Abstract
Transcriptional gene silencing has been reported with siRNA targeting the promoter region of genes. We tested several siRNAs directed against the human VEGF promoter. Of these, siVFp(−992) exhibited ≥50% suppression of VEGF production in two human cell lines. To determine the specificity of this siRNA-mediated suppression, plasmids were prepared to express a luciferase reporter under the control of VEGF promoters featuring wild-type, mutated, or deleted target sequences. siRNA transfection assays established sequence-specific inhibition of luciferase from the reporter plasmid featuring the wild-type VEGF promoter. However, siVFp(−992) also suppressed the luciferase expression from the plasmids with mutated or deleted target sites, suggesting that silencing was due to a sequence-specific off-target phenomenon, and this was supported by subsequent microarray and bioinformatics analyses. To determine if our concerns regarding the specificity of promoter targeting siRNAs were relevant to other systems where RNA-mediated transcriptional silencing had been previously reported, we tested a published small RNA sequence directed to the HIVSF2-LTR promoter. siRNA transfection assays performed in human cells expressing a luciferase reporter gene under the control of the HIVSF2-LTR promoter revealed significant suppression whether the target sequence was intact or mutated, or when the entire HIVSF2-LTR was replaced by an irrelevant promoter. These data stress the need to examine target specificity when conducting investigations into transcriptional gene regulation with siRNA.
Keywords: Off-target silencing, promoter-specific siRNA, RNA interference, transcriptional gene silencing
INTRODUCTION
RNA interference (RNAi) is a ubiquitous and evolutionarily conserved pathway that effects post-transcriptional gene silencing in the cytoplasmic fraction of a cell by double-stranded RNA (dsRNA) targeting mRNA either for sequence-specific degradation or inhibition of translation (Fire et al. 1998). In addition, it has been proposed that RNAi may manifest in the nucleus as a modulator of gene activity at the transcription level. Early studies demonstrated transcriptional arrest of gene expression by dsRNA homologous to promoter elements in plants (Mette et al. 2000), fission yeast (Volpe et al. 2002), and Drosophila (Pal-Bhadra et al. 2004) via a mechanism that involves epigenetic modifications of genomic DNA. Recent work has proposed that a similar mechanism may exist in mammalian cells to regulate the transcription of genes in both a suppressive and inducive manner.
Transcriptional gene silencing in human cells has been reported with synthetic short-interfering RNA (siRNA) targeted to genomic DNA at specific sites within promoters expressing Ubiquitin C (UBC), E-cadherin (CDH1), Ras association domain family member (RASSF1A), Chemokine receptor type 5 (CCR5), and Elongation Factor-1 (EF1A) genes (Castanotto et al. 2005; Ting et al. 2005; Kim et al. 2006; Weinberg et al. 2006; Hawkins et al. 2009). Similarly, transcriptional silencing of a luciferase reporter transgene was achieved with an RNA targeted to the U3 region of an HIVSF2-LTR promoter-driven construct integrated in human cells (Weinberg et al. 2006). Conversely, Li and coworkers demonstrated transcriptional activation with siRNA targeted to promoters for the human vascular endothelial growth factor (VEGF), cyclin-dependent kinase inhibitor (P21), and CDH1 genes, while a previous study described induction of a neuron-specific gene by an endogenously expressed small dsRNA (Kuwabara et al. 2004; Li et al. 2006). In the majority of studies describing siRNA-mediated transcriptional regulation in human cells, the promoter region of genes implicated in human disease was targeted. Initial assessments determined that promoter-targeting siRNA offered a high level of suppression and exquisite sequence specificity similar to those functioning via cytoplasmic RNAi (Elbashir et al. 2001c), as single-nucleotide mismatches dramatically altered activity. It is noteworthy, however, that numerous investigations for cytoplasmic RNAi have demonstrated so-called “off-target effects” for RNA targeting transcripts in a sequence-dependent manner (Jackson et al. 2003, 2006; Scacheri et al. 2004; Birmingham et al. 2006; Tschuch et al. 2008). Off-target gene silencing occurs when duplex or single-stranded RNAs cross-hybridize with sequences other than the intended target site (Jackson et al. 2003; Birmingham et al. 2006). The converse (i.e., activation) can also occur as exemplified by the recent finding in which a small RNA intended for transcriptional silencing of a promoter-expressed reporter gene appeared to suppress the expression of other genes (Weinberg et al. 2007). Microarray analysis was utilized to identify a gene of previously unknown function that when repressed caused a concomitant up-regulation of the reporter gene.
To evaluate RNA-mediated transcriptional gene silencing in human cells, we designed siRNA to interact with specific sites within the promoter from a well-characterized gene. This gene encodes VEGF, which specifically functions as a mitogen in endothelial cells and is primarily responsible for angiogenesis and neovascularization (Xie et al. 2004). VEGF expression is controlled at many levels by a number of different factors in response to a range of external stimuli. We identified a VEGF model system as appropriate for this study as its promoter has been well characterized, as has its transcriptional regulation. Furthermore, VEGF secretion is regulated by transcription in response to constitutive factors as well as inducible stimuli, such as hypoxia (Xie et al. 2004). Here we show that an siRNA targeted to the human VEGF promoter was responsible for the sequence-specific off-target suppression of VEGF secretion as well as the VEGF promoter-driven expression of a luciferase reporter gene. A microarray and bioinformatics approach was employed to test the hypothesis that the suppression of unintended genes was responsible for this sequence-specific off-target silencing. To determine the relevance of our findings to previously published studies of transcriptional silencing (Kuwabara et al. 2004; Ting et al. 2005; Li et al. 2006; Weinberg et al. 2006), we then tested an siRNA targeted to the U3 region of the HIVSF2-LTR promoter (Weinberg et al. 2006) and found a similar lack of target specificity in relation to the intended promoter target.
RESULTS
dsRNAs targeting the VEGF promoter inhibit gene expression
To determine whether dsRNAs could induce transcriptional gene regulation at the endogenous VEGF promoter, a series of siRNAs (Table 1) was transfected into HeLa and ARPE-19 cells. Nine candidate siRNAs with sequence homology with specific target sites within the human VEGF promoter were designed according to rules for rational siRNA design. A VEGF promoter targeting siRNA, siVFp(−706), which was shown to induce VEGF gene expression (Li et al. 2006), was also included.
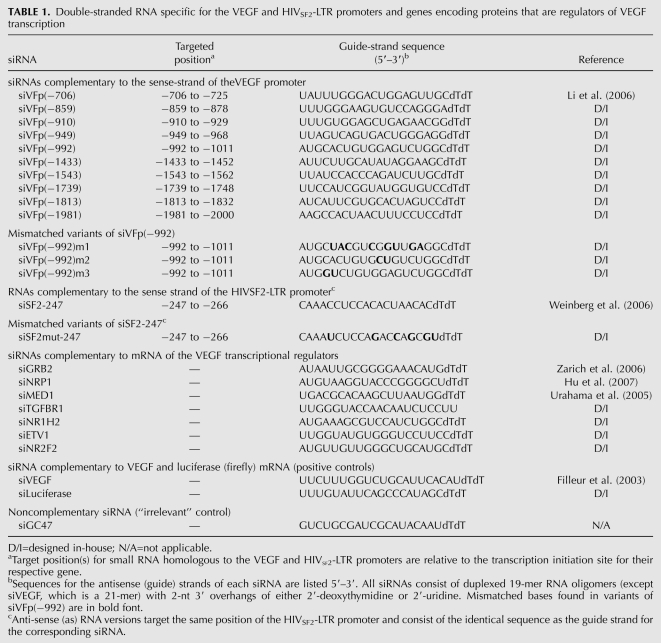
TABLE 1.
Double-stranded RNA specific for the VEGF and HIVSF2-LTR promoters and genes encoding proteins that are regulators of VEGF transcription
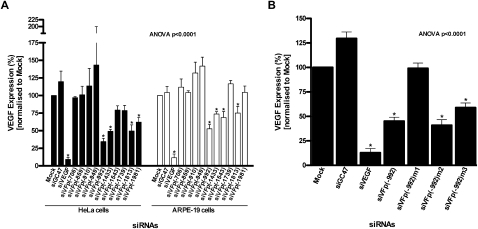
None of the siRNAs designed to target the VEGF promoter displayed any significant level of toxicity (Supplemental Fig. S1; Supplemental Methods). In HeLa cells, siVFp(−992), siVFp(−1433), and siVFp(−1813) demonstrated >50% reduction of VEGF gene expression at 50 nM concentration compared with “Mock” transfected cells (Fig. 1A). In ARPE-19 cells, the overall suppression by the VEGF promoter-specific siRNAs was not as pronounced as that seen in the HeLa cells; however, the pattern of activity was similar. When compared with Mock transfected ARPE-19 cells, siVFp(−992) displayed ≥50% inhibition of VEGF expression, while siVFp(−910), siVFp(−1433), siVFp(−1543), and siVFp(−1813) showed lesser yet significant knockdown (Fig. 1A). Notably, an “irrelevant” siRNA control (siGC47) that lacks complementarity with either the VEGF promoter or mRNA had no significant effect in either cell line, while the positive control, siVEGF, which was targeted to VEGF mRNA, displayed potent suppression of VEGF. Interestingly, siVFp(−706) demonstrated no significant induction of VEGF expression in either HeLa or ARPE-19 cells.
FIGURE 1.
VEGF expression in human cells treated with siRNA targeting the endogenous VEGF promoter. (A) VEGF expression in HeLa and ARPE19 cells, transfected with siRNAs designed to specific sites within the endogenous VEGF gene promoter. (B) VEGF gene expression in HeLa cells transfected with siVFp(−992) and its variants. VEGF expression from cells transfected with each siRNA is normalized to “Mock” transfected cells and presented as the percentage mean ± SD from three independent experiments, with three replicate samples per experiment; (*) p < 0.05.
Critical sequence requirements for dsRNA targeting the VEGF promoter
The sequence specificity for siRNAs targeting the VEGF promoter was established using a series of mismatched variants of siVFp(−992). When comparing the activity of all the VEGF promoter-specific siRNAs, siVFp(−992) displayed the greatest knockdown of endogenous VEGF expression in both HeLa and ARPE-19 cells and was, henceforth, involved in all further manipulations. The siVFp(−992)m1 variant contained a series of sequence mismatches designed to abrogate the inhibitory activity shown by siVFp(−992), whereas siVFp(−992)m2 and siVFp(−992)m3 contained mutations commonly included in the design of siRNA to elucidate their mode of function with respect to the RNAi pathway.
The mismatched variants were transfected into HeLa cells, and their activity against endogenous VEGF was compared with that of siVFp(−992). As observed previously in HeLa cells, siVFp(−992) demonstrated significant inhibition of VEGF production (>50%) at 50 nM concentration compared with Mock transfected cells, whereas silencing was completely abrogated in cells transfected with variant siVFp(−992)m1 (Fig. 1B). In addition, siVFp(−992)m2, which contained mismatches in the central region of the siRNA, retained significant suppressive activity. In contrast, siVFp(−992)m3, which contained mismatches in the “seed” region of the molecule (base position 2–8 from the 5′-end), showed only partial suppression of VEGF. Taken together, these results confirmed the sequence specificity for siVFp(−992) and demonstrate that mismatches in the central region of the molecule appear to be tolerated, whereas partial abrogation of suppression when mismatches are introduced into the “seed” region suggests that this part of the molecule may be important for anti-promoter functionality.
dsRNA-mediated inhibition is not through specific targeting of promoters
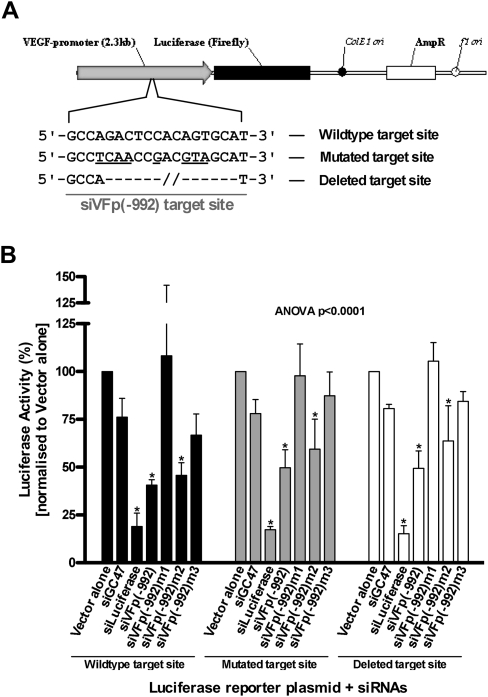
To establish whether siVFp(−992) specifically targets the VEGF promoter, the siRNA was co-transfected into HeLa cells with reporter plasmids pVEGFprom2.3-Luc and pGL4.73. The pVEGFprom2.3-Luc plasmid was constructed to contain the human VEGF promoter, cloned to express a reporter gene coding for a variant of firefly (Photinus pyralis, GL3) luciferase (Fig. 2A), whereas pGL4.73 contained a reporter gene coding for sea pansy (Renilla reniformis) luciferase and served as an internal control. Sequence alignment searches (NCBI BLASTN alignment tool) revealed no significant complementarity between siVFp(−992) and either luciferase mRNA (data not shown). VEGF promoter-mediated firefly luciferase expression was quantitated as a percentage normalized to Renilla luciferase expression. In HeLa cells co-transfected with dsRNA, the pattern of activity observed for siVFp(−992) and its mismatched variants was consistent with that seen when targeting the endogenous VEGF promoter (Fig. 2B). Furthermore, there was no significant activity observed for mismatch variant siVFp(−992)m3 when targeting the VEGF promoter-expressed reporter plasmid, providing further evidence that the seed region is important for siVFp(−992)-mediated inhibition. These data confirmed that siVFp(−992)-mediated suppression of VEGF expression was not due to nonspecific targeting of VEGF mRNA and supported the hypothesis that siVFp(−992) was specifically targeting the VEGF promoter. It is worth noting, however, that siVFp(−1433), siVFp(−1543), and siVFp(−1813), which were shown to repress endogenous VEGF secretion, demonstrated no inhibition of VEGF promoter-driven luciferase expression in HeLa cells (data not shown), suggesting that suppression of VEGF expression by these siRNAs was not occurring through a promoter or transcription-related mechanism.
FIGURE 2.
Luciferase assay performed in human cells co-transfected with VEGF promoter containing reporter plasmid and siRNA targeting the VEGF promoter. (A) Schematic representation of the luciferase reporter plasmids engineered to contain either wild-type (pVEGFprom2.3-Luc), mutated (pVEGFprom2.3.VFp992mut-Luc), or deleted (pVEGFprom2.3.VFp992del-Luc) promoter target sequences. Mismatched bases are underlined, and deleted bases are indicated with a hyphen. (B) Luciferase (firefly) gene activity from HeLa cells transfected with each reporter plasmid when treated with siVFp(−992) or its variants. Luciferase activity is normalized to that from cells transfected with vector without siRNA (Vector alone) and is presented as the percentage mean ± SD from three independent experiments, with three replicate samples per experiment; (*) p < 0.05.
To confirm that siVFp(−992)-mediated inhibition was occurring through specific interaction with the siVFp(−992) target site, plasmid pVEGFprom2.3-Luc was engineered to produce reporter constructs carrying either a mutated or deleted siVFp(−992) target motif within the VEGF promoter (Fig. 2A). As an indicator of promoter functionality, luciferase expression was induced four- to sixfold from the wild-type or altered VEGF promoter in reporter plasmids transfected in either HeLa or ARPE-19 cells and treated with hypoxic-mimic, deferoxamine (130 μM/24 h duration), when compared with untreated cells (data not shown). In HeLa cells, co-transfection of the reporters carrying either the mutated (pVEGFprom2.3.VFp992mut-Luc) or the deleted target site (pVEGFprom2.3.VFp992del-Luc) with siVFp(−992) resulted in a ∼50% reduction of luciferase expression (Fig. 2B) as compared with control. Moreover, the pattern of activity observed for siVFp(−992) and its mismatched variants was almost identical to that seen in corresponding transfections involving the reporter plasmid with wild-type target sequence (pVEGFprom2.3-Luc). Furthermore, a similar pattern of suppression was observed for siVFp(−992) and its mismatched variants with deferoxamine-induced luciferase expression from VEGF promoter experiments performed in HeLa cells (data not shown). Importantly, siVFp(−992)m1, which possessed perfect base complementarity to the mutated siVFp(−992) target sequence demonstrated no significant inhibition of luciferase. These data suggest that siVFp(−992) mediated inhibition of VEGF expression was not occurring through specific targeting of the VEGF promoter.
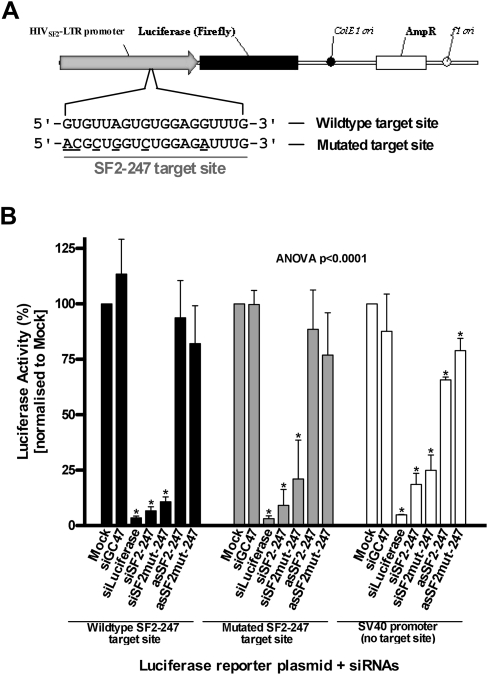
To characterize the target specificity of other promoter-targeting RNA, siSF2-247 was designed with the identical sequence to a small RNA investigated by Weinberg et al. (2006) and reported to suppress Tat-induced transcription of HIVSF2-LTR promoter-driven luciferase expression in human cells. To facilitate testing of siSF2-247, HeLa cells were prepared for stable expression of a luciferase reporter gene under the control of an HIVSF2-LTR promoter that was engineered with either a wild-type or mutated SF2-247 target sequence (Fig. 3A). As an indicator for Tat responsiveness of the HIVSF2-LTR promoter, with either wild-type or mutated target motif, luciferase expression from HeLa-HIVSF2-LTR cells was induced 10–20-fold in the presence of Tat-expression plasmid pSV40.Tat-86, whereas cells with an SV40 promoter-driven luciferase transgene demonstrated no responsiveness to Tat (data not shown). None of the small RNAs designed to target the HIVSF2-LTR promoter displayed any significant level of toxicity (Supplemental Fig. S2; Supplemental Methods). In HeLa cells possessing the wild-type HIVSF2-LTR promoter, Tat-induced luciferase expression was suppressed >90% by transfection with 50 nM siSF2-247 as compared with control cells (Fig. 3B). However, significant inhibition of Tat-mediated luciferase expression by mismatched variant siSF2mut-247 (>90%) suggested that this suppression was not sequence-specific. Furthermore, a similar pattern of inhibition was observed for siSF2-247 and its mismatched variant when targeting the HIVSF2-LTR promoter containing the mutated target site, or an SV40 promoter-driven reporter (Fig. 3B). Interestingly, an antisense RNA (asSF2-247) that was also targeted to position −247 of the HIVSF2-LTR promoter demonstrated no significant suppression of Tat-induced luciferase expression (Fig. 3B). Taken together, these data suggest that the siSF2-247-mediated suppression of Tat-induced luciferase expression was not occurring through specific targeting of the HIVSF2-LTR promoter.
FIGURE 3.
Luciferase assay performed in human cells containing an integrated HIVSF2-LTR promoter driven reporter gene and co-transfected with siRNA targeting the HIVSF2-LTR promoter and Tat expression plasmid, pSV40.Tat-86. (A) Schematic representation of the luciferase reporter plasmids engineered to contain either wild-type (pGL4.20.SF2-LTR-Luc) or mutated (pGL4.20.SF2mut-LTR-Luc) promoter target sequences within the HIVSF2-LTR promoter. Mismatched bases are underlined. (B) Luciferase (firefly) gene activity from HeLa cells containing an integrated reporter gene when treated with siSF2-247 or its variants. Tat-induced firefly luciferase activity is normalized to that from cells not transfected with siRNA (“Mock”) and is presented as the percentage mean ± SD from three independent experiments, with three replicate samples per experiment; (*) p < 0.05.
Microarray and bioinformatics analyses of genes suppressed by siVFp(−992)
Microarray analysis was performed to identify candidate genes that could be involved in the off-target suppression of VEGF production by siVFp(−992). Total RNA isolated from HeLa cells transiently transfected with either siVFp(−992), mismatched variant siVFp(−992)m1, or lipofectamine alone (“Mock”) were subjected to hybridization with an Affymetrix Human Genome (HG) U133 Plus 2.0 GeneChip. OmniViz software was employed to identify probe sets corresponding to genes down-regulated by siVFp(−992) to a minimum of log2(0.7). This threshold was set at a relatively relaxed limit to allow for the inclusion of genes/probe sets that may have demonstrated a subtle decrease of RNA levels by siVFp(−992) compared with inhibition observed for the corresponding protein. Analysis revealed 709 probe sets representing a total of 497 genes that showed reduced expression in cells treated with siVFp(−992). A greater number of probe sets were present than genes owing to the fact that some genes were represented by multiple probes sets. Probe sets corresponding to RNA transcripts decreased by log2(0.7) or more in the presence of mismatched variant siVFp(−992)m1 were excluded, and the remaining genes were sorted on the basis of their involvement in either transcription, regulation, and angiogenesis. A total of 52 candidate genes, including VEGF itself, qualified for further bioinformatics analysis.
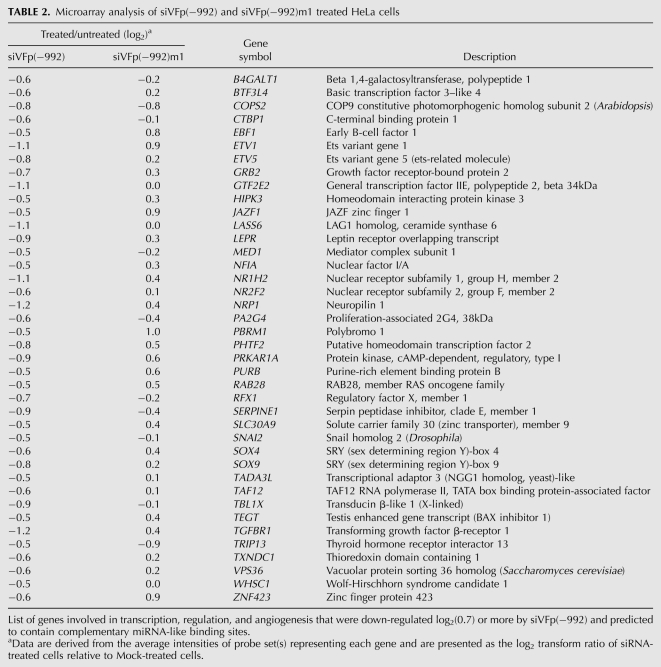
The subset of genes that were down-regulated by siVFp(−992) according to microarray analysis, and involved in transcription, regulation, and angiogenesis, were further assessed for the presence of 3′UTR miRNA-like interaction motifs. We refer to these motifs as “miRNA-like” so as to distinguish them from known miRNA target sites for endogenous miRNAs. Sequence alignment analysis with siVFp(−992) revealed 40 genes that contained putative miRNA-like target motifs within the 3′-UTR of their expressed transcript (Table 2). The methodology, predicted alignments for siVFp(−992) and miRNA-like binding sites are outlined in supplemental materials (see Supplemental Table S2). Gene Ontologies were examined for each gene for possible direct or indirect involvement in regulation of VEGF. Genes were assessed on the functional significance of the proteins they encode in pathways that regulate VEGF transcription with specific, upstream regulators considered highly, while those that perform a more general function in basal transcription received a lower ranking. Seven of the 40 genes predicted to have putative siVFp(−992) miRNA-like binding sites were identified by Gene Ontology assessment as known regulators of VEGF transcription and considered as candidates for siVFp(−992) off-targeting. These genes and the proteins they encode include growth factor receptor bound protein-2 (GRB2); neuropilin-1 (NRP1); mediator complex subunit-1 (MED1); transforming growth factor receptor beta-receptor-1 (TGFBR1); nuclear receptor subfamily-1, group H, member-2 (NR1H2); ets variant gene-1 (ETV1); and the nuclear receptor subfamily-2, group F, member-2 (NR2F2).
TABLE 2.
Microarray analysis of siVFp(−992) and siVFp(−992)m1 treated HeLa cells
Gene knockdown experiments and Western analysis of VEGF transcriptional regulators reveal a candidate for siVFp(−992) off-targeted suppression
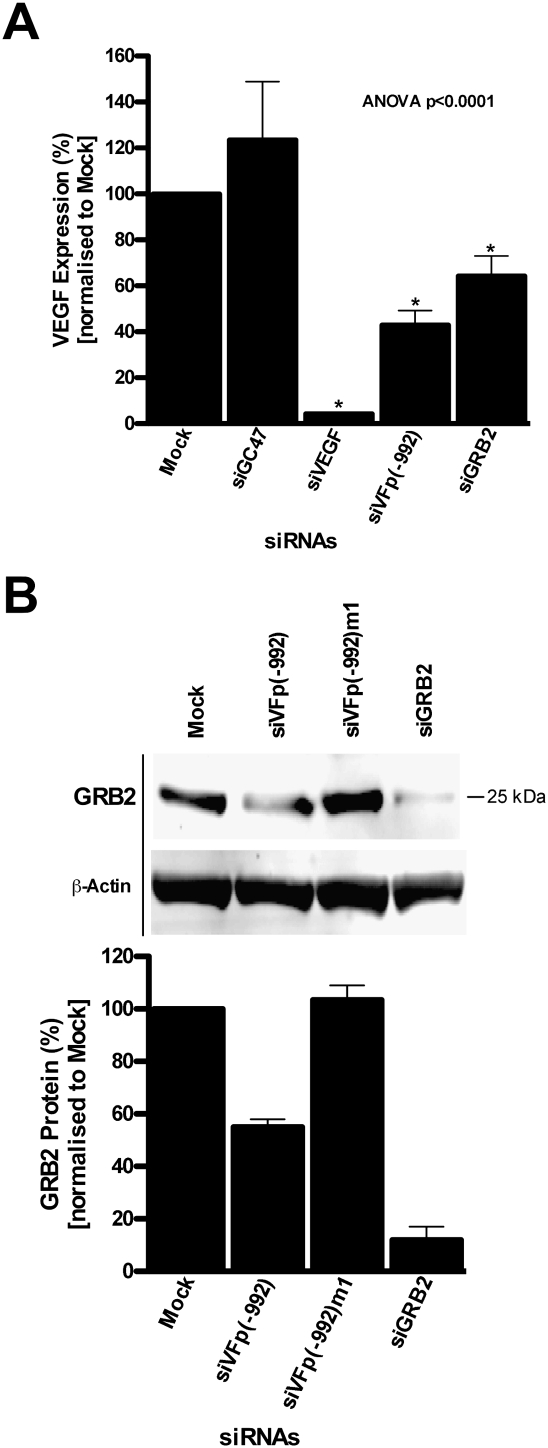
Gene knockdown experiments were performed to confirm whether candidates identified by microarray and bioinformatics analysis as putative targets for siVFp(−992) silencing were involved in regulation of VEGF expression. Candidate genes were targeted by siRNAs homologous to the coding region of each transcript and assessed for their effect on endogenous VEGF production. In HeLa cells, knockdown of GRB2 was significant in decreasing VEGF expression (Fig. 4A). Protein expression analysis suggested GRB2 as the target for siVFp(−992)-mediated silencing. Western analysis on total protein extracted from HeLa cells transfected with siVFp(−992) revealed a 45% decrease of GRB2 expression compared with Mock transfected cells (Fig. 4B). Significantly, the siVFp(−992) mismatch variant, siVFp(−992)m1, did not suppress GRB2 expression. It is worth noting that the effect of siGRB2-mediated inhibition on VEGF expression (Fig. 4A) is less than the knockdown of GRB2 protein by the same siRNA (Fig. 4B). We consider this suggests that GRB2 may not be the only target for siVFp(−992), and it is likely that inhibition of VEGF expression is the result of the unintended knockdown of additional genes. SiRNA knockdown assays revealed that the other siRNAs targeting known regulators of VEGF had little or no effect on VEGF production with the exception of siETV1; however, subsequent Western analysis determined that ETV1 was not a target for siVFp(−992) (data not shown). Taken together, these data provide strong support for the hypothesis that suppression of VEGF expression by siVFp(−992) is caused by the sequence-specific off-targeting of one or more regulators of VEGF transcription, including GRB2.
FIGURE 4.
Protein expression analysis from human cells transfected with siRNA complementary to the VEGF gene promoter or GRB2 mRNA. (A) VEGF expression in HeLa cells transfected with siGRB2 or control siRNA. VEGF expression from cells transfected with each siRNA is normalized to “Mock” transfected cells and presented as the percentage mean ± SD from three independent experiments, with three replicate samples per experiment; (*) p < 0.05. (B) Western blot analysis for the detection and quantitation of GRB2 protein extracted from HeLa cells treated with siVFp(−992) or control siRNA. A representative blot is shown with samples loaded per lane, as follows: (lane 1) Mock transfected cells; (lane 2) siVFp(−992); (lane 3) siVFp(−992)m1; and (lane 4) siGRB2. The intensity of each band for GRB2 was quantified by densitometry and normalized to that for β-actin. GRB2 protein levels from HeLa cells transfected with each siRNA are shown as a ratio relative to Mock transfected cells and presented as the percentage mean ± range from two independent experiments, with two replicate samples per experiment.
DISCUSSION
RNAi provides an effective method of silencing genes with possible application to the therapeutic setting. Several studies have recently explored the possible utility of siRNA in controlling gene transcription in mammalian cells (Kuwabara et al. 2004; Ting et al. 2005; Kim et al. 2006; Li et al. 2006; Weinberg et al. 2006; Hawkins et al. 2009). In this study, we have applied the same design principles to produce candidate siRNA inhibitors of VEGF transcription. To this end, we have investigated the sequence specificity and target specificity of dsRNA in experiments measuring endogenous VEGF production and the expression of a luciferase reporter gene under the control of the VEGF promoter.
One of several siRNAs designed to target the VEGF promoter, siVFp(−992), provided sequence-dependent suppression of VEGF production in two unrelated human cell lines. We felt it important to test the siRNAs in more than one cell line to mitigate the risk that the observed effects were an idiosyncrasy of a particular cell line (for example, the result of differences in dsRNA sensing by Toll-like receptors) or experimental condition. As a case in point, the siRNAs targeting from position −706 of the VEGF promoter and previously reported by Li et al. (2006) to induce expression of endogenous VEGF in HeLa cells did not significantly affect endogenous VEGF expression in either ARPE-19 or HeLa cell lines under the conditions we used.
Our candidate dsRNA, siVFp(−992), also inhibited VEGF promoter-driven expression of a luciferase reporter in HeLa cells, suggesting that transcriptional regulation of VEGF could be achieved with dsRNA. However, subsequent assays with reporters for which the promoter harbored a target site that was mutated or deleted revealed that silencing was independent of the target sequence and, therefore, could not be inferred to be mediated through the proposed mechanism. Comparison of the results obtained with siVFp(−992) and its variants indicated that the seed region sequence was important in the observed suppression of VEGF and VEGF promoter-driven luciferase. This suggested that sequence-specific off-target silencing by siVFp(−992) may have been occurring through an miRNA-like interaction with an unintended target mRNA, similar to that found in studies of conventional cytoplasmic RNAi (Jackson et al. 2003, 2006). To identify candidate genes involved in sequence-specific off-targeting, HeLa cells treated with either siVFp(−992) or mismatched variant siVFp(−992)m1 were subjected to gene expression analysis by microarray. Previous studies have used genome-wide profiling by microarray to determine transcripts affected by siRNA-mediated off-target suppression (Jackson et al. 2003; Birmingham et al. 2006; Weinberg et al. 2007). The effects of siVFp(−992) and its variants in VEGF promoter reporter experiments were highly similar to those seen with endogenous VEGF production; therefore, we reasoned that the unintended target(s) was involved directly or indirectly in the transcriptional regulation of VEGF. From the bioinformatics analysis of the microarray data, 40 repressed target transcripts contained putative siVFp(−992) miRNA-like target sites within their 3′-UTRs. Of these, seven were genes with Gene Ontology data implicating them in pathways controlling VEGF transcription. Subsequent knockdown experiments with siRNAs targeting each of these found that one, GRB2, was involved in controlling VEGF expression in HeLa cells. Validation experiments involving Western analysis determined that GRB2 was suppressed by siVFp(−992) but not siVFp(−992)m1 and suggested that this protein, possibly among others, may have been an unintended target for siVFp(−992) silencing. The implication that additional proteins may also be the unintended targets for siVFp(−992) is supported by the observation that down-regulation of endogenous VEGF production, resulting from siGRB2 reduction of GRB2 expression, is not consistent with the observed level of knockdown for GRB2 protein by siGRB2. GRB2 is an adaptor protein that binds growth factor receptor tyrosine kinase through its SH2 domain and associates with proteins involved in the Ras-ERK/MAPK signal cascade, which, in turn, regulates genes involved in a wide range of biological processes including embryonic development, cell growth, proliferation and migration, angiogenesis, and oncogenesis (Lowenstein et al. 1992; Furge et al. 2000; Dhillon et al. 2007; McKay and Morrison 2007). GRB2 stimulates the MAPK signal transduction pathway when bound to the phosphorylated epidermal growth factor receptor leading to Ras activation and ultimately ERK 1/2 phosphorylation of HIF-1α and Sp1, which induce transcription of VEGF (Richard et al. 1999; Furge et al. 2000). Interestingly, GRB2 antagonists inhibit VEGF stimulated angiogenesis in several human cell lines (Soriano et al. 2004), confirming the biological role of GRB2 in VEGF signaling and angiogenesis. Further work to evaluate the extent to which targeting of GRB2 is responsible for the sequence-specific off-targeting of VEGF could involve mutation of the siRNA target sites and rescue of expression experiments. In any case, the data presented strengthen the hypothesis that the effects of siVFp(−992) on VEGF are mediated by sequence-specific interaction with a nontargeted mRNA.
In this study, we explored the nature of sequence and target specificity of dsRNAs designed to interact with a promoter target sequence. Traditionally, studies of siRNAs have demonstrated sequence specificity using mismatched, scrambled versions of their siRNA, or molecules that targeted “irrelevant” genes (Fire et al. 1998; Hamilton and Baulcombe 1999; Zamore et al. 2000; Bernstein et al. 2001; Elbashir et al. 2001a,b,c). Unfortunately, these controls reveal little about the specificity for the intended target motif. Indeed, the emergence of publications reporting off-target interactions between the siRNA and unintended targets (Jackson et al. 2003) heralded the need for a more stringent set of controls in loss-of-function studies involving RNAi. An editorial published in Nature Cell Biology outlined the most suitable controls required when displaying RNAi data based on the general consensus reached at the Horizon Symposium in RNA, 2003 (Anonymous 2003). Mismatched or scrambled siRNAs were noted as acceptable standards to include in RNAi-related experiments but were considered of limited value. The authors concluded that the “ultimate control” to confirm specificity of an RNAi phenotype involved rescue of expression by the target gene in a form that was refractory to the siRNA, ideally within the physiological range of expression for the target gene. This involved the introduction of one or more silent mutations within the target motif in an effort to establish target-specificity for the siRNA being tested. While the considerations and recommendations provided by the editorial were primarily related to post-transcriptional RNAi, we consider them equally relevant to studies of RNA-induced transcriptional gene regulation. While studies of putative RNA-induced transcriptional gene regulation have employed siRNAs carrying mismatched or scrambled bases to establish sequence specificity (Ting et al. 2005; Li et al. 2006; Janowski et al. 2007), the majority of these investigations have not examined target specificity by mutating or altering the intended target sites, and therefore, not ruled out the possibility that the observed effects are not mediated through promoter targeting. With this in mind, we felt it important to investigate the target specificity for promoter-targeting RNA that had been previously reported to mediate transcriptional gene regulation and at the same time confirm that our observations were not exclusive to the VEGF promoter. To this end, we tested an siRNA shown previously to inhibit HIVSF2-LTR promoter-driven luciferase expression (Weinberg et al. 2006) and determined that the observed effect was not occurring through the specific targeting of the intended target motif. Indeed, a more recent study by the same investigators revealed off-targeted gene activation by a small RNA directed to a sequence proximal to the original target site in the HIVSF2-LTR promoter (Weinberg et al. 2007). Microarray analysis was used to identify a gene of previously unknown function that when repressed caused a concomitant up-regulation of the LTR-driven reporter gene. Hence, our concern regarding the role of sequence-specific off-targeting in studies of RNA-mediated transcriptional regulation is justified.
To our knowledge, our study is the first to investigate the possible targeting of promoter sequences using dsRNA that has included the examination of target specificity. Our findings underscore the need for such studies, particularly in the investigation of such a novel area as the transcriptional regulation of genes by siRNA.
MATERIALS AND METHODS
Design and preparation of dsRNAs
All dsRNAs were chemically synthesized by Sigma-Proligo. They were provided as deprotected and desalted, annealed duplexed RNA oligonucleotides (siRNAs), with each strand composed of 21–23 nucleotides (nt) that included two (2′-deoxy)thymidines (dTdT) or uridines (UU) on the 3′-end. Sequences for the antisense strand (“guide” strand) are listed in Table 1. siRNAs designed to target the VEGF gene promoter are identified relative to the transcription start site. Ten candidate siRNAs homologous to sites within the VEGF promoter were designed and selected for testing. Nine siRNAs were generated using the Block-IT RNAi Designer (Invitrogen), while an additional siRNA, targeting from position −706 relative to the transcription start site of the VEGF promoter, was designed with the identical sequence to that used previously by Li et al. (2006). An siRNA and antisense RNA targeting at position −247 relative to the transcription start site of the LTR promoter from HIV-1 strain SF2 were designed with the identical sequence to that used previously by Weinberg et al. (2006). The sequences for siRNAs homologous to target genes that encode known regulators of VEGF signaling were adapted from the literature or generated using the Block-IT RNAi Designer (Table 1). An “irrelevant” control siRNA (siGC47), which lacks homology with any known human sequences, was included. Conversely, a positive control (siVEGF) was designed with the identical sequence to an siRNA used by Filleur et al. (2003) and was employed to specifically target VEGF mRNA.
Cell culture and dsRNA transfection
HeLa and ARPE-19 cell lines were used in this study as they both produce measurable quantities of VEGF. HeLa cells are immortalized human epithelial cells derived from cervical carcinoma, whereas ARPE-19's are of human retinal pigmented epithelium origin. Both cell lines were maintained at 37°C in a humidified atmosphere containing 5% CO2 and cultured in DMEM (GIBCO) supplemented with 10% (v/v) heat-inactivated (56°C, 30 min) FBS (JRH Biosciences), 4.5 g/L glucose, and 4 mM L-glutamine. Cells were passaged routinely (twice weekly) to maintain exponential growth. Twenty-four hours prior to transfection, cells at 60%–90% confluency were trypsinized, diluted in fresh culture medium, and seeded at cell densities described for each assay. All transfections were performed in triplicate for 48 h of duration with 50 nM siRNA in Opti-MEM media (GIBCO) containing Lipofectamine 2000 (Invitrogen) used according to the manufacturer's instructions. Cell viability was determined by Cell Titer Blue assay (Promega), as described by the manufacturer's protocol.
Construction of reporter plasmids and luciferase assay
Reporter plasmid constructs containing either the human VEGF and HIVSF2-LTR promoters, and a Tat-expression plasmid were based on the pGL3 and pGL4 series (Promega) expressing a firefly luciferase gene. Preparation of constructs containing the VEGF promoter and primer sequences for PCR amplification of VEGF promoters are described in the supplemental material (Supplemental Methods; Supplemental Table S1, respectively). Preparation of plasmids expressing the HIVNL43-Tat gene or reporter constructs containing the HIVSF2-LTR promoter and production of HeLa cell lines with an HIVSF2-LTR promoter for stable expression of the firefly luciferase reporter gene, are also described in the supplemental materials (Supplemental Methods).
The human VEGF promoter was cloned into the pGL3-Promoter plasmid immediately upstream of a firefly luciferase reporter gene to produce construct pVEGFprom2.3-Luc (Fig. 2A). DNA fragments carrying either a mutated or a deleted siVFp992 target site were subcloned into pVEGFprom2.3-Luc to produce constructs pVEGFprom2.3.VFp992mut-Luc and pVEGFprom2.3.VFp992del-Luc (Fig. 2A), respectively. Luciferase activity from transient transfections of HeLa cells with reporter constructs containing the VEGF promoter was determined with the Dual Luciferase Assay system (Promega), according to the manufacturer's instructions. The luminescent signal was quantitated with the FLUOstar Optima plate reader (BMG Labtech). Luciferase expression was measured from cells transfected with plasmid constructs carrying the firefly luciferase reporter gene. As an internal control, plasmid pGL4.73 (Promega), which contained the Renilla luciferase reporter gene, was co-transfected. Briefly, HeLa cells were plated at 5 × 103 cells per well in opaque-walled 96-well plates (Greiner) and incubated for 24 h. Cells were transfected sequentially, beginning with 50 nM siRNA and then followed 16–20 h later with 80 ng of firefly luciferase reporter construct and 1 ng of pGL4.73, per well. Both transfections were performed in Opti-MEM media containing 0.2 μg of Lipofectamine 2000. Cells were assayed for luciferase activity 24 h after transfection with reporter plasmid.
The HIVSF2-LTR promoter with either the wild-type or mutated SF2-247 target sequence was cloned into the pGL4.20 plasmid immediately upstream of a firefly luciferase reporter gene to produce construct pGL4.20.SF2-LTR-Luc and pGL4.20.SFmut2-LTR-Luc (Fig. 3A), respectively. A Tat expression construct was prepared to facilitate Tat-induced expression of luciferase in HeLa cells containing an integrated transgene featuring the HIVSF2-LTR promoter. The HIV Tat gene was amplified from plasmid pCSN-Tatx12, which contains both exons of the HIV-1 Tat gene derived from strain NL4.3. The Tat gene was cloned immediately downstream from the SV40 promoter found in plasmid pGL3-Control to produce construct pSV40.Tat-86. Luciferase activity from HeLa cells carrying an integrated reporter construct was determined with the Luciferase (firefly) Assay system (Promega), according to the manufacturer's instructions. Briefly, HeLa cells with integrated reporter plasmid were plated at 5 × 103 cells per well in opaque-walled 96-well plates and incubated for 24 . Cells were transfected sequentially, beginning with 50 nM siRNA and then followed 16–20 h later with 112 ng of pSV40.Tat-86, per well. Both transfections were performed in Opti-MEM media containing 0.2 μg of Lipofectamine 2000. Cells were assayed for luciferase activity 24 h after transfection with Tat expression plasmid.
RNA isolation, microarray, and computational analyses
HeLa cells were seeded in 150-cm2 (T150) tissue culture flasks (Corning) at 2.35 × 106 cells/flask and incubated for 24 h. Cells were transiently transfected with 50 nM siVFp(−992), variant siVFp(−992)m1, or without siRNA (Mock), in Opti-MEM media containing 94 μg of Lipofectamine 2000 per flask. Cells were harvested 48 h after transfection, washed twice with sterile PBS (GIBCO), and total RNA was extracted using the RNeasy Mini Kit (QIAGEN), according to the manufacturer's instructions. Total RNA (15 μg) was provided for microarray analysis, which was performed at the Australian Genome Research Facility. Gene expression profiles for each sample were determined using an Affymetrix Human Genome (HG) U133 Plus 2.0 GeneChip. The HG-U133 Plus 2.0 GeneChip contains ∼54,000 human probe sets used to analyze the expression of more than 47,000 RNA transcripts and splice variants. The measures of experimental quality for both sample RNA and assay indicated a high overall standard, as reflected by the percentage of genes present across all samples (44.91% ± 1.32%), and the 3′–5′ probe set intensity ratios for glyceraldeyde-3-phosphate dehydrogenase (GAPDH) and β-actin (1.03 ± 0.02 and 1.15 ± 0.07, respectively). Gene transcripts corresponding to probe sets that yielded raw intensity values below the background threshold (50 intensity units) were considered “absent” and disregarded from further analysis.
Raw intensities from all “present” probe sets were normalized by quantile with the aid of RMAexpress software (http://rmaexpress.bmbolstad.com/) (Bolstad et al. 2003). Normalized data were log2-transformed as a ratio of test (siRNA transfected cells) to control (Mock transfected cells) and subjected to an unsupervised hierarchical cluster analysis using OmniViz, version 6.0 software (BioWisdom). The latter was employed to identify a subset of genes down-regulated by siVFp(−992) by a log2(0.7) factor or more rather than determine the relationship between different clusters of genes. Genes/probe sets with Gene Ontology classifications containing “transcription,” “regulation,” and “angiogenesis” were selected from this list of down-regulated transcripts. Their 3′-UTR sequences were subjected to a sequence alignment search for siVFp(−992) miRNA-like target sites using the RNAhybrid alignment tool (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/) (Rehmsmeier et al. 2004). Putative siVFp(−992) miRNA-like target sites were assessed according to previously established criteria for miRNA–target interactions and prediction of miRNA target motifs (Brennecke et al. 2005; Didiano and Hobert 2006; Grimson et al. 2007). Identification and selection of potential miRNA binding sites for siVFp(−992) are described in Supplemental Methods. Gene Ontologies and pathways for genes down-regulated by siVFp(−992) and containing putative miRNA binding motifs were examined using the MetaCore gene pathway analysis and data mining database (GeneGo, Inc.) in association with Entrez Gene and OMIM (http://www.ncbi.nlm.nih.gov/sites/entrez?db=OMIM&cmd) to establish any direct or indirect relationship between the protein encoded by each candidate gene and VEGF regulation.
Enzyme linked immunosorbent assay (ELISA) and Western blotting
HeLa cells (4 × 103 cells/well) or ARPE-19 cells (5 × 103 cells/well) were seeded in 96-well plates (Greiner) and incubated for 24 h. Cells were transfected with 50 nM siRNA, or without siRNA (Mock), in Opti-MEM media containing 0.2 μg of Lipofectamine 2000, per well. Mock transfections were performed with Opti-MEM media containing 0.2 μg of Lipofectamine 2000. Forty-eight hours after transfection, the supernatant from each well was collected for a sandwich ELISA performed with the commercially available Quantikine Human VEGF Immunoassay kit (R&D Systems), according to the manufacturer's instructions.
Western analysis was performed on total protein lysates extracted from HeLa cells transfected with siRNA. HeLa cells were seeded in 6-well culture plates (BD Biosciences) at 1.5 × 105 cells/well and incubated for 24 h. Cells were transfected with or without (Mock) 50 nM siRNA in Opti-MEM media containing 6 μg of Lipofectamine 2000 per well. Transfected cells were washed twice with sterile PBS 48 h after transfection and total protein extracted using RIPA buffer (150 mM NaCl, 0.1% [w/v] sodium dodecyl sulphate, 1% [v/v] Nonidet, P-40, 0.5% [w/v] sodium deoxycholate, 50 mM Tris-HCl at pH 8.0, containing EDTA-free protease inhibitors). Total protein (20–30 μg) was denatured for 10 min at 80°C in 1× sample loading buffer (Invitrogen) containing 1× reducing agent (Invitrogen) and separated by electrophoresis on a 4%–12% NuPAGE bis-tris acrylamide gel (Invitrogen) run in 1× MOPS-SDS running buffer (Invitrogen). Following electrophoresis, proteins were transferred to Hybond-LFP PVDF membranes (Amersham) for 2 h at 30 V in 1× transfer buffer (Invitrogen) containing 10% (v/v) methanol. Membranes were blocked with 3% BSA in TBST buffers (10 mM Tris at pH 8, 30 mM NaCl, 0.05% [v/v] Tween 20) overnight at 4°C. Membranes were incubated for 2 h at room temperature with rabbit-α-GRB2 (Abcam) and mouse-α-β-actin (BD Biosciences) primary antibodies diluted in 3% BSA-TBST at concentrations recommended by the manufacturer. The membranes were washed three times for 10 min each with TBST buffer and then incubated for 1 h at room temperature with goat-α-rabbit IgG-Cy5 (GE Healthcare) and goat-α-mouse IgG-Cy3 (GE Healthcare) secondary antibodies diluted 1/2000 in 5% skim milk-TBST. Membranes were washed again three times for 10 min each at room temperature in TBST buffer and fluorescence visualized using the Typhoon 9410 Scanner (GE Healthcare). Protein abundance was quantitated with ImageQuant (GE Healthcare) sofware (Version 5.0).
Microarray data
Raw data from the microarray analysis performed in this study have been submitted to ArrayExpress (MIAME) (http://www.ebi.ac.uk/microarray-as/ae/). To gain access to these data, provide the following login details: Login: Reviewer_E-MEXP-2221; Password: 1245346547492.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Dr. Glen McIntyre and Dr. Tanya Applegate from Johnson & Johnson Research Pty. Ltd. (Sydney, Australia) for providing plasmid pCSN-Tatx12. We also thank Dr. Michael Poidinger from Johnson & Johnson Research Pty. Ltd. (Sydney, Australia) for technical assistance and advice with the bioinformatics analysis.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1808510.
REFERENCES
- Anonymous. Whither RNAi? Nat Cell Biol. 2003;5:489–490. doi: 10.1038/ncb0603-490. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA–target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanotto D, Tommasi S, Li M, Li H, Yanow S, Pfeifer GP, Rossi JJ. Short hairpin RNA-directed cytosine (CpG) methylation of the RASSF1A gene promoter in HeLa cells. Mol Ther. 2005;12:179–183. doi: 10.1016/j.ymthe.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signaling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA–target interactions. Nat Struct Mol Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001a;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001b;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001c;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filleur S, Courtin A, Ait-Si-Ali S, Guglielmi J, Merle C, Harel-Bellan A, Clezardin P, Cabon F. SiRNA-mediated inhibition of vascular endothelial growth factor severely limits tumor resistance to antiangiogenic thrombospondin-1 and slows tumor vascularization and growth. Cancer Res. 2003;63:3919–3922. [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Furge KA, Zhang YW, Vande Woude GF. Met receptor tyrosine kinase: Enhanced signaling through adapter proteins. Oncogene. 2000;19:5582–5589. doi: 10.1038/sj.onc.1203859. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hawkins PG, Santoso S, Adams C, Anest V, Morris KV. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009;37:2984–2995. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Guo P, Bar-Joseph I, Imanishi Y, Jarzynka MJ, Bogler O, Mikkelsen T, Hirose T, Nishikawa R, Cheng SY. Neuropilin-1 promotes human glioma progression through potentiating the activity of the HGF/SF autocrine pathway. Oncogene. 2007;26:5577–5586. doi: 10.1038/sj.onc.1210348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- Kuwabara T, Hsieh J, Nakashima K, Taira K, Gage FH. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–793. doi: 10.1016/s0092-8674(04)00248-x. [DOI] [PubMed] [Google Scholar]
- Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenstein EJ, Daly RJ, Batzer AG, Li W, Margolis B, Lammers R, Ullrich A, Skolnik EY, Bar-Sagi D, Schlessinger J. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70:431–442. doi: 10.1016/0092-8674(92)90167-b. [DOI] [PubMed] [Google Scholar]
- McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–3121. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 2000;19:5194–5201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard DE, Berra E, Gothie E, Roux D, Pouyssegur J. p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1α (HIF-1α) and enhance the transcriptional activity of HIF-1. J Biol Chem. 1999;274:32631–32637. doi: 10.1074/jbc.274.46.32631. [DOI] [PubMed] [Google Scholar]
- Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wofsberg TG, Umayam L, Lee JC, Hughes CM, Shanmugam KS, Bhattacharjee A, Meyerson M, et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano JV, Liu N, Gao Y, Yao ZJ, Ishibashi T, Underhill C, Burke TR, Jr, Bottaro DP. Inhibition of angiogenesis by growth factor receptor bound protein 2-Src homology 2 domain bound antagonists. Mol Cancer Ther. 2004;3:1289–1299. [PubMed] [Google Scholar]
- Ting AH, Schuebel KE, Herman JG, Baylin SB. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nat Genet. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschuch C, Schulz A, Pscherer A, Werft W, Benner A, Hotz-Wagenblatt A, Barrionuevo LS, Lichter P, Mertens D. Off-target effects of siRNA specific for GFP. BMC Mol Biol. 2008;9:60. doi: 10.1186/1471-2199-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urahama N, Ito M, Sada A, Yakushijin K, Yamamoto K, Okamura A, Minagawa K, Hato A, Chihara K, Roeder RG, et al. The role of transcriptional coactivator TRAP220 in myelomonocytic differentiation. Genes Cells. 2005;10:1127–1137. doi: 10.1111/j.1365-2443.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Weinberg MS, Villeneuve LM, Ehsani A, Amarzguioui M, Aagaard L, Chen ZX, Riggs AD, Rossi JJ, Morris KV. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2006;12:256–262. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg MS, Barichievy S, Schaffer L, Han J, Morris KV. An RNA targeted to the HIV-1 LTR promoter modulates indiscriminate off-target gene activation. Nucleic Acids Res. 2007;35:7303–7312. doi: 10.1093/nar/gkm847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Wei D, Shi Q, Huang S. Constitutive and inducible expression and regulation of vascular endothelial growth factor. Cytokine Growth Factor Rev. 2004;15:297–324. doi: 10.1016/j.cytogfr.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Zarich N, Oliva JL, Martinez N, Jorge R, Ballester A, Gutierrez-Eisman S, Garcia-Vargas S, Rojas JM. Grb2 is a negative modulator of the intrinsic Ras-GEF activity of hSos1. Mol Biol Cell. 2006;17:3591–3597. doi: 10.1091/mbc.E05-12-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]