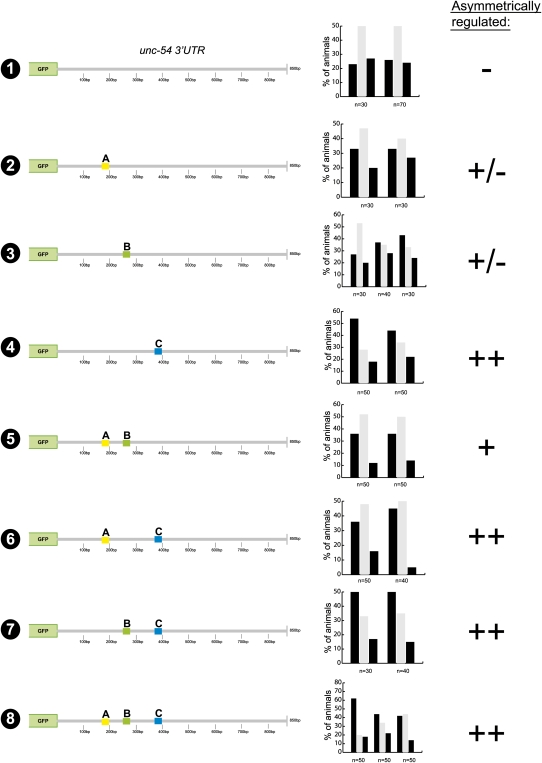
FIGURE 4.
Identification of three cis-regulatory elements required for asymmetric regulation of the die-1 3′UTR in ASEL versus ASER. For explanation of the symbols, see the Figure 3 legend. Gray boxes are the conserved mir-273 sites shown in Figure 1C.
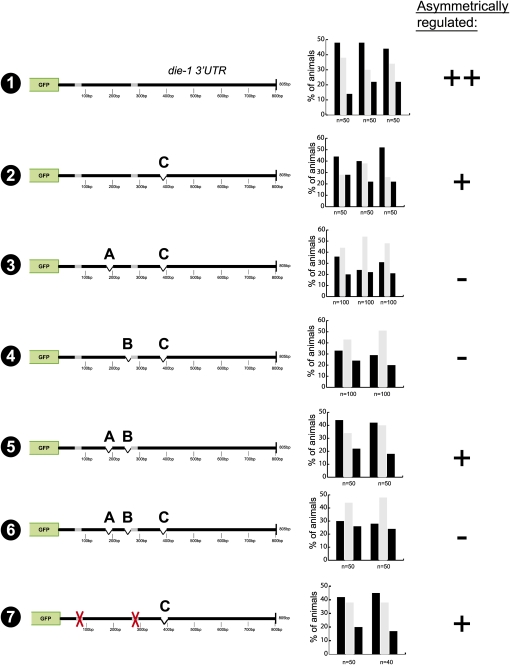
In order to further investigate the redundant and combinatorial nature of this regulation, we generated synthetic variations of the die-1 3′UTR with various combinations of the three identified 25-bp elements, in which one of the elements replaces one or both of the other identified elements (Fig. 5). This analysis shows that 3′UTRs that contain the C element (375–400 bp) always display regulation, indicating that the C element is sufficient to confer regulation in the absence of the other sequence motifs. Three copies of the A or B element display weak or no regulation. However, 3′UTRs containing at least one copy of the A element plus the B element display regulation. This is consistent with the initial scanning analysis in which loss of element C alone (preserving A and B) still displays regulation. In sum, this analysis points to a regulatory mechanism entailing both independent and combinatorial regulation, with the C element acting independently and redundantly with a combination of the A and B elements.
FIGURE 5.
die-1 3′UTR sequence motif replacement experiments indicate that sequence motifs act in a redundant and combinatorial manner. For explanation of the symbols, see the Figure 3 legend.
Given the apparently redundant and combinatorial nature of die-1 3′UTR regulation, we asked whether the two sites for the ASER-expressed and regulatory-loop controlled miRNA mir-273 may work in a redundant manner with any of the newly identified regulatory elements. A role for the mir-273 sites is not revealed in the absence of the A and B elements since a construct in which the entire proximal 300 bp of the die-1 3′UTR is deleted—including the A and B elements and the two mir-273 sites, but still retaining the C element—still is regulated (Fig. 3, construct 4). An impact of the double deletion of both mir-273 sites is also not revealed in the absence of the C element (Fig. 4, construct 7).
Cis-regulatory elements are sufficient to confer regulation to a heterologous context
We further probed the three defined cis-regulatory elements by assessing whether these elements also work in a similar manner in a heterologous 3′UTR context and are therefore sufficient to confer regulation. To this end, we transferred the three 25-bp elements independently, and in combination, into the heterologous and normally unregulated unc-54 3′UTR. The motifs were inserted with a similar distance away from the stop codon as they are in the wild-type die-1 3′UTR, and we also maintained the relative distance between the sequence motifs (Fig. 6). Of the three sequence elements, only the C element is sufficient to confer regulation when inserted into the unc-54 3′UTR alone (Fig. 6, constructs 2–4). Elements A and B are able to transfer regulation when inserted into the unc-54 3′UTR together, but not alone (Fig. 6, construct 5). Insertion of element A, element B, or both in combination does not enhance the level of regulation above that displayed by the insertion of element C alone (Fig. 6, cf. constructs 6, 7, 8 and construct 4). Overall, our results in the heterologous unc-54 3′UTR context are consistent with those found in the mutational experiments within the endogenous die-1 3′UTR context, indicating that all relevant regulatory elements required for die-1 regulation have been identified.
FIGURE 6.
Sequence motifs confer regulation to unc-54 3′UTR. For explanation of symbols, see the Figure 3 legend.
Sequence features of the three regulatory elements
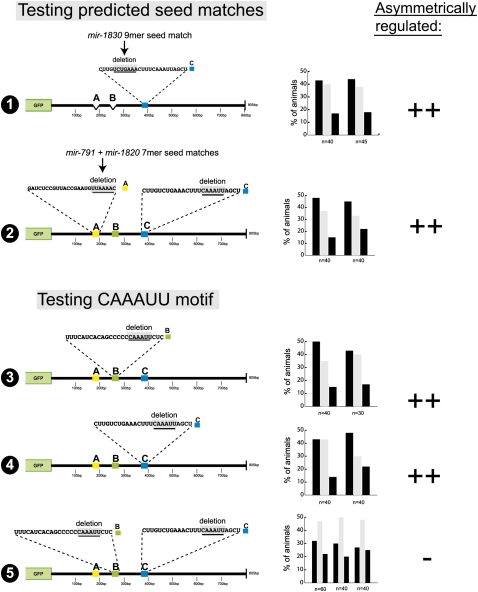
We analyzed the sequence content of the three regulatory elements in more detail by first examining whether any of them contain seed matches to any of the presently known C. elegans miRNAs currently deposited in miRBase (http://microrna.sanger.ac.uk/sequences/). Only the A and C elements contain >6-bp seed matches to miRNAs, namely, mir-791, mir-1820, and mir-1830. However, small 6-bp deletions that remove these seed matches in the A and C elements do not display the loss of 3′UTR regulation observed upon deleting the entire element (Fig. 7, constructs 1, 2).
FIGURE 7.
Mutational analysis of individual sequence motifs with the three cis-regulatory elements. Note that in constructs 1 and 2 the seed mutations were generated in the context of a mutation in redundantly acting elements because it is only in such a “sensitized” construct that loss of an individual element would have an effect (see Fig. 4). The C deletion in construct 2 (testing the seed match in site A) is not a complete 25-bp deletion, but a CAAAUU motif deletion, which mimics the complete 25-bp deletion. Also note that compared to all other constructs in this article, construct 5 defines the most minimal changes that lead to a complete loss of die-1 3′UTR regulation, namely, the loss of two 6-bp motifs of the same sequence. For explanation of symbols, see the Figure 3 legend.
We also compared the sequence of the A, B, and C elements and noted no similarity between A and the other two elements. However, the B and C elements share a core CAAAUU motif (Fig. 1C). We mutated this motif in the B and C elements and found the motif to be required for B and C element function (Fig. 7, constructs 3–5). These data indicate that the B and C elements may be regulated by a common trans-acting factor. No known miRNA is predicted to pair with this motif in a canonical manner.
Differential impact of 3′UTR regulation on die-1 and cog-1 regulation
Not only do regulatory elements in the die-1 3′UTR act redundantly with one another, but the 3′UTR regulation of the die-1 locus is also redundant with other, non-3′UTR–mediated gene regulatory mechanisms. The first hint in this direction was the observation that the die-1 3′UTR reporter, even though down-regulated in ASER, is not completely turned off (Fig. 2A). This is in striking contrast to the 3′UTR of the ASER-inducer cog-1, which is more prominently turned off in ASEL (Fig. 2A; Didiano and Hobert 2006, 2008). That is, even though both 3′UTRs are asymmetrically regulated in a reciprocal manner, the extent of down-regulation of the 3′UTR appears to be distinct. This dichotomy was consistently observed across multiple lines and multiple distinct concentrations of the respective 3′UTR sensor.
This differential down-regulation is in striking contrast to the expression of the DIE-1 and COG-1 protein. Using fosmid-recombineering technology (Tursun et al. 2009), we tagged genomic die-1 and cog-1 loci with fluorescent reporter genes and corroborated the previous conclusion that in adult animals DIE-1 protein expression is restricted to ASEL and COG-1 is restricted to ASER (Chang et al. 2003, 2004); this restriction is absolute, i.e., no residual levels of the tagged protein can be observed in ASEL or ASER in adult animals (Fig. 8A,B). These reporter genes are fully functional and rescue the cog-1 and die-1 mutant phenotypes, respectively (data not shown). The left/right asymmetric regulation of cog-1 and die-1 is functionally relevant since ectopic expression of cog-1 in ASEL converts ASEL into ASER and overexpression of die-1 converts ASER to ASEL (Chang et al. 2003; Johnston et al. 2005; Sarin et al. 2009; data not shown).
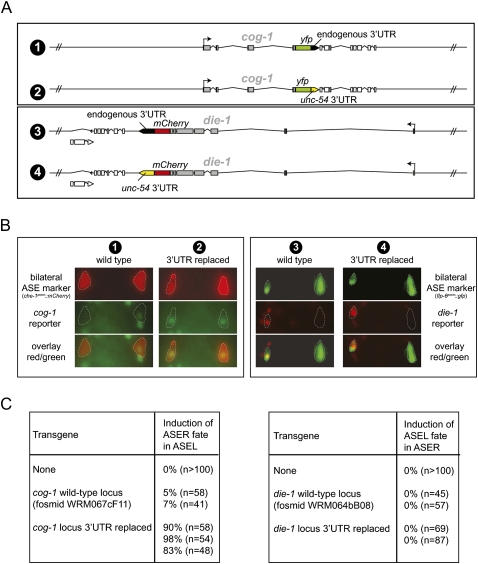
FIGURE 8.
Differential contribution of the 3′UTR for regulation of expression and activity of the die-1 and cog-1 gene. (A) Schematic representation of the fosmid clones used for expression (panel B) and functional (panel C) analysis of die-1 and cog-1. Fosmid clones are around about 40 kbp in size. Fluorescent reporters were first recombineered into the respective fosmids. In a subsequent recombineering step, 3′UTRs were replaced with the unc-54 3′UTR as schematically indicated. Fosmids were all injected at similar concentration to generate transgenic animals. (B) Testing the effect of 3′UTR replacement on the left/right asymmetric expression of die-1 and cog-1. Representative yfp/mCherry expression of transgenic animals expressing the reporter genes shown in panel A. At least two independent transgenic lines were tested for expression, and expression was found to be almost fully penetrant in that, for the cog-1 reporters, the construct with wild-type 3′UTR was asymmetric while the reporter with the unc-54 3′UTRs was not. For the die-1 reporters, both always showed left/right asymmetric expression regardless of the 3′UTR tested. Bilateral markers (flp-6prom∷cameleon = ntIs13 and che-1prom∷mCherry = otIs232) were used to unambiguously identify the ASE neurons (highlighted with stippled lines). Numbers above each panel correspond to the constructs shown in panel A. (C) Testing the effect of 3′UTR replacement on functional derepression of die-1 and cog-1 gene activity. Based on previous work, derepression of die-1 function in ASER is expected to result in a transformation of ASER to ASEL (Johnston et al. 2005), while derepression of cog-1 function in ASEL is expected to result in a transformation of ASEL to ASER (Chang et al. 2003; Johnston et al. 2005; Sarin et al. 2009). Transgenic animals containing reporters with wild-type 3′UTRs or replaced 3′UTR (as shown in panel A) were assayed for their ability to convert cell fate, as assessed with the ASER fate marker gcy-5∷gfp (ntIs1), which is expressed in ASEL if ASER fate is induced (left table) and repressed in ASER if ASEL fate is induced (right table). Each row represents an independent transgenic line. For die-1, we also performed the same experiment with fosmids in which the locus was not tagged with a fluorescent reporter. In that case zero of four lines with the wild-type 3′UTR induced ASEL fate in ASER, and one of four lines with the replaced 3′UTR induced ASEL in ASER in 25% (n = 89) of animals.
However, we find that the functional importance of 3′UTR regulation to achieve the left/right asymmetric activity of cog-1 (in ASER) and die-1 (in ASEL) is strikingly different. We arrived at this conclusion by replacing the 3′UTR of the cog-1 and die-1 loci, in the context of the rescuing fosmids schematically shown in Figure 8A, with the 3′UTR of the unc-54 locus, a 3′UTR that is not differentially regulated in ASEL versus ASER (Fig. 2A,B). We assessed ASEL/R expression through yfp tagging of the respective fosmids; furthermore, we assessed function of these fosmid clones by injecting them into wild-type animals in which ASEL and ASER fate are monitored through terminal cell fate markers. We find that in the case of the cog-1 locus, the replacement of its 3′UTR results in a complete derepression of COG-1∷YFP expression in ASEL (Fig. 8B). This derepression results in a highly penetrant conversion of ASEL fate into ASER fate (Fig. 8C). This finding is consistent with the effects of a dominant cog-1 allele containing a 3′UTR deletion, which causes COG-1 to be derepressed in ASEL (Sarin et al. 2007).
In contrast, replacement of the die-1 3′UTR with the unc-54 3′UTR does not result in depression of DIE-1∷YFP expression in ASER (Fig. 8B). Consequently, there is also no ectopic ASEL fate induction in ASER in transgenic animals expression the 3′UTR-replaced fosmid (Fig. 8C). As mentioned above, animals that are forced to express DIE-1 in ASER can, in principle, result in an ASER to ASEL fate conversion (Johnston et al. 2005; data not shown). Thus, the failure of the 3′UTR-replaced construct to do so corroborates the notion that loss of the 3′UTR does not result in the derepression of functionally significant amounts of DIE-1 in ASER.
In conclusion, these experiments demonstrate that COG-1 expression and activity is primarily restricted to ASER via its 3′UTR, but DIE-1 expression and activity is restricted to ASEL not by its 3′UTR alone, even though the 3′UTR is asymmetrically regulated. In other words, cog-1 left/right asymmetry is primarily and nonredundantly regulated via its 3′UTR, while die-1 left/right asymmetry is regulated by redundant mechanisms, composed of 3′UTR regulation, and other, as yet unknown, means (such as, for example, transcriptional or post-translational regulation).
DISCUSSION
We have employed here a powerful in vivo system to probe the activity of a 3′UTR that is regulated in an intriguing biological context. The dissection of functional elements in a 3′UTR lags far behind the dissection of functional elements embedded in DNA sequences and that, like 3′UTR elements, are important to shape the gene expression profiles of individual cell types. Our analysis demonstrates that core principles of the regulatory logic of DNA-based cis-regulatory elements (i.e., transcriptional regulatory event) can be found in 3′UTR regulation as well, namely, combinatorial activity of cis-regulatory elements and redundancy (Fig. 9; Hobert 2008). Conceptually, the die-1 3′UTR can be divided into two functional modules. The first module contains the A and the B element, and the second module contains the C element. Each module is sufficient to confer regulation in a heterologous context, yet each module acts redundantly with one another in their normal context. Module one is composed of two elements that, themselves, act in a combinatorial manner. The A and B elements are alone not sufficient to confer regulation, but together they can. The A and B elements, furthermore, act independently of the C element. That is, when C is removed, both A and B become important and removal of either (in the absence of C) results in a loss of regulation. Transcriptional control elements often work by similar principles. To give just one out of many examples, separable cis-regulatory elements in the transcriptional control region of the mouse CD4 locus provide redundant cis-regulatory information (Duncan et al. 1996).
FIGURE 9.
Summary. (A) Redundant and combinatorial regulation by three cis-regulatory motifs from the die-1 3′UTR. (B) Revised regulatory loop controlling ASEL/R fate specification. Note that manipulation of cog-1 or lsy-6 activity results in a complete symmetrization of die-1 expression. We have shown here that die-1 regulation occurs on the level of its 3′UTR but also on a non-3′UTR-dependent mechanism, with both mechanisms being cog-1 dependent.
Redundancy of control mechanisms is not only observed on the level of individual regulatory elements in the 3′UTR but also occurs in regard to the overall types of regulatory events that restrict die-1 expression to ASEL. This was revealed by our 3′UTR replacement experiment, which demonstrated that left/right asymmetric die-1 expression is still observed after 3′UTR removal. In other words, the 3′UTR of die-1 is sufficient (at least to a certain extent) to confer left/right asymmetric expression to a reporter protein but is not required to confer asymmetric expression of the DIE-1 protein. It will be interesting to see whether the redundant, non-3′UTR–dependent mechanism that restricts DIE-1 to ASEL (and that appears to be transcriptional) (L Cochella and O Hobert, unpubl.) behaves in a similar way. That is, even though this mechanism (e.g., transcriptional) is apparently sufficient, as shown with our 3′UTR-replaced fosmid reporter; it may or may not be required. For example, if this mechanism were eliminated (e.g., through the deletion of negative transcriptional regulatory motifs that repress DIE-1 expression in ASER), one may not observe much of an effect on asymmetric DIE-1 protein accumulation, as the 3′UTR-dependent mechanism may be able to substitute for the loss of the putative transcriptional mechanism. Such redundant mechanisms of regulating protein expression and function are common. For example, the asymmetric distribution of the SKN-1 transcription factor in distinct blastomeres of the early C. elegans embryo is controlled by two redundant mechanism, one involving protein degradation, the other mRNA translation or stability (Page et al. 2007).
An alternative explanation other than plain redundancy is that the non-3′UTR and the 3′UTR-mediated regulatory mechanisms are not “on par” in terms of their relative importance. The 3′UTR mechanism may have a role so minor that it is too subtle to allow experimental detection under standard conditions. One such difficult-to-identify role would be one in which the 3′UTR provides robustness to die-1 repression in ASEL. One prominent example of this sort of “fail-safe” mechanism is the regulation of Hoxb8 expression by miR-196 in chick, where the Hoxb8 expression domain is primarily determined at the transcriptional level but is sharpened by miR-196 activity (Hornstein et al. 2005). In a robustness-type scenario, the effects of the loss of 3′UTR regulation may only become apparent under relatively extreme external circumstances, as evidenced by the example of miR-7 in flies. The miR-7-mediated regulation of genes involved in photoreceptor development is not apparent in mirR-7 knockout animals under uniform laboratory condition but does become apparent when animals are subject to fluctuating environmental temperature conditions (Li et al. 2009). If the role of the die-1 3′UTR were indeed to confer robustness, then the bistable regulatory network that controls ASE asymmetry would provide a microcosm of what have been proposed to be the two predominant modes of 3′UTR regulatory mechanisms—a robustness-conferring 3′UTR mechanism (die-1) and a “switch-type” 3′UTR regulation (cog-1).
Several more lessons are to be learned from our study. One is that overexpression of miRNAs may overestimate the importance of an individual miRNA species, as exemplified by our assessment of mir-273 involvement. Notably, our case for mir-273 involvement appeared strong not just because of the overexpression results (in which we converted ASEL to ASER) (Chang et al. 2004) but also because mir-273 and other family members are expressed in the right place (ASER) at the right time (Chang 2004; Chang et al. 2004) and regulated by the same factor (cog-1) that regulates the asymmetric activity of the die-1 3′UTR (Johnston et al. 2005). Given that we have clearly shown here that both the 3′UTR itself can be regulated by redundant regulatory mechanisms and that 3′UTR regulation per se is also redundant, we can still not entirely rule out that the mir-51 family (of which mir-273 is a member) plays a role in die-1 3′UTR regulation. Our mir mutant analysis does not rule this out, as we had to use terminal ASE fate markers (a measure of overall die-1 gene activity) rather than the die-1 3′UTR sensor as a readout for the removal of the relevant mir gene (the sensor cannot be scored in embryos when septuple mir mutant animals arrest development). Yet a mir-51-family function is, if it exists, clearly not alone required for die-1 3′UTR regulation.
Another cautionary note resulting from of our studies relates to using sequence conservation as a guide to identify functionally relevant elements in a 3′UTR, a strategy widely used for miRNA/target predictions (Watanabe et al. 2007). There are only two extended patches of sequence conservation in the die-1 3′UTR (the mir-273 sites), and neither appears to be required for 3′UTR regulation, at least not in the cellular context we examined. In contrast, the elements that we do find to be important for regulation show very little conservation. This is reminiscent of our findings in the cog-1 3′UTR, in which, with the exception of the lsy-6 target sites, evolutionary conservation did not correlate with sequences verified as functionally relevant by in vivo mutational analysis (Didiano and Hobert 2008). Moreover, like in the die-1 3′UTR, mutations of conserved sequence motifs in the cog-1 3′UTR had no effect on the observed regulation. Even though one should be cautious in interpreting and generalizing these findings, they serve as a clear illustration of the importance of experimental validation of in silico predictions.
A key question left open by our present work is the identity of the trans-acting factors that control the A, B, and C elements. Those factors may be regulatory RNAs or RNA binding proteins (RBPs) or a combination of both. No known miRNA binds to any of the three experimentally identified sites with canonical seed matches, yet a known miRNA may pair to these sites in a noncanonical manner. Also, as more miRNAs are continuously being identified, it is possible that cognate miRNA(s) remain to be identified. Alternatively, other types of small RNAs may control these elements. Indeed, we found at least one annotated 21U RNA, the C. elegans equivalent of piRNAs (Wang and Reinke 2008), to match to the C element. As no mutant allele is currently available for this RNA, it is not yet possible to assess the importance of this RNA in die-1 3′UTR regulation.
We examined the alternative hypothesis of binding of RBPs to any of these sites by conducting an RNAi screen of all >300 RBPs that are recognizable in the C. elegans genome based on the presence of canonical RNA binding domains. We identified no RBP whose RNAi-mediated down-regulation derepresses die-1 activity in ASEL (Supplemental Fig. 3). As RNAi may not be efficient enough to down-regulate a functionally relevant RBP and as the 3′UTR of die-1 is just one of apparently several mechanisms that down-regulate die-1 in 3′UTR, this negative result has relatively little meaning. The latter argument also does not allow us to distinguish between RBP versus miRNA regulation of the die-1 3′UTR by simply disabling global miRNA function (by removal of Argonautes, Dicer, or Drosha).
Numerous miRNA target prediction algorithms have proposed that a large fraction of 3′UTRs are regulated. However, due to sheer volume, most of this has not of yet been experimentally verified or refuted. Nevertheless, numerous examples have demonstrated that whether it is miRNA-dependent or not, 3′UTR-mediated post-transcriptional regulation is essential for many biological processes (Kuersten and Goodwin 2003; Wilkie et al. 2003; de Moor et al. 2005). The presence of hundreds of RBPs in animal genomes and thousands of small regulatory RNAs of various classes provides a vast pool of trans-acting factors that organisms have at their disposal to target 3′UTR sequences for regulation. Apart from the already much appreciated and pervasive role of 3′UTR regulation in controlling expression of maternally provided mRNAs in early embryogenesis (Kuersten and Goodwin 2003; de Moor et al. 2005), it is conceivable that 3′UTR regulation may also be prominently employed in controlling neuronal laterality. Neuronal laterality is a state that is considered to be ontogenetically and phylogenetically derived from a bilateral ground state (Hobert et al. 2002; Hugdahl and Davidson 2003). 3′UTR regulation may be superimposed onto an ancestral bilateral expression pattern, brought about by transcriptional regulation, to induce and/or sharpen the lateralization of expression patterns (Hobert 2008). 3′UTR regulation may also explain why so few genes have so far been found to be asymmetrically expressed in vertebrates. Most vertebrate expression analysis is done through in situ detection of mRNAs (e.g., Gray et al. 2004; Lein et al. 2007), which measures only mRNA abundance but not translational regulation. Lateralized translational regulation in functionally lateralized brain regions may therefore have escaped notice.
MATERIALS AND METHODS
3′UTR sensor system
The 3′UTR sensor system employed in this analysis of the die-1 3′UTR is based on the expression of gfp under the ASE bilateral promoter (ceh-36) with either control (unc-54) or experimental (die-1) 3′UTRs, and is the same as that utilized in two previous publications examining regulation of the cog-1 3′UTR by the lsy-6 miRNA (Didiano and Hobert 2006, 2008). The ceh-36 promoter is expressed equally in both the ASEL and ASER neurons, as well as another pair of bilateral neurons (AWCL/R), which are used as internal controls for mosaic expression. Therefore, the only variable in the system is the regulatory influence of the 3′UTR tested; this allows each individual transgenic animal to be binned into one of three categories: gfp expression in ASEL > ASER, ASEL = ASER, or ASEL < ASER (scoring is always done in young adult animals). Previous analysis indicates that differences in fluorescent intensity greater than 25% are discernable by eye. After binning the relative expression levels into the three different categories described above, we calculate an asymmetry index (A.I.), which is defined as number of animals displaying [(L > R) – (R > L)]/ total number of animals. This index ranges from −1.0 to 1.0, with either representing complete asymmetry. The A.I. is then averaged across multiple transgenic lines; note that usually there is very little variation of regulation between individual lines of the same construct. The A.I. average is then transformed into a more easily scanned +/− system, with the following, arbitrarily set ranges: A.I. > 0.25 is indicated as “++”; 0.25 > A.I. > 0.15 is indicated as “+”; 0.15 > A.I. > 0.1 is indicated as “+/−”; and A.I. < 0.1 is indicated as “−.”
We noted that multicopy overexpression of the die-1 3′UTR gfp sensor fails to report the correct left/right asymmetric expression of die-1; rather, the sensor is bilaterally expressed (Fig. 2B). Similarly, an analysis of the hid/bantam 3′UTR regulatory relationship found that miRNA-mediated down-regulation is sensitive to 3′ UTR sensor transgene expression concentrations (Nahvi et al. 2009). Overexpressed sensor transgenes may overpower a limiting amount of trans-acting factors that control the 3′UTR. To reduce the risk of these overexpression effects, we altered the protocols compared to the Chang et al. (2004) analysis (which reported on the initial characterization of the die-1 3′UTR) in four ways: (1) All constructs are cloned into the pPD95.75 vector from the Fire Vector Kit as opposed to using the PCR-fusion method from Chang et al. (2004). Vector-based arrays tend to be less multicopy in nature than PCR amplicon-based arrays (Etchberger and Hobert 2008) and can therefore be expected to produce less mRNA copies in a cell. (2) We utilize here a shorter and less highly expressed version of the ceh-36 promoter than that used previously (“ceh-36p2” = 1847 bp, instead of the 5030-bp “ceh-36p1” used by Chang et al. (2004). (3) Injection concentrations were adjusted down to 5 ng/μL. (4) DNA injection concentrations are determined by photo-spectrometer (NanoDrop ND-1000 spectrophotometer) rather than based on gel electrophoresis band-intensity comparisons, which tend to produce more variable results. All these four points may have contributed to the differences in results of the mir-273 site deletions reporter here and by Chang et al. (2004). Note that these altered, improved conditions were also applied in the extensive analysis of the cog-1 3′UTR that we previously described (Didiano and Hobert 2006, 2008).
Generation of 3′UTR sensor constructs and transgenic lines
All die-1 3′UTR sensor constructs are cloned into the same promoter/vector backbone, consisting of 1.8 kb of the ceh-36 promoter driving gfp from the pPD95.75 expression vector. All full-length cloned (nontruncated) die-1 3′UTRs are longer than the endogenous die-1 3′UTR (805 bp; as assessed by an EST clone) by ∼300 bp; however, this extra sequence is most likely removed from the mRNA transcript during poly-adenylation. All die-1 3′UTR sensors are cloned into the EcoR1/Eag1 restriction sites of the gfp expression vector pPD95.75, which replaces the vectors unc-54 3′UTR. With the exception of those constructs described below, mutations were introduced by PCR fusion (Hobert 2002) and then subcloned into the same position. Constructs containing multiple mutations were generated by PCR fusion on templates in which the other mutation(s) were already introduced.
All 3′UTR sensor constructs were injected as plasmids into wild-type N2 worms at 5 ng/μL (if not indicated otherwise in figures) with 100 ng/μL rol-6 as injected marker. Stable transgenic lines were selected in the F2 generation.
Generation of fosmid based reporters
The cog-1 and die-1 fosmid based reporters were generated following the procedure described by Tursun et al. (2009). Fosmids WRM064bB08 (die-1) and WRM067cF11 (cog-1) were recombined in Escherichia coli strain SW105 to introduce a fluorescent protein in frame at the C terminus of each protein of interest. After confirmation of the correct recombination products, subsequent recombinations were performed on the fluorescent protein-containing fosmids to exchange the endogenous 3′UTR for the unregulated unc-54 3′UTR. All constructs were confirmed by sequencing the recombination junctions and the region of interest, digested with restriction enzymes cutting the fosmid backbone, and injected as complex arrays with either rol-6 or elt-2∷dsRed as coinjection markers and bacterial genomic DNA as carrier.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Q. Chen for expert technical assistance in generation transgenic lines and Ezequiel Alvarez-Saavedra, Eric Miska, and Robert H. Horvitz for kindly providing miRNA deletion alleles prior to publication. We acknowledge funding by the NIH (R01NS039996-05; R01NS050266-03). D.D. was funded by an NIH predoctoral fellowship, B.T. is funded by a Francis Goelet Postdoctoral Fellowship, and L.C. is funded by a postdoctoral fellowship from the Helen Hay Whitney Foundation. O.H. is an Investigator of the HHMI.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1931510.
REFERENCES
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Chang SL. Columbia University; New York: 2004. “Development of left/right asymmetry in chemosensory neurons in Caenorhabditis elegans.”. PhD thesis, [Google Scholar]
- Chang S, Johnston RJ, Jr, Hobert O. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes & Dev. 2003;17:2123–2137. doi: 10.1101/gad.1117903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Johnston RJ, Frokjaer-Jensen C, Lockery S, Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- de Moor CH, Meijer H, Lissenden S. Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin Cell Dev Biol. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Molecular architecture of a miRNA-regulated 3′ UTR. RNA. 2008;14:1297–1317. doi: 10.1261/rna.1082708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan DD, Adlam M, Siu G. Asymmetric redundancy in CD4 silencer function. Immunity. 1996;4:301–311. doi: 10.1016/s1074-7613(00)80438-0. [DOI] [PubMed] [Google Scholar]
- Etchberger JF, Hobert O. Vector-free DNA constructs improve transgene expression in C. elegans. Nat Methods. 2008;5:3. doi: 10.1038/nmeth0108-3. [DOI] [PubMed] [Google Scholar]
- Gray PA, Fu H, Luo P, Zhao Q, Yu J, Ferrari A, Tenzen T, Yuk DI, Tsung EF, Cai Z, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- Ha I, Wightman B, Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes & Dev. 1996;10:3041–3050. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hobert O. Architecture of a microRNA-controlled gene regulatory network that diversifies neuronal cell fates. Cold Spring Harb Symp Quant Biol. 2006;71:181–188. doi: 10.1101/sqb.2006.71.006. [DOI] [PubMed] [Google Scholar]
- Hobert O. Gene regulation by transcription factors and microRNAs. Science. 2008;319:1785–1786. doi: 10.1126/science.1151651. [DOI] [PubMed] [Google Scholar]
- Hobert O, Johnston RJ, Jr, Chang S. Left–right asymmetry in the nervous system: The Caenorhabditis elegans model. Nat Rev Neurosci. 2002;3:629–640. doi: 10.1038/nrn897. [DOI] [PubMed] [Google Scholar]
- Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- Hugdahl K, Davidson RJ. The asymmetrical brain. MIT Press; Cambridge, MA: 2003. [Google Scholar]
- Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Jr, Chang S, Etchberger JF, Ortiz CO, Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc Natl Acad Sci. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuersten S, Goodwin EB. The power of the 3′ UTR: Translational control and development. Nat Rev Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137:273–282. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahvi A, Shoemaker CJ, Green R. An expanded seed sequence definition accounts for full regulation of the hid 3′ UTR by bantam miRNA. RNA. 2009;15:814–822. doi: 10.1261/rna.1565109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Ortiz CO, Faumont S, Takayama J, Ahmed HK, Goldsmith AD, Pocock R, McCormick KE, Kunimoto H, Iino Y, Lockery S, et al. Lateralized gustatory behavior of C. elegans is controlled by specific receptor-type guanylyl cyclases. Curr Biol. 2009;19:996–1004. doi: 10.1016/j.cub.2009.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page BD, Diede SJ, Tenlen JR, Ferguson EL. EEL-1, a Hect E3 ubiquitin ligase, controls asymmetry and persistence of the SKN-1 transcription factor in the early C. elegans embryo. Development. 2007;134:2303–2314. doi: 10.1242/dev.02855. [DOI] [PubMed] [Google Scholar]
- Ruvkun G, Wightman B, Ha I. The 20 years it took to recognize the importance of tiny RNAs. Cell. 2004;116:S93–S96. doi: 10.1016/s0092-8674(04)00034-0. 2 p following S96. [DOI] [PubMed] [Google Scholar]
- Sarin S, O'Meara MM, Flowers EB, Antonio C, Poole RJ, Didiano D, Johnston RJ, Jr, Chang S, Narula S, Hobert O. Genetic screens for Caenorhabditis elegans mutants defective in left/right asymmetric neuronal fate specification. Genetics. 2007;176:2109–2130. doi: 10.1534/genetics.107.075648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S, Antonio C, Tursun B, Hobert O. The C. elegans Tailless/TLX transcription factor nhr-67 controls neuronal identity and left/right asymmetric fate diversification. Development. 2009;136:2933–2944. doi: 10.1242/dev.040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B, Cochella L, Carrera I, Hobert O. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS One. 2009;4:e4625. doi: 10.1371/journal.pone.0004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes & Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Reinke V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr Biol. 2008;18:861–867. doi: 10.1016/j.cub.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Tomita M, Kanai A. Computational methods for microRNA target prediction. Methods Enzymol. 2007;427:65–86. doi: 10.1016/S0076-6879(07)27004-1. [DOI] [PubMed] [Google Scholar]
- Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]