Abstract
Multiple reaction monitoring (MRM) is a highly sensitive method of targeted mass spectrometry (MS) that can be used to selectively detect and quantify peptides based on the screening of specified precursor peptide-to-fragment ion transitions. MRM-MS sensitivity depends critically on the tuning of instrument parameters, such as collision energy and cone voltage, for the generation of maximal product ion signal. Although generalized equations and values exist for such instrument parameters, there is no clear indication that optimal signal can be reliably produced for all types of MRM transitions using such an algorithmic approach. To address this issue, we have devised a workflow functional on both Waters Quattro Premier and ABI 4000 QTRAP triple quadrupole instruments that allows rapid determination of the optimal value of any programmable instrument parameter for each MRM transition. Here, we demonstrate the strategy for the optimizations of collision energy and cone voltage, but the method could be applied to other instrument parameters, such as declustering potential, as well. The workflow makes use of the incremental adjustment of the precursor and product m/z values at the hundredth decimal place to create a series of MRM targets at different collision energies that can be cycled through in rapid succession within a single run, avoiding any run-to-run variability in execution or comparison. Results are easily visualized and quantified using the MRM software package Mr. M to determine the optimal instrument parameters for each transition.
Keywords: multiple reaction monitoring (MRM), selective reaction monitoring (SRM), optimization, collision energy, cone voltage, triple quadrupole, targeted proteomics
Introduction
Multiple reaction monitoring (MRM) is a targeted mass spectrometry (MS) technique that allows the detection and quantification of specific molecules in a complex mixture. MRM-MS has been cited as an alternative to antibody-based studies for biomarker verification due to its throughput, selectivity, and sensitivity.1,2 MRM-MS has been used successfully to target small molecules,3–5 drug metabolites,6,7 and phosphopeptides.8 Quantitative measurements have also been performed in human serum with limits of detection reported in the nanogram per milliliter (ng/mL) or subnanogram per milliliter (subng/mL) range.9,10
MRM-MS employs tandem quadrupoles to select for specified transitions from precursor peptides to product ions. Individual peptides in the entering ion beam are selected based on mass-to-charge ratio (m/z) by the first quadrupole mass filter and fragmented by collision-induced dissociation (CID) in the second quadrupole. The resulting fragments are then transferred to the third quadrupole, which selectively detects product ions at specified mass-to-charge ratios. The high selectivity of the tandem filters and the long dwell time afforded by the method permit considerable sensitivity and multiplexing.
MRM-MS sensitivity is dependent upon the appropriate tuning of instrument parameters such as collision energy (CE), cone voltage (CV), and declustering potential in order to generate optimal peptide fragmentation and maximal transmission of the desired product ions. The determination of the optimal values for such parameters, however, is still quite empiric. The typical approach for triple quadrupoles employs a data-derived function for collision energy that is linearly proportional to precursor m/z and uses a constant, global value for cone voltage.11,12 While useful as a general rule, this approach may fail to produce the maximum signal under the wide variety of conditions encountered in MRM experiments. Because bond formation depends on peptide residue content and proton mobility,13–16 particular residues or residue combinations in tryptic peptides may not generate the maximum response when fragmented under generalized conditions. In addition, b-type ions tend to undergo secondary fragmentation during transit through the second quadrupole and may require collision energies that are individually optimized.17,18 Peptides with added basicity and limited proton mobility, such as those resulting from missed tryptic cleavage or use of a proteolytic enzyme other than trypsin, may also require collision energies that are far from those employed for the typical tryptic peptide. Finally, the instrument parameters for optimal signal response may not be stable over time for a given ion; variations in gas pressure or drift in instrument voltages may alter even an optimized equation, requiring periodic calibrations.
We have performed two MRM-MS experiments to demonstrate the ease of optimization of any programmable instrument parameter for a given set of precursor–product pairs in a single run. In both experiments, we used a set of 90 transitions from 22 triply charged peptides from a standard protein mixture to investigate how well the peptides obeyed the generalized instrument parameters. In the first experiment, we varied the collision energy by ±6 V relative to the equation-derived value, and in the second experiment, we varied the cone voltage by ±6 V relative to a constant default value. Since multiple entries for a single precursor–product pair are normally not allowed in an MRM experiment, this variation of parameter values was accomplished by subtly adjusting the precursor and product m/z values enough to make a single precursor–product target repeated at multiple collision energies or cone voltages appear to the instrument as multiple targets. With this approach, we enabled the analysis of multiple values for a given instrument parameter in a single run. Data analysis was performed using the MRM software package Mr. M (Single Organism Software Inc., Portland, OR) to easily and quickly determine the optimal operational parameters.
Methods
Preparation of ISB Standard Protein Mixture
A mixture of 18 proteins (Supplemental Table 1) was prepared as described.19 Briefly, 1 nmol of each protein was dissolved in 20 mM, pH 8.0, ammonium bicarbonate with 0.05% SDS added to a final concentration of 1 μM, reduced with 2.5 mM TCEP at 50 °C for 30 min, and alkylated for 1 h with 10 mM iodoacetimide. The proteins were then digested by overnight incubation at 37 °C with sequencing-grade trypsin (Promega, Madison, WI) at a 1:40 (w/w) ratio. Samples were dried in a Speed Vac and cleaned up using a Waters (Milford, MA) Oasis MCX cartridge per the manufacturer’s instruction. The final eluate was evaporated and resuspended in 1 mL of 0.1% formic acid and 1% acetonitrile (ACN), in HPLC-grade water (VWR, West Chester, PA).
Selection of MRM-MS Targets
A published MS/MS data set for the ISB standard protein mixture19 was searched using SEQUEST (version 27) against a database containing the 18 standard proteins of interest and known contaminating proteins appended to a Haemophilus influenzae decoy database (parameters file and database available upon request). The resulting peptide identifications were assigned probabilities using PeptideProphet,20 and all spectra for peptides with probability ≥0.9 were included in the creation of a consensus spectral library using SpectraST.21,22 From this spectral library, SpectraST was used to generate a list of potential MRM targets, which was then filtered to contain only singly charged y-ion fragments greater than 3 amino acids in length generated from triply charged precursor peptides with no methionine residues, no N-terminal glutamine residues, and no modified residues except for carbamiodomethyl cysteine. It was also ensured that at least three transitions per peptide were included in the list to facilitate confident peptide identification. From this list, the top 22 performing peptides were selected for the final analyses, corresponding to 12 different proteins and a total of 90 transitions. This full MRM transition list is provided in Supplemental Table 2.
Reprogramming the Precursor and Product m/z Values
The initial list of 90 transitions was modified to incorporate seven versions of each transition—corresponding to seven different collision energies or cone voltages—for a total of 630 MRM targets. This was done using a simple Perl script (available upon request) that rounds each precursor and product m/z to the nearest tenth, allowing the second decimal place of each value to code for transition and collision energy (or cone voltage), respectively. The script then programs the appropriate collision energy (or cone voltage) for each MRM target based on user-specified values. In this case, the collision energies varied from 6 V less to 6 V more than the equation-derived collision energy in steps of 2 V. The generalized CE equation, which was provided by Waters and developed for doubly charged peptides, is given as:
The cone voltage variation run included voltages varied from 6 V less to 6 V more than the previously determined optimal value of 36 V in steps of 2 V. To illustrate these modification steps, an excerpt from the tab-delimited output of this script is provided and described in Table 1; the full transition list used for both the CE and CV optimization runs is given in Supplemental Table 3.
Table 1.
Excerpt of the Modified Transition List Used To Program the CE Optimization Experiment, Containing All Entries from the Peptide TPHPALTEAKa
| peptide | original Q1 m/z | original Q3 m/z | ion type | default CV | default CE | adjusted Q1 m/z | adjusted Q3 m/z | adjusted CE |
|---|---|---|---|---|---|---|---|---|
| TPHPALTEAK | 355.53 | 448.24 | y4 | 36 | 13.4 | 355.51 | 448.21 | 7.4 |
| TPHPALTEAK | 355.53 | 448.24 | y4 | 36 | 13.4 | 355.51 | 448.22 | 9.4 |
| TPHPALTEAK | 355.53 | 448.24 | y4 | 36 | 13.4 | 355.51 | 448.23 | 11.4 |
| TPHPALTEAK | 355.53 | 448.24 | y4 | 36 | 13.4 | 355.51 | 448.24 | 13.4 |
| TPHPALTEAK | 355.53 | 448.24 | y4 | 36 | 13.4 | 355.51 | 448.25 | 15.4 |
| TPHPALTEAK | 355.53 | 448.24 | y4 | 36 | 13.4 | 355.51 | 448.26 | 17.4 |
| TPHPALTEAK | 355.53 | 448.24 | y4 | 36 | 13.4 | 355.51 | 448.27 | 19.4 |
| TPHPALTEAK | 355.53 | 561.32 | y5 | 36 | 13.4 | 355.52 | 561.31 | 7.4 |
| TPHPALTEAK | 355.53 | 561.32 | y5 | 36 | 13.4 | 355.52 | 561.32 | 9.4 |
| TPHPALTEAK | 355.53 | 561.32 | y5 | 36 | 13.4 | 355.52 | 561.33 | 11.4 |
| TPHPALTEAK | 355.53 | 561.32 | y5 | 36 | 13.4 | 355.52 | 561.34 | 13.4 |
| TPHPALTEAK | 355.53 | 561.32 | y5 | 36 | 13.4 | 355.52 | 561.35 | 15.4 |
| TPHPALTEAK | 355.53 | 561.32 | y5 | 36 | 13.4 | 355.52 | 561.36 | 17.4 |
| TPHPALTEAK | 355.53 | 561.32 | y5 | 36 | 13.4 | 355.52 | 561.37 | 19.4 |
| TPHPALTEAK | 355.53 | 729.41 | y7 | 36 | 13.4 | 355.53 | 729.41 | 7.4 |
| TPHPALTEAK | 355.53 | 729.41 | y7 | 36 | 13.4 | 355.53 | 729.42 | 9.4 |
| TPHPALTEAK | 355.53 | 729.41 | y7 | 36 | 13.4 | 355.53 | 729.43 | 11.4 |
| TPHPALTEAK | 355.53 | 729.41 | y7 | 36 | 13.4 | 355.53 | 729.44 | 13.4 |
| TPHPALTEAK | 355.53 | 729.41 | y7 | 36 | 13.4 | 355.53 | 729.45 | 15.4 |
| TPHPALTEAK | 355.53 | 729.41 | y7 | 36 | 13.4 | 355.53 | 729.46 | 17.4 |
| TPHPALTEAK | 355.53 | 729.41 | y7 | 36 | 13.4 | 355.53 | 729.47 | 19.4 |
The adjusted values are shown in bold font. The Perl script adjusted Q1 and Q3 m/z values by rounding the original m/z values to the nearest tenth and using the second decimal place of each value to code for transition and collision energy, respectively. For example, the adjusted Q1 m/z value of 355.51 is used to signify the 355.53 → 448.24 transition group, and the adjusted Q3 m/z value of 448.21 codes for the given transition targeted with the first collision energy value (default CE −6 V), which in this case is 7.4 V. Each increment of 0.01 m/z encodes an increase in CE of 2 V.
Mass Spectrometry
Data were acquired on a Waters Quattro Premier triple quadrupole coupled with a Waters nanoAcquity UltraPerformance LC (UPLC) pump fitted with a Waters Symmetry 5 μm particle diameter C18 180 μm × 20 mm trap column and a 1.7 μm particle BEH130 C18 100 μm × 100 mm analytical column. After loading and washing for 5 min with 0.1% formic acid in water (buffer A), peptides were eluted using a linear gradient of 1–35% 0.1% formic acid in ACN (buffer B) over 30 min at a flow rate of 300 nL/minute. Both the collision energy variation and cone voltage variation lists were split into two runs each to ensure high-quality, quantifiable elution curves would be acquired, resulting in a total of four MS runs. For all runs, the MS instrument was operated in the positive mode. MS source conditions for all experiments were evaluated for best response under positive mode nanoESI conditions by infusing a standard solution on a regular basis. MS source parameters were as follows: capillary voltage, 2.9 kV; cone voltage, 36 V (unless otherwise specified); source temperature, 90 °C; and cone gas flow rate, 40 L/h at 4 psi. Nitrogen (99.998% purity, Airgas, Seattle, WA) and argon (99.999% purity, Airgas) were used as the cone and collision gases, respectively. The dwell time for each transition was 20 ms with a 5-ms interscan delay, a 5-ms interchannel delay, and a scan width of 0 Da. Each run was scheduled using 32 segments and previously determined retention times ±1 min. Data acquisition for all experiments was carried out by Masslynx V4.1 software. Acquired data files are available upon request.
Data Analysis with Mr. M
Data collected from the four MRM-MS runs were visualized, manually inspected, and quantified using the Mr. M software package provided by Single Organism Software Inc., Portland, OR (see Supplemental Figure 1 for program layout). The MRM transition lists from the aforementioned Perl script and the Masslynx raw files from the four MRM-MS runs were loaded into Mr. M, and each transition was manually reviewed to ensure correct start and end times of peak elution for accurate area under the curve (AUC) calculation. If after manual review a transition was deemed unobservable, the AUC for the transition was reported as zero. AUC was calculated in units of intensity · seconds using each point sampled within the elution period of each measured transition to perform the quantification. Tab-delimited files reporting the observed AUC for each transition were automatically generated and used to determine the optimal CE and CV values.
Results and Discussion
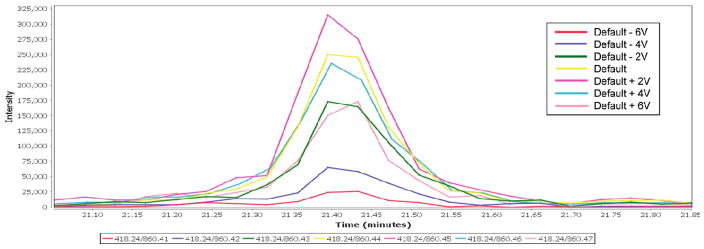
We demonstrate the implementation of a method for collision energy or cone voltage optimization on the Waters Quattro Premier triple quadrupole mass spectrometer with possible application to instruments from other manufacturers. We sought to create an MRM-MS run that successively collected data from the same transition while varying a voltage parameter such as collision energy or cone voltage. Ideally, we would have liked to program the instrument to successfully ramp the collision energy or cone voltage for the same precursor–product pair; however, during a standard analysis, the CE and CV cannot vary for a single pairing. To defeat this restriction, we used an in-house Perl script that alters the m/z value of each precursor peptide and product ion by 0.01 to represent each change in transition and each change in voltage, respectively. This effort yields a “family” of product ion scans from each precursor–product pair that differ in product ion m/z by less than 1 m/z (see Table 1). For each of these entries, the desired CE or CV value can be entered, enabling an effective CE or CV ramp. Because the mass selection window of the triple quadrupole is ~1 m/z, the subtle differences in the precursor and product ion masses have little impact on the quantity of product ion reaching the detector. The end result of this programming strategy is to generate an MS run in which the same precursor–product pair is sequentially dwelled upon with an increasing CE or CV value. Because the switching time of the mass spectrometer (~10 ms) is much faster than the time scale of chromatographic peak elution (~15 s), the sequential scans produce superimposable elution peaks that can easily be compared with one another graphically or in a tabular format using the analysis portion of the MRM software package Mr. M. An example of such an elution peak plot from Mr. M is given in Figure 1.
Figure 1.
Elution peaks (plotted as intensity vs elution time) for the transition 418.2 → 860.4 acquired at seven different collision energies. The second decimal place of the product m/z values listed in the figure codes for the collision energy used, with 860.41 corresponding to the lowest CE tested (default CE −6 V) and 860.47 corresponding to the highest CE tested (CE +6 V). For this particular transition, the plot clearly shows that the signal is maximal at 2 V higher than the default CE (magenta line) and decreases monotonically for both higher and lower CEs.
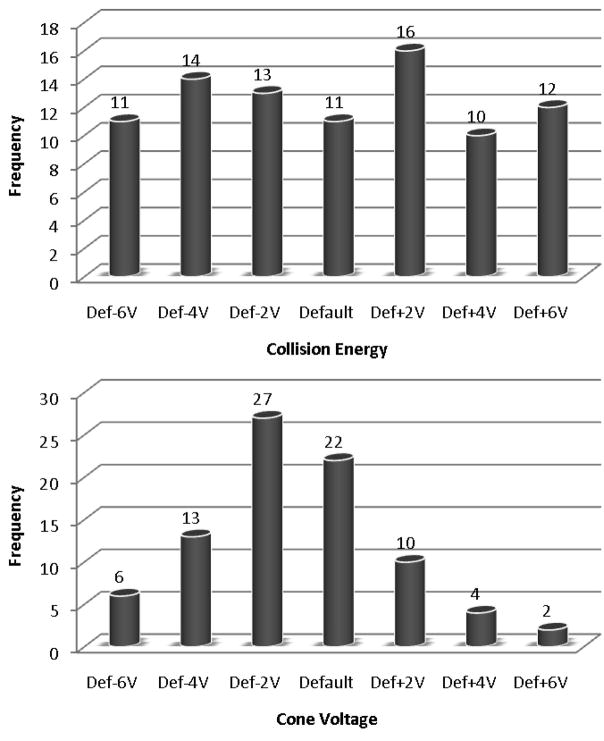
Using this technique, we have programmed CE and CV optimizations of 22 triply charged peptides from a standard mixture of commercial proteins. Triply charged peptides were chosen for this demonstration because default CE values were known to work poorly for this class of targets, but this optimization method could be applied to any set of transitions. In the first optimization, the collision energy was varied across a precursor m/z-dependent range of 12 V, and in the second optimization, the cone voltage was varied across a constant range of 12 V. Each analysis took 1 h to perform and produced optimal CE and CV values for each transition. Excerpts from the tabular data from these analyses are provided in Tables 2 and 3, respectively, and the full data sets are reported in Supplemental Tables 4 and 6, respectively. This information is also reported in terms of the percent gain (or loss) in sensitivity for each CE or CV value in Supplemental Tables 5 and 7. These tables show that the variation of CE produced more substantial gains in signal than the variation in CV, with 59% of transitions experiencing an increase in AUC greater than 30% for the best-performing CE relative to the default CE, while only 35% of transitions experienced this level of increase for CV. A graphical summary of the data is also provided in Figure 2. Most importantly, this tabular data clearly identifies the optimal CE value and the optimal CV value to use for each individual transition, allowing all of these transitions to be targeted in subsequent analyses with maximal sensitivity.
Table 2.
Excerpt from the Tabulated Results from the CE Optimization Experimenta
| peptide | Q1 m/z | Q3 m/z | default CE | AUC def −6 V | AUC def −4 V | AUC def −2 V | AUC default | AUC def +2 V | AUC def +4 V | AUC def +6 V | max AUC | best CE | % AUC gain at best CE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TPHPALTEAK | 355.53 | 448.24 | 13.4 | 175.8 | 1874.6 | 6049.2 | 7466.0 | 7334.2 | 5263.4 | 3703.2 | 7466.0 | Default | 0.0 |
| TPHPALTEAK | 355.53 | 561.32 | 13.4 | 360.8 | 1911.8 | 3415.8 | 3613.9 | 2720.1 | 1611.0 | 966.8 | 3613.9 | Default | 0.0 |
| TPHPALTEAK | 355.53 | 729.41 | 13.4 | 365.2 | 1946.8 | 5394.0 | 10367.1 | 10694.3 | 6385.5 | 2310.9 | 10694.3 | Default +2 V | 3.2 |
| WC[160]TISTHEANK | 449.54 | 461.24 | 16.6 | 125.6 | 247.2 | 1021.6 | 2100.8 | 1775.3 | 2094.8 | 1335.8 | 2100.8 | Default | 0.0 |
| WC[160]TISTHEANK | 449.54 | 598.30 | 16.6 | 10.0 | 134.7 | 403.8 | 1592.6 | 1968.4 | 2065.0 | 1368.2 | 2065.0 | Default +4 V | 29.7 |
| WC[160]TISTHEANK | 449.54 | 786.37 | 16.6 | 96.8 | 437.7 | 1814.0 | 2836.7 | 3591.7 | 2934.5 | 1094.3 | 3591.7 | Default +2 V | 26.6 |
| AVVQDPALKPLALVYGEATSR | 733.41 | 434.24 | 26.25 | 148325.7 | 140175.1 | 147672.8 | 149722.0 | 160255.1 | 180514.7 | 188246.9 | 188246.9 | Default +6 V | 25.7 |
| AVVQDPALKPLALVYGEATSR | 733.41 | 563.28 | 26.25 | 157344.7 | 143305.0 | 130574.7 | 118566.2 | 102495.3 | 96345.9 | 84241.0 | 157344.7 | Default −6 V | 32.7 |
| AVVQDPALKPLALVYGEATSR | 733.41 | 620.30 | 26.25 | 792037.0 | 714166.7 | 667376.4 | 596919.4 | 538276.2 | 467596.2 | 393553.0 | 792037.0 | Default −6 V | 32.7 |
| AVVQDPALKPLALVYGEATSR | 733.41 | 783.36 | 26.25 | 794143.7 | 701925.4 | 670617.9 | 571015.4 | 508404.3 | 421550.2 | 342849.4 | 794143.7 | Default −6 V | 39.1 |
| AVVQDPALKPLALVYGEATSR | 733.41 | 882.44 | 26.25 | 161400.8 | 164003.5 | 161493.5 | 154642.9 | 153000.7 | 134212.1 | 124132.4 | 164003.5 | Default −4 V | 6.1 |
The AUC calculations were performed by the MRM software package Mr. M, and the CE that produced the largest AUC was taken to be the best-performing CE for the given transition. The percent gain in AUC represents the increase in AUC for the best-performing CE relative to the default CE.
Table 3.
Excerpt from the Tabulated Results from the CV Optimization Experimenta
| peptide | Q1 m/z | Q3 m/z | default CV | default CV | default CV | AUC def −2 V | AUC default | AUC def +2 V | AUC def +2 V | AUC def +6 V | max AUC | best CV | % AUC gain at best CV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TPHPALTEAK | 355.53 | 448.24 | 36 | 1575.0 | 2621.0 | 3605.1 | 3764.7 | 3552.3 | 3775.7 | 3480.7 | 3775.7 | Default +4 V | 0.3 |
| TPHPALTEAK | 355.53 | 561.32 | 36 | 1359.2 | 1609.6 | 1922.3 | 1702.8 | 1630.8 | 1892.5 | 1481.6 | 1922.3 | Default −2 V | 12.9 |
| TPHPALTEAK | 355.53 | 729.41 | 36 | 4328.5 | 5235.2 | 5102.2 | 5014.6 | 4653.8 | 4927.4 | 3742.2 | 5235.2 | Default −4 V | 4.4 |
| WC[160]TISTHEANK | 449.54 | 461.24 | 36 | 1109.8 | 1010.4 | 1145.1 | 1115.9 | 991.5 | 822.2 | 635.9 | 1145.1 | Default −2 V | 2.6 |
| WC[160]TISTHEANK | 449.54 | 598.30 | 36 | 791.0 | 507.6 | 545.3 | 814.5 | 828.1 | 405.2 | 471.4 | 828.1 | Default +2 V | 1.7 |
| WC[160]TISTHEANK | 449.54 | 786.37 | 36 | 1037.7 | 1472.7 | 1726.9 | 2283.3 | 1313.4 | 1529.4 | 806.8 | 2283.3 | Default | 0.0 |
| AVVQDPALKPLALVYGEATSR | 733.41 | 434.24 | 36 | 7249.9 | 7305.5 | 8052.4 | 8030.0 | 8136.8 | 5980.1 | 7246.1 | 8136.8 | Default +2 V | 1.3 |
| AVVQDPALKPLALVYGEATSR | 733.41 | 563.28 | 36 | 5310.8 | 5920.4 | 5729.5 | 5378.3 | 6143.5 | 5195.3 | 4956.1 | 6143.5 | Default +2 V | 14.2 |
| AVVQDPALKPLALVYGEATSR | 733.41 | 620.30 | 36 | 23479.0 | 26911.3 | 24702.9 | 27168.1 | 26404.7 | 22470.2 | 24323.1 | 27168.1 | Default | 0.0 |
| AVVQDPALKPLALVYGEATSR | 733.41 | 783.36 | 36 | 27002.5 | 27145.7 | 24043.7 | 26749.6 | 25157.1 | 23732.7 | 23591.2 | 27145.7 | Default −4 V | 1.5 |
| AVVQDPALKPLALVYGEATSR | 733.41 | 882.44 | 36 | 6775.5 | 7654.5 | 7950.4 | 7954.0 | 7068.5 | 6519.1 | 5395.6 | 7954.0 | Default | 0.0 |
The AUC calculations were performed by the MRM software package Mr. M, and the CV that produced the largest AUC was taken to be the best-performing CV for the given transition. The percent gain in AUC represents the increase in AUC for the best-performing CV relative to the default CV.
Figure 2.
Histograms illustrating the number of times each CE (top) and CV (bottom) was the best-performing CE or CV for a targeted transition. The best-performing CE or CV for each transition was defined as the value that produced the maximum AUC for the given transition.
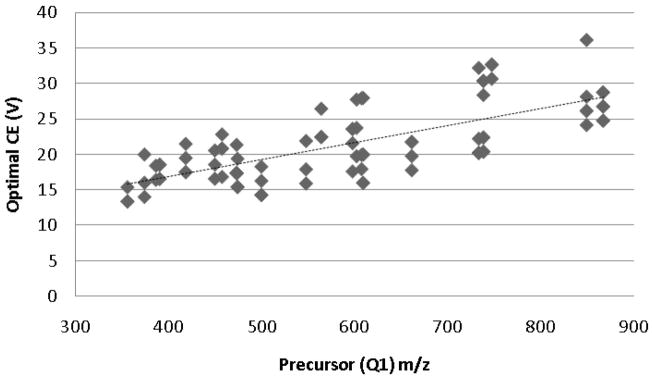
In addition to using the optimized CE and CV values for each transition directly, one can also use these results to determine generalized equations or values for certain classes of peptides or transitions to allow the extrapolation of optimal parameter values for transitions not explicitly optimized in the analysis. In this case, we can use the optimization results to try to determine CE and CV equations or values for triply charged peptides. The data from our optimization runs suggest that cone voltage is fairly generalizable for this class of peptides, as the large majority of transitions were optimized at the default CV (36 V) or 2 V less than the default (34 V). The collision energy, however, is more difficult to generalize, as all seven of the relative collision energies tested were optimal for approximately the same number of transitions. This is not unexpected, since the default CE equation is linearly dependent upon precursor peptide m/z. The optimal CE values determined by our analysis for each transition were therefore plotted against precursor (Q1) m/z (Figure 3), and the data was fit with a linear curve, yielding the equation
Figure 3.
Plot of the experimentally determined optimal CE for each transition versus the corresponding precursor peptide m/z value. The dashed line (y = 0.0241x + 7.2239) represents a linear fit to the data (R2 = 0.5238).
with an R2 value of 0.5238. Three major conclusions can be drawn from this CE generalization: (1) the newly derived equation for collision energy exhibits a linear dependence upon precursor m/z, as expected; (2) this equation is substantially different than the default CE equation, suggesting that triply charged peptides indeed require optimization for CE that is different from that used for the more typical doubly charged peptides; and (3) there is considerable variance in the optimal collision energies, suggesting that further studies are required to identify the peptide-specific parameters that determine which transitions follow the equation and which require individual optimization. Further optimization experiments can then be performed—using the same method described here—to elucidate more precise CE equations for any identified subsets of transitions.
Our strategy streamlines the determination of optimal instrument parameters for individual MRM transitions. Although a simplistic, well-defined protein mixture was used for the demonstration of this method, it works just as well for peptides within a complex background, provided the concentration of the given peptide is within the detection limits of the mass spectrometer. We have illustrated this functionality in a supplementary analysis in which we performed a collision energy optimization of 40 transitions from 10 yeast peptides in a yeast lysate (see the Supplemental Optimization in the Supporting Information). Using the optimization technique described herein, we determined an optimal CE value for each of the 40 transitions, proving that the method is applicable to biologically relevant samples. Additionally, although this optimization method was developed for the Waters Quattro Premier, we have also successfully performed this same technique using the ABI 4000 QTRAP (methods and data available in the Supplemental Analysis included in the Supporting Information), and it can potentially be applied to other platforms as well. Finally, a commercial MRM reviewing application (Mr. M) was used for all data analysis; however, our recently published MRMer software12 will also allow evaluation and output of the results, as will the software provided by the instrument vendors.
The primary advantages offered by this optimization method are threefold. First, this technique allows the investigator to perform a CE or CV ramp for one or many precursor–product transitions in a single run, thereby eliminating run-to-run variability. Second, the method permits such an optimization using a protein mixture or complex sample, rather than requiring the infusion of a purified or synthesized peptide. Lastly, the ease of data interpretation and analysis afforded by programs such as Mr. M makes this optimization fast and efficient.
Conclusions
We have defined a method for performing effective collision energy and cone voltage ramps for many precursor-product pairs within a single MRM run and without the need for a pure peptide. With this strategy, one can easily determine the optimal instrument parameters for experimental conditions and transitions of interest. This method may speed the development of targeted analysis for both basic and translational proteomic workflows.
Supplementary Material
Acknowledgments
This work was supported by grant P50GM076547 and 5R21CA126216 (to D.B.M) and contract N01-HV-28179 from the National Heart, Lung, and Blood Institute.
Footnotes
Supporting Information Available: Supplemental Figure 1, a screen shot of the MRM software package Mr. M; Supplemental Table 1, a list of the 18 proteins constituting the ISB standard protein mix; Supplemental Table 2, the initial MRM transition list, containing 90 transitions, used in the CE and CV optimization experiments; Supplemental Table 3, the full transition list, including m/z modifications, used to program the CE and CV optimization experiments; Supplemental Table 4, the tabulated results (in the form of AUC calculations) for the CE optimization experiment; Supplemental Table 5, the same tabulated CE optimization results, this time in terms of the percent gain (or loss) in sensitivity for each CE; Supplemental Table 6, the tabulated results (in the form of AUC calculations) for the CV optimization experiment; Supplemental Table 7, the same tabulated CV optimization results, this time in terms of the percent gain (or loss) in sensitivity for each CV; Supplemental Optimization, the methods and results for the collision energy optimization of yeast peptides in a complex background of yeast lysate; and Supplemental Analysis, the methods and results for the collision energy optimization performed on an ABI 4000 QTRAP. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Arnott D, Kishiyama A, Luis EA, Ludlum SG, Marsters JC, Jr, Stults JT. Selective detection of membrane proteins without antibodies: a mass spectrometric version of the Western blot. Mol Cell Proteomics. 2002;1(2):148–156. doi: 10.1074/mcp.m100027-mcp200. [DOI] [PubMed] [Google Scholar]
- 2.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5(4):573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Kostiainen R, Kotiaho T, Kuuranne T, Auriola S. Liquid chromatography/atmospheric pressure ionization-mass spectrometry in drug metabolism studies. J Mass Spectrom. 2003;38(4):357–372. doi: 10.1002/jms.481. [DOI] [PubMed] [Google Scholar]
- 4.Sannino A, Bolzoni L, Bandini M. Application of liquid chromatography with electrospray tandem mass spectrometry to the determination of a new generation of pesticides in processed fruits and vegetables. J Chromatogr, A. 2004;1036(2):161–169. doi: 10.1016/j.chroma.2004.02.078. [DOI] [PubMed] [Google Scholar]
- 5.Tai SS, Bunk DM, White Et, Welch MJ. Development and evaluation of a reference measurement procedure for the determination of total 3,3′,5-triiodothyronine in human serum using isotope-dilution liquid chromatography-tandem mass spectrometry. Anal Chem. 2004;76(17):5092–5096. doi: 10.1021/ac049516h. [DOI] [PubMed] [Google Scholar]
- 6.Lee MS, Kerns EH. LC/MS applications in drug development. Mass Spectrom Rev. 1999;18(3–4):187–279. doi: 10.1002/(SICI)1098-2787(1999)18:3/4<187::AID-MAS2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 7.Tiller PR, Cunniff J, Land AP, Schwartz J, Jardine I, Wakefield M, Lopez L, Newton JF, Burton RD, Folk BM, Buhrman DL, Price P, Wu D. Drug quantitation on a benchtop liquid chromatography-tandem mass spectrometry system. J Chromatogr, A. 1997;771(1–2):119–125. doi: 10.1016/s0021-9673(97)00147-7. [DOI] [PubMed] [Google Scholar]
- 8.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci USA. 2007;104(14):5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6(12):2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, Krek W, Aebersold R, Domon B. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics. 2007;6(10):1809–1817. doi: 10.1074/mcp.M700132-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin DB, Holzman T, May D, Peterson A, Eastham A, Eng J, McIntosh M. MRMer, an interactive open source and cross-platform system for data extraction and visualization of multiple reaction monitoring experiments. Mol Cell Proteomics. 2008;7(11):2270–2278. doi: 10.1074/mcp.M700504-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y, Triscari JM, Tseng GC, Pasa-Tolic L, Lipton MS, Smith RD, Wysocki VH. Statistical characterization of the charge state and residue dependence of low-energy CID peptide dissociation patterns. Anal Chem. 2005;77(18):5800–5813. doi: 10.1021/ac0480949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dongre AR, Jones JL, Somogyi A, Wysocki VH. Influence of peptide composition, gas-phase basicity, and chemical modification on fragmentation efficiency: evidence for the mobile proton model. J Am Chem Soc. 1996;118(35):8365–8374. [Google Scholar]
- 15.Kapp EA, Schutz F, Reid GE, Eddes JS, Moritz RL, O’Hair RA, Speed TP, Simpson RJ. Mining a tandem mass spectrometry database to determine the trends and global factors influencing peptide fragmentation. Anal Chem. 2003;75(22):6251–6264. doi: 10.1021/ac034616t. [DOI] [PubMed] [Google Scholar]
- 16.Wysocki VH, Tsaprailis G, Smith LL, Breci LA. Mobile and localized protons: a framework for understanding peptide dissociation. J Mass Spectrom. 2000;35(12):1399–1406. doi: 10.1002/1096-9888(200012)35:12<1399::AID-JMS86>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 17.Paizs B, Suhai S. Towards understanding the tandem mass spectra of protonated oligopeptides. 1: mechanism of amide bond cleavage. J Am Soc Mass Spectrom. 2004;15(1):103–113. doi: 10.1016/j.jasms.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 18.Paizs B, Suhai S. Fragmentation pathways of protonated peptides. Mass Spectrom Rev. 2005;24:508–548. doi: 10.1002/mas.20024. [DOI] [PubMed] [Google Scholar]
- 19.Klimek J, Eddes JS, Hohmann L, Jackson J, Peterson A, Letarte S, Gafken PR, Katz JE, Mallick P, Lee H, Schmidt A, Ossola R, Eng JK, Aebersold R, Martin DB. The standard protein mix database: a diverse data set to assist in the production of improved peptide and protein identification software tools. J Proteome Res. 2008;7:96–103. doi: 10.1021/pr070244j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005;1:2005.0017. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam H, Deutsch EW, Eddes JS, Eng JK, King N, Stein SE, Aebersold R. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics. 2007;7(5):655–667. doi: 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- 22.Lam H, Deutsch EW, Eddes JS, Eng JK, Stein SE, Aebersold R. Building consensus spectral libraries for peptide identification in proteomics. Nat Methods. 2008;5:873–875. doi: 10.1038/nmeth.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.