Abstract
Here, we present a new recurrent RNA arrangement, the so-called adenosine wedge (A-wedge), which is found in three places of the ribosomal RNA in both ribosomal subunits. The arrangement has a hierarchical structure, consisting of elements previously described as recurrent motifs, namely, the along-groove packing motif, the A-minor and the hook-turn. Within the A-wedge, these elements are involved in different types of cause–effect relationships, providing together for the particular tertiary structure of the motif.
Keywords: RNA structure, RNA motif, ribosomal RNA, A-minor, along-groove packing motif
DESCRIPTION OF THE ADENOSINE WEDGE MOTIF
An essential part of the knowledge on RNA structure is collected in the form of recurrent motifs, which are found in different molecules or in different parts of the same molecule and have identical or very similar tertiary structure (Batey et al. 1999; Moore 1999; Noller 2005). The fact that recurrent motifs are found in different structural contexts where they have virtually the same conformation presumes at least some level of autonomy of their folding. Therefore, analysis of recurrent motifs is important for understanding the principles governing the formation of RNA tertiary structure. Here we present a new motif, the so-called adenosine wedge (A-wedge), which, among the available RNA structures, has been identified in three locations, all in the ribosomal RNA. The motif can be considered as an arrangement of three previously identified recurrent motifs, the along-groove packing motif (AGPM) (Gagnon and Steinberg 2002; Mokdad et al. 2006), the A-minor motif (Doherty et al. 2001; Nissen et al. 2001), and the hook-turn motif (Szép et al. 2003).
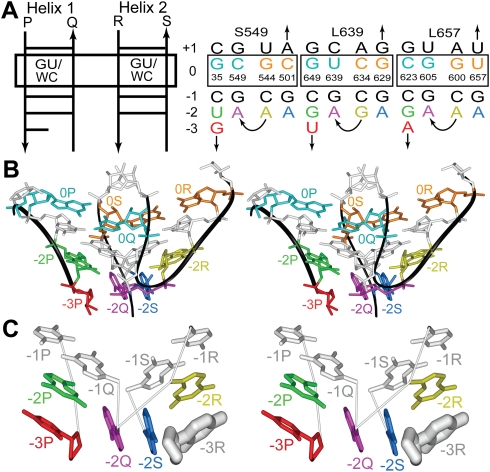
The main body of the A-wedge motif is formed by AGPM, which refers to the arrangement of two closely packed double helices positioned such that the backbone of one helix interacts with the minor groove of the other helix and vice versa (Fig. 1, left). AGPM has been found in more than a dozen locations in ribosomal RNA, always having practically the same conformation well superposable with the conformations of other cases of the motif (Gagnon and Steinberg 2002; Mokdad et al. 2006). The nomenclature of the helices, strands, levels, and individual nucleotides of AGPM is given in Figure 2A (left). The structure of AGPM is characterized by an axial symmetry with the axis being about perpendicular to the planes of the base pairs at the 0-level. This symmetry has been discussed in detail previously (Gagnon and Steinberg 2002); its axis is shown in Figure 1 (left) by the dash-dotted line. When any of the two helices is rotated around this axis for 180°, it almost completely coincides with the other helix. However, there are places in the AGPM structure that have different conformations in both helices and thus cannot be superposed in this way. In particular, at the 0-level, where the two helices interact most closely, a Watson–Crick (WC) base pair in one helix is often packed against a GU base pair in the other helix (Fig. 1, right). Compared to the situation when both base pairs at the 0-level are WC (this happens, for example, in motif S549, whose nucleotide sequence is shown in Fig. 2A), the coexistence of GU and WC makes the shapes of the two helices complementary to each other, thus extending the area of their close packing (Gagnon and Steinberg 2002).
FIGURE 1.
The along-groove packing motif (AGPM). (Left) Schematic representation of AGPM. Trapezoids stand for base pairs opened toward the minor grooves. Arrows represent backbones directed 5′→3′. The external (P, R) and internal (Q, S) strands are labeled. The internal strand of each helix is packed along the minor groove of the opposite helix. Rotation of one helix for 180° around the symmetry axis (dash-dotted line) superposes it with the other helix. (Right) Juxtaposition of the base pairs at the 0-level (for the definition of layers, see Fig. 2). Dashed lines stand for hydrogen bonds. In most known cases of AGPM, one base pair is WC, while the other one is GU. The characteristic geometry of the GU base pair makes the shapes of the minor grooves of the two helices complementary to each other (Gagnon and Steinberg 2002).
FIGURE 2.
The adenosine wedge motif. (A) The secondary structures of the A-wedge-containing AGPMs found in the E. coli rRNA (Schuwirth et al. 2005). The template (left) is used for presentation of the secondary structures of the particular AGPMs (right). The names of the helices (Helix 1 and Helix 2) are provided. The 5′→3′ directions of individual strands are indicated by arrows. Strands P, Q, R, and S correspond to those in Figure 1 (left). In each helix, nucleotides and base pairs are shown as horizontal lines positioned at layers numbered from −3 to +1. For strands Q and S, the 5′→3′ direction of the polynucleotide chain corresponds to the move from a lower to a higher level, while for strands P and R, it is opposite. The base pairs at the 0-level are boxed. GU/WC stands for the allowed identities of the corresponding base pair (GU or WC). The juxtaposition of base pairs at the 0-level is shown in Figure 1 (right). At the −3-level, all structures contain only one external nucleotide −3P. The name of each motif starts with letter “S” or “L,” depending on the subunit, small or large, in which it is found, followed by the number in the rRNA polynucleotide chain of the internal nucleotide of the GU or equivalent base pair at the 0-level (Gagnon and Steinberg 2002). In each strand, only the nucleotide of the 0-level is numbered according to the standard E. coli rRNA numeration (Schuwirth et al. 2005). (B) Stereo view of the tertiary structure of AGPM L639 (PDB entry code 2aw4) (Schuwirth et al. 2005). Each nucleotide is colored as in (A, right). For nucleotide −3P, only the backbone and the ribose are shown. (C) A simplified representation of AGPM L639. Each nucleotide is shown as a base, while the lines between nucleotides stand for covalent connections. The riboses are provided only for nucleotides −3P and −3R. −3R (thick white) stands for the imaginary position of this nucleotide if strand R continued to the −3-level in the regular A-RNA conformation. The simultaneous presence of −3R and −3P would have made the space between their riboses too narrow to accommodate adenosines [−2Q;−2S].
In the ribosome structure, the A-wedge motif is associated with three cases of AGPM, S549, L639, and L657 (Fig. 2A, right). The essential element of the A-wedge that makes it different from other cases of AGPM consists in the particular position of nucleotides −2Q and −2S, which are both adenosines in all three cases of the motif existing in all available high-resolution ribosome structures from Escherichia coli (Schuwirth et al. 2005), Thermus thermophilus (Selmer et al. 2006), and Haloarcula marismortui (Ban et al. 2000). Instead of being involved in base pairing with, respectively, −2P and −2R, these adenosines form a stack that is arranged in the area between the two double helices roughly perpendicular to nucleotides of both helices (Fig. 2B,C). Such arrangement could be seen as a wedge formed by the adenosine stack [−2Q;−2S] that is packed in the crack between the two helices. The A-wedge motifs found in all ribosome structures have almost identical conformations and can be superposed with RMSD 0.36 Å (Supplemental Fig. 1). For clarity, we describe the cases found in the E. coli ribosome, unless otherwise indicated.
Within the A-wedge motif, adenosines −2Q and −2S make contacts with nucleotides −2P and −2R, respectively. Both contacts represent A-minor motifs and include internucleotide van der Waals interactions and hydrogen bonds (Figs. 3, 4; Supplemental Table 1). In particular, two symmetrical hydrogen bonds are observed between atoms N1 of both adenosines −2Q and −2S and the ribose O2′–H groups of, respectively, −2P and −2R (Fig. 3). These hydrogen bonds are specific to the Type-I A-minor interaction, as it is described in Nissen et al. (2001). Thus, a unique feature of the A-wedge motif consists in the fact that although the two adenosines −2Q and −2S stack on each other, they form A-minor interactions with two different double helices. Other interactions that include adenosines [−2Q;−2S] are discussed below.
FIGURE 3.
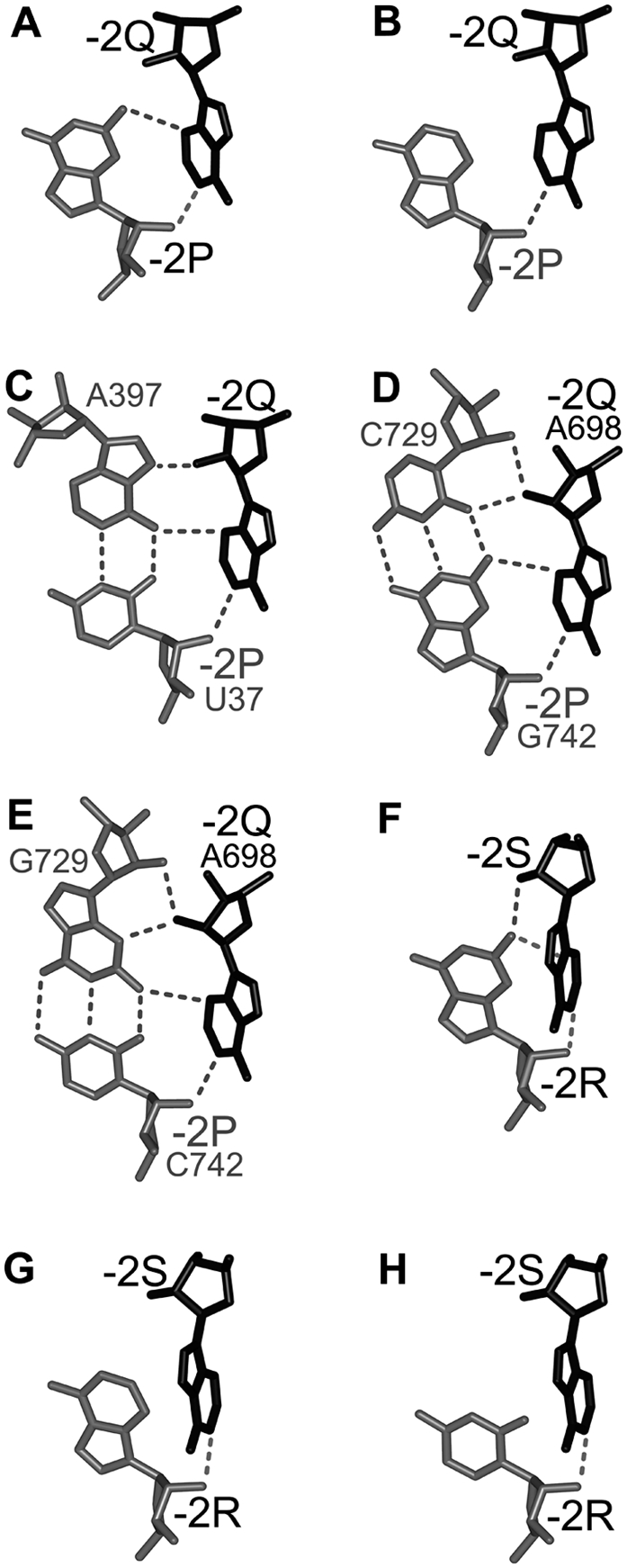
The interaction between nucleotides −2P and −2Q (A–E) and between −2R and −2S (F–H) in different cases of the A-wedge motif (see text). (Black) Nucleotides −2Q and −2S; (gray) other nucleotides; (dashed lines) hydrogen bonds. The numbers of nucleotides in rRNA are indicated, when necessary. The angle between bases −2R and −2S (F–H) is sharper than between bases −2P and −2Q (A–E). Correspondingly, the interbase H-bonds between −2R and −2S are less favorable than between −2P and −2Q.
FIGURE 4.
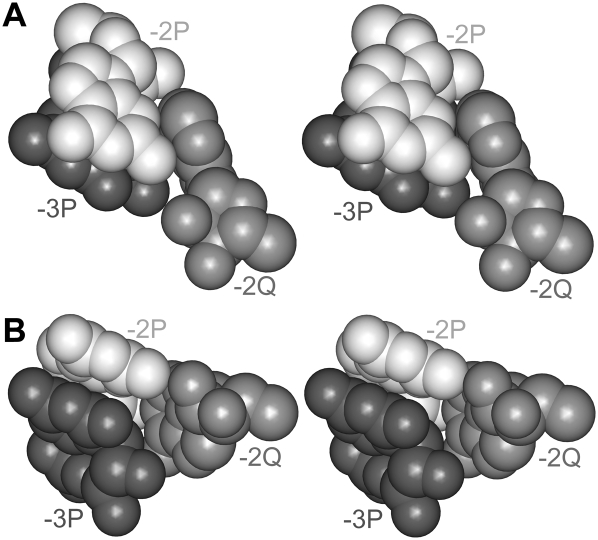
Two stereo views of the [−3P;−2Q;−2P] triangle. Nucleotides −2P (white) and −3P (black) stack to each other. Adenosine −2Q (gray) forms A-minor interaction with −2P and stacks to the ribose of −3P. A color version is provided in Supplemental Figure 2.
ASYMMETRY OF THE ADENOSINE WEDGE MOTIF
Based on the given description of the A-wedge motif, one could suggest that the adenosine stack [−2Q;−2S] is arranged symmetrically with respect to both double helices. In reality, however, this arrangement is characterized by the presence of an essential asymmetrical component. Above, we have already mentioned the asymmetry between the two double helices within AGPM that relates to the presence of the GU-versus-WC base pairs at the 0-level (Fig. 1, right). As we told, this type of asymmetry guarantees the close packing of the two helices and is observed in most cases of AGPM. However, the asymmetry related to the structure of the A-wedge motif has a different origin; it is attributed only to those cases of AGPM that form the A-wedge and is completely independent of the GU-versus-WC asymmetry at the 0-level.
First, we noticed a difference in the structure of the two presumed symmetrical external strands P and R. While nucleotide −2P and the following nucleotide −3P in all three cases of the A-wedge motif stack to each other, nucleotide −2R, which is symmetrical to −2P, never interacts with the following nucleotide. In all three cases of the motif, the nucleotide following −2R occupies position −2Q, which makes the two strands R and Q directly connected (Fig. 2A, right). In fact, this asymmetry between the external strands P and R was used for the initial definition of which of the two helices should be named Helix 1 and Helix 2 (Fig. 2A, left). Below we demonstrate that the presence of nucleotide −3P and the absence of nucleotide −3R are essential for the structure of the A-wedge motif and lead to other asymmetrical aspects observed in the A-wedge arrangement.
DISPLACEMENT OF THE ADENOSINE STACK
To understand why the absence of one of the two external nucleotides at the −3-level (nucleotide −3R) is critical for the integrity of the A-wedge motif, one can imagine what would have happened if both −3P and −3R were present in the structure. Analysis shows that in such a situation, the space between the riboses of −3P and −3R would have been too narrow to accommodate the adenosine stack [−2Q;−2S] (Fig. 2C). Therefore, the removal of nucleotide −3R allows the adenosine stack to fit into the space without collision. Such fitting also forces the adenosine stack to be displaced by about 2 Å from the symmetrical position between the two helices toward strand R and to take the space originally allocated for −3R. Due to this displacement, the adenosine stack avoids the collision with nucleotide −3P, the only remaining external nucleotide at the −3-level.
The described displacement of the adenosine stack [−2Q;−2S] toward strand R leads to other asymmetric aspects of the A-wedge motif. In particular, it brings nucleotide −2Q close enough to −2R to allow the covalent connection between them (Fig. 2). The existence of this connection provides a stabilizing effect for the whole arrangement. Structurally, this connection represents a bent of the polynucleotide chain by almost 180°, which can be described in terms of the recently identified hook-turn motif (Szép et al. 2003). The displacement of adenosines [−2Q;−2S] also extends the distance between nucleotides −2P and −2S, the symmetrical equivalents of −2R and −2Q, thus making the symmetrical connection between −2P and −2S impossible.
INCLINATION OF THE ADENOSINE STACK
Although adenosine stack [−2Q;−2S] has been displaced from the center of the interhelix space toward strand R, both adenosines, as mentioned above, still maintain interactions with the symmetrically positioned external nucleotides −2P and −2R. Keeping the latter interactions after the displacement of the adenosine stack would require that the bases of nucleotides −2Q and −2S become inclined by about 35°–40° in the direction opposite to that in which these nucleotides originally moved (Fig. 2C). Such inclination of the two adenosine bases introduces the asymmetry in their interactions with nucleotides −2P and −2R.
In Helix 1, the inclination of −2Q and −2S opens the angle between the bases of −2Q and −2P, thus facilitating the H-bonding between them (Fig. 2C). In motifs L639 and L657, in addition to the discussed above hydrogen bond between atom N1 of −2Q and the O2′–H group of −2P, the inclination of the bases would optimize the geometry of another hydrogen bond between atom N3 of adenosine −2Q and the amino group of guanosine −2P (Fig. 3A). The two hydrogen bonds correspond to the previously described Type-I A-minor interaction pattern (Nissen et al. 2001). In motif S549, position −2P is occupied by uridine, which is unable to form a hydrogen bond with adenosine −2Q equivalent to that observed in motifs L639 and L657. However, because this uridine is involved in a reverse-WC base pair with adenosine 397 of 16S rRNA, the amino group of adenosine 397 forms the equivalent hydrogen bond with atom N3 of adenosine −2Q (Fig. 3C). The inclination of −2Q would thus favor the formation of this hydrogen bond.
While opening the angle between the bases of −2Q and −2P, the above-mentioned inclination of adenosines [−2Q;−2S] simultaneously sharpens the angle between the symmetrically positioned bases of −2S and −2R (Fig. 2C). This aspect would provide a damaging effect on the quality of potential hydrogen bonds between the bases of −2S and −2R. Correspondingly, out of the three cases of the A-wedge motif existing in the E. coli ribosome, only in L639 position −2R is occupied by guanosine (Fig. 2A, right), which forms a presumably weak hydrogen bond with adenosine −2S (Fig. 3F). In the other two motifs S549 and L657, nucleotide −2R is adenosine, which allows it to form a van der Waals interaction with the base of −2S without H-bonding (Fig. 3G).
In addition to the interactions discussed above, adenosines [−2Q;−2S] form several more hydrogen bonds with the surrounding nucleotides. Although we are unable to discuss here all of these hydrogen bonds in detail, we show them on different panels in Figure 3 and list them in Supplemental Table 1.
ROLE OF NUCLEOTIDE −3P
The inclined position of the adenosine stack [−2Q;−2S] is additionally stabilized by the interaction of the −2Q base with the ribose of −3P (Figs. 2B,C, 4). As mentioned above, nucleotide −3P exists in all three cases of the A-wedge motif found in the E. coli ribosome, where it always stacks to the previous nucleotide −2P. Such position of nucleotide −3P makes its ribose oriented toward the base of −2Q, thus promoting the interaction between the two. This interaction additionally stabilizes the inclined position of adenosine −2Q, as well as of its stacked partner adenosine −2S with respect to the rest of the A-wedge motif. Thus, not only the universal absence of nucleotide −3R, but also the universal presence of its symmetrical analog −3P seems to be important for the integrity of the whole A-wedge arrangement.
VARIATIONS IN THE STRUCTURE OF THE ADENOSINE WEDGE MOTIF
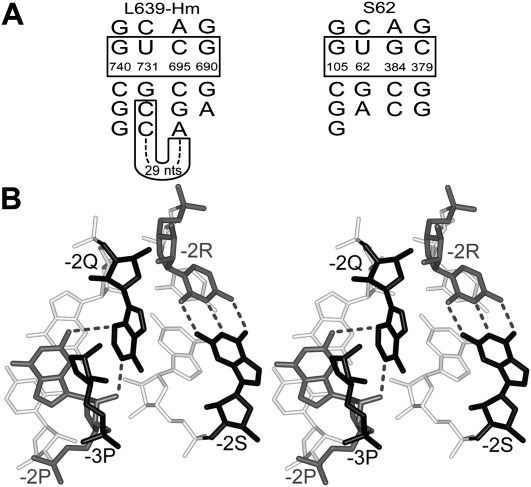
In the structure of motif L639 existing in the 50S subunit of H. marismortui (Ban et al. 2000), there is a 31-nucleotide insertion 699–729 between positions −2Q and −1Q (Fig. 5A, left). All other elements of this arrangement stay at the same places as in the standard structure of the A-wedge motif (data not shown). The last two nucleotides of the insertion are cytidines C728–C729, which forms WC base pairs with guanosines G742 (position −2P) and G743 (position −3P). As a result, G742 (−2P) forms hydrogen bonds simultaneously with two nucleotides A698 and C729, which approach −2P from different sides (Fig. 3D). The same insertion can be found in all available nucleotide sequences of 23S rRNA from archaea (Wuyts et al. 2004).
FIGURE 5.
Variations of the structure of the A-wedge motif. (A) The nucleotide sequences of two modifications of the A-wedge motif observed in motifs L639 from the H. marismortui 23S rRNA (L639-Hm) (Ban et al. 2000) and S62 from the E. coli 16S rRNA (Schuwirth et al. 2005). Each secondary structure is drawn using the same AGPM template as in Figure 2A. In each case, the nucleotide numbering is taken from the original publication of the corresponding structure. (B) Stereo view of the tertiary structure of the “half-wedge” observed in AGPM S62. For nucleotide −3P, only the ribose and backbone are shown. (White) Base pairs at the −1-level; (gray) nucleotides −2P and −2R; (black) nucleotides −3P, −2Q, and −2S. The positions of nucleotides −2P, −3P, and −2Q in S62 are superposable with those in the standard A-wedge arrangement with RMSD 0.59 Å (data not shown). Unlike in the standard A-wedge structure, nucleotide −2S does not stack to −2Q, but instead, forms a WC base pair with nucleotide −2R. A color version is provided in Supplemental Figure 3.
Among cases of AGPM not associated with the A-wedge there is one, S62, which contains some wedge elements (Fig. 5A, right). In this motif, all nucleotides of Helix 1 are positioned exactly as in the three cases of the A-wedge motif, but Helix 2 is different: All three base pairs of this helix at levels 0, −1, and −2 are WC. The two helices are not covalently connected and nucleotides −2Q and −2R do not contact each other. In this case, the arrangement of nucleotides −2P, −3P, and −2Q can thus be seen as a half of the A-wedge motif. This particular case demonstrates that for the maintenance of the specific position of adenosine −2Q, its interaction with nucleotides −2P and −3P is sufficient, while its interactions with nucleotides of Helix 2 are not required.
CONSERVATION OF THE ELEMENTS FORMING THE ADENOSINE WEDGE MOTIF
Analysis of the 12,697 and 436 available prokaryotic nucleotide sequences of, respectively, 16S and 23S rRNAs (Wuyts et al. 2004) shows that the structural elements responsible for the integrity of the A-wedge motif are highly conserved among prokaryotes. Thus, for each of the three motifs S549, L639, and L657, each of the two adenosines −2Q and −2S is present in more than 99% of the sequences (Supplemental Table 2).
Also, in most nucleotide sequences, the identity of nucleotide −2P is such that it allows the proper fixation of adenosine −2Q. In particular, in motifs L639 and L657, position −2P is occupied by guanosine in 93.2% of all 23S rRNA sequences, and the exceptions almost exclusively contain adenosine (3.3%) or cytidine (3.3%) (Supplemental Table 2). The presence of guanosine in position −2P allows the maintenance of its hydrogen bond with adenosine −2Q (Fig. 3A). In the 3.3% of the sequences when nucleotide −2P is adenosine, it forms a van der Waals contact with adenosine −2Q, as in motif L657 of the 50S subunit from H. marismortui (PDB entry code 1s72), (Fig. 3B; Ban et al. 2000). Finally, all instances of cytidine in position −2P are found exclusively in motif L639 from archaea. As mentioned above, in all nucleotide sequences of archaeal 23S rRNA, there is an insertion between positions −2Q and −1Q, and the last nucleotide of this insertion forms a base pair with nucleotide 742 in position −2P (Fig. 5A, left). Analysis shows that base pair 742–729 is WC in all 37 available nucleotide sequences of archaeal 23S rRNA. In nine cases, this base pair is G742–C729 (Fig. 3D), while in the other 28 cases it is C742–G729 (Fig. 3E). Therefore, in all cases when nucleotide −2P is cytidine, it forms a CG base pair with guanosine G729. The latter guanosine will be able to form a hydrogen bond with atom N3 of adenosine −2Q in the same way as the guanosine located in position −2P. Finally, in motif S549 the UA combination between uridine in position −2P and adenosine 397 is present in more than 99% of the sequences, thus allowing the maintenance of the hydrogen bond between adenosine −2Q and adenosine 397 (Fig. 3C; Supplemental Table 2).
As mentioned above, the inclination of the adenosine stack [−2Q;−2S] toward nucleotide −2P would weaken the hydrogen bond between adenosine −2S and guanosine −2R (Fig. 3F). Correspondingly, the presence of guanosine in position −2R is relatively low, reaching 62.7% in motifs L639 and L657 and only 33.7% in S549. At the same time, the percentage of adenosine in position −2R becomes relatively high, reaching 35.4% in L639 and L657 and almost 51% in S549 (Fig. 3G). For motif S549, position −2R is occupied by a pyrimidine in 15% of the sequences. The presence of such pyrimidine would weaken the contact of its base with −2S even more, thus providing additional flexibility to this region (Fig. 3H; Supplemental Table 2). Thus, the most conserved nucleotides of the A-wedge arrangement are −2Q, −2S, and −2P, while the level of conservation of −2R is notably lower. We thus suggest that a relatively low conservation of −2R reflects the fact that because of the inability of the −2R base to form stable hydrogen bonds with adenosine −2S, variations of the identity of −2R cannot substantially affect the stability of the whole arrangement and thus play a comparably minor role in the integrity of the motif.
THE [−3P;−2Q;−2P] TRIANGLE
Analysis of the structures of the A-wedge motif and of its modifications leads us to the suggestion that the arrangement encompassing three nucleotides −3P, −2Q, and −2P (Fig. 4) plays a special role in the integrity of the motif. Indeed, this arrangement constitutes the only part of the structure that is present in all variations of the A-wedge motif. Moreover, the example of motif S62 shows that the formation of arrangement [−3P;−2Q;−2P] does not require a participation of any element from the opposite helix. When nucleotide −2P is guanosine (Fig. 3A), which happens in most cases of motifs L639 and L657, all three nucleotides of the [−3P;−2Q;−2P] arrangement become involved in tight pairwise interactions with the other two nucleotides: −3P stacks to −2P and donates the ribose for stacking with −2Q, while −2Q and −2P form two strong hydrogen bonds and several van der Waals contacts. When nucleotide −2P is a pyrimidine (Fig. 3C,E), it always forms a base pair with either adenosine, as in motif S549, or with guanosine, as in motif L639 in some archaea. Such a base pair would effectively substitute guanosine −2P in all interactions that it makes with both −3P and −2Q. Arrangements of three elements tightly interacting with each other can be called triangles.
While triangle [−3P;−2Q;−2P] seems to be stable on its own and does not require the participation of elements of the other helix, the symmetric three-nucleotide arrangement in Helix 2 cannot be formed at all. Indeed, Helix 2 does not contain nucleotide −3R, which would have allowed the formation of a triangle in Helix 2 equivalent to [−3P;−2Q;−2P]. Also, the interactions between −2S and −2R within the A-wedge arrangement are not optimal and can exist only within the particular context provided by nucleotides of Helix 1. These arguments leads us to a suggestion that triangle [−3P;−2Q;−2P] plays the central role in the formation of the whole A-wedge arrangement. Our preliminary analysis shows that triangles similar to this one can also be observed in other parts of the ribosomal RNA structure unrelated to the A-wedge (data not shown). Whether such nucleotide triangles can play important roles in the formation of other arrangements as well, would need further analysis.
CONSENSUS SECONDARY STRUCTURE
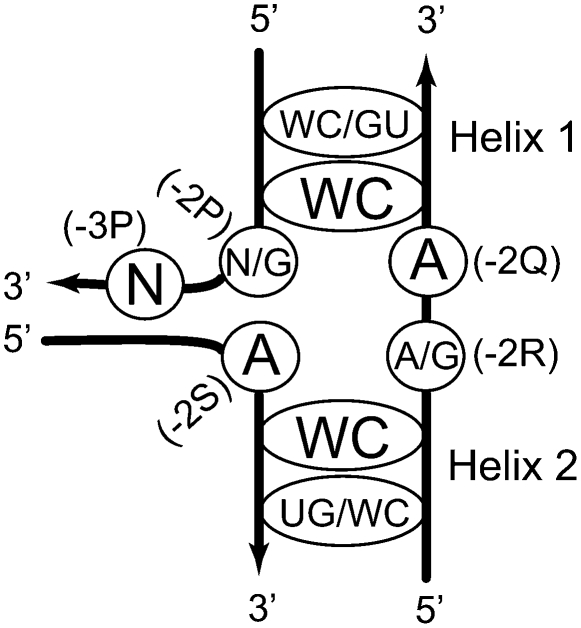
Based on the presented analysis, we propose a consensus secondary structure (CSS) of the A-wedge motif that could be used for identification of such arrangements in RNA molecules with yet unknown tertiary structure. The CSS shown in Figure 6 contains two aspects of uncertainty, which could affect its usability as a search machine. First, the requirements for the identity of nucleotide −2P are very different whether it forms a base pair or not. In the first case, position −2P can be occupied by practically any nucleotide, while in the second case the identity of −2P is strongly biased toward guanosine. Another uncertainty pertains to the packing of Helices 1 and 2. Although in most AGPMs, the helices are closely packed, which is manifested by the coexistence of GU and WC base pairs at the 0-level, exceptions are also known. One of such exceptions is motif S549, in which both base pairs at the 0-level are WC (Fig. 2A, right). We thus made two variations of CSS, a more restrictive (CSS-R) and a more inclusive (CSS-I). In CSS-R, nucleotide −2P must be G, while the 0-level must contain a GU and a WC base pair. In CSS-I, no requirements were imposed on the identity of −2P, while at the 0-level, one base pair must be WC, while the other must be either WC or GU.
FIGURE 6.
The consensus secondary structure of the A-wedge motif. The nucleotides directly involved in the formation of the wedge arrangement are circled. (N) Any nucleotide. The WC and GU base pairs within each helix are shown by the ellipses. The WC/GU and UG/WC identities correspond to the base pairs at the 0-level. The WC identities correspond to the base pairs at the −1-level. Additional explanations are given in the text.
Testing of the CSS-I on the set of the AGPMs existing in the E. coli ribosome provided neither false negatives nor false positives: The A-wedge motif is found in those and only in those AGPMs that fit the CSS-I. In a similar experiment, CSS-R provided only two A-wedges L639 and L657, while motif S549 was missed. Testing the CSS-R on the secondary structures of both 16S and 23S rRNAs from E. coli provided three hits, two of which represented the real A-wedges L639 and L657, while the third one was a false-positive. Alternatively, when the CSS-I was tested on the 16S and 23S rRNAs, the number of hits increased to 11, eight of which were false-positive. This exercise shows that for stable motifs having no essential interaction with other parts of the molecule, the prediction power of CSS-R is rather high. However, when a motif is less stable and/or forms important interactions with other regions, CSS-R would, probably, miss it. Such motifs could still be detected with CSS-I, but only together with a high number of false-positives.
CONCLUDING REMARKS
The A-wedge motif represents an RNA arrangement of four repetitive elements, AGPM, the A-minor, the hook-turn, and the nucleotide triangle, which are involved in complex cause–effect relationships between themselves. The scaffold for the A-wedge motif consists of two symmetrically positioned double helices forming AGPM. However, the fitting of the adenosine stack [−2Q;−2S] into the space between these helices faces steric problems and thus allows only one of the two external nucleotides at the −3-level (−3P) to be stacked to the previous nucleotide −2P. The presence of nucleotide −3P and the absence of the corresponding nucleotide in position −3R unleash, in their turn, a chain of cause–effect relationships that leads to the asymmetry of the motif. The core element of the A-wedge motif, the [−3P;−2Q;−2P] triangle, has a completely asymmetric structure.
The tight fitting of the adenosine stack [−2Q;−2S] into the niche between the two double helices would provide a strong solidifying effect on the whole AGPM arrangement. The elevated stability of the A-wedge motif would enable it to stay on its own without involvement of any other part of ribosomal RNA, which correlates with the fact that all three cases of the motif are located on the ribosome surface and form only marginal contacts with other rRNA domains. The self-sufficiency of the A-wedge motif may also relate to the way it emerged during the ribosome evolution. Recently, we proposed a hierarchical model for the evolution of 23S rRNA (Bokov and Steinberg 2009). In this model, both helices of each of the two A-wedge motifs L639 and L657 emerged simultaneously, providing together for a self-contained entity. This aspect distinguishes motifs L639 and L657 from other AGPMs existing in 23S rRNA, in which the two helices emerged at different moments. Whether, indeed, the A-wedge motif can form and maintain its conformation independently of other parts of the ribosome structure and thus constitutes a folding domain will require further experimental analysis.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.
ACKNOWLEDGMENTS
This work was supported by grant MOP-89923 to S.V.S. from the Canadian Institutes of Health Research.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1550310.
REFERENCES
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Batey RT, Rambo RP, Doudna JA. Tertiary motifs in RNA structure and folding. Angew Chem Int Ed Engl. 1999;38:2326–2343. doi: 10.1002/(sici)1521-3773(19990816)38:16<2326::aid-anie2326>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Bokov K, Steinberg SV. A hierarchical model for evolution of 23S ribosomal RNA. Nature. 2009;457:977–980. doi: 10.1038/nature07749. [DOI] [PubMed] [Google Scholar]
- Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat Struct Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- Gagnon MG, Steinberg SV. GU receptors of double helices mediate tRNA movement in the ribosome. RNA. 2002;8:873–877. doi: 10.1017/s135583820202602x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokdad A, Krasovska MV, Sponer J, Leontis NB. Structural and evolutionary classification of G/U wobble basepairs in the ribosome. Nucleic Acids Res. 2006;34:1326–1341. doi: 10.1093/nar/gkl025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PB. Structural motifs in RNA. Annu Rev Biochem. 1999;68:287–300. doi: 10.1146/annurev.biochem.68.1.287. [DOI] [PubMed] [Google Scholar]
- Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc Natl Acad Sci. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF. RNA structure: Reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Szép S, Wang J, Moore PB. The crystal structure of a 26-nucleotide RNA containing a hook-turn. RNA. 2003;9:44–51. doi: 10.1261/rna.2107303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts J, Perriere G, Van De Peer Y. The European ribosomal RNA database. Nucleic Acids Res. 2004;32:D101–D103. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]