Abstract
Notch signaling plays an essential role in diverse biological processes during development and in pathogenesis of diseases ranging from cancer to cerebrovascular disorders. Precise regulation of Notch signaling is essential for normal function and requires both timely activation and inactivation of the intracellular domain (ICD) of Notch receptors. In addition, inappropriate buildup of Notch3 ectodomain is a hallmark pathological feature of the stroke and dementia disorder cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL). Thus, a clear understanding of mechanisms of Notch protein turnover is essential for understanding normal and pathological mechanisms of Notch function. Previous studies showed that the degradation of ICDs of Notch1 and Notch4 is controlled by the ubiquitin-proteasome system (UPS), though more recent work demonstrated that Notch1 ICD is also controlled by lysosomal degradation. The mechanism of degradation of Notch3 has not yet been identified. Here we report that the degradation of ICD of Notch3 (N3-ICD) is mediated by lysosomes. Lysosome inhibitors chloroquine and NH4Cl led to the accumulation of transfected N3-ICD in 293 cells and endogenous N3-ICD in C2C12, H460, and HeLa cell lines; in addition, inhibition of lysosome function by chloroquine and NH4Cl delayed the degradation of N3-ICD. In contrast, N3-ICD was not affected by proteasome inhibitors MG132 and lactacystin. Furthermore, we find that the Notch3 extracellular domain (N3-ECD) is also subject to lysosome-dependent degradation. In sum, our experiments demonstrate a critical role for lysosomes in the degradation of Notch3, which distinguishes it from Notch1 and Notch4.
Keywords: Notch3, lysosome, proteasome, degradation, ectodomain
1. Introduction
Notch signaling pathways are essential for cell fate determination during development and critical effectors of disease pathogenesis. To activate Notch signaling, Notch receptors (Notch1 to Notch4) undergo a series of proteolytic processing events. Initially, Notch is targeted to the endoplasmic reticulum and Golgi apparatus, where it undergoes proteolytic processing (at the S1 site; (Blaumueller et al., 1997; Logeat et al., 1998)). Upon binding to Notch ligands, Notch undergoes extracellular cleavage at the S2 site (Brou et al., 2000; Mumm et al., 2000). The C-terminal product of this event is an intermediate that undergoes further proteolysis within the transmembrane domain (S3 site; (Okochi et al., 2002; Saxena et al., 2001)) to release the Notch intracellular domain (NICD), which translocates to the nucleus and regulates transcriptional activity of target genes, such as the hairy/enhancer of split (HES) genes (Artavanis-Tsakonas S, 1999; Iso et al., 2003).
Degradation of Notch protein is important for at least two reasons. First, NICD levels determine the potency of cell signaling; proteolysis of NICD may correlate with attenuation of Notch activation of target genes. Second, ectodomain degradation may be a key modulator of signaling and may also play a direct role in disease pathogenesis, either limiting Notch signaling or exerting non-canonical (N3-ICD-independent functions). During mammalian development, quantitative levels of Notch signaling exert profound effects on phenotype. For example, changes in hair color progressively change with each stepwise reduction in the number of Notch1/2 alleles active in knockout mice (Schouwey et al., 2007). Notch3 is overexpressed in ovarian (Park et al., 2006), lung (Dang et al., 2000), and breast (Yamaguchi et al., 2008) cancers; both ICD and ectodomain degradation could, in theory, attenuate signaling through Notch and impair tumor growth. Additionally, accumulation of Notch3 ectodomain has been reported in the stroke and dementia disorder CADASIL (Joutel et al., 2000), which is caused by stereotypical mutations in the NOTCH3 gene (Joutel et al., 1996). Enhanced clearance of the Notch3 ectodomain could ameliorate stroke and cognitive deficits in this disease.
Previous studies have focused primarily on Notch1 degradation and have demonstrated a role of the ubiquitin-proteasome system (UPS). E3 ubiquitin ligases Fbw7/Sel-10, c-Cbl1 and Itch are capable of catalyzing ubiquitylation of Notch1 (Gupta-Rossi et al., 2001; McGill and McGlade, 2003; Oberg et al., 2001; Qiu et al., 2000). Inhibition of proteasomes in cell cultures transiently overexpressing Notch1 ICD results in enhanced protein levels (Gupta-Rossi et al., 2001; McGill and McGlade, 2003; Oberg et al., 2001; Qiu et al., 2000), suggesting a role or the UPS in regulating levels of activated Notch1. Although a large body of work supports the ubiquitylation and proteasome-mediated degradation of Notch1, these studies have not examined the levels of endogenously produced Notch1 ICD in the presence of UPS inhibition, which is hard to evaluate because of levels of Notch1 ICD production.
In addition, more recent investigations have suggested that ubiquitylation driven lysosomal degradation may account for proteolysis of Notch1 ICD (Jehn et al., 2002). Jehn et al. showed that N1-ICD is ubiquitylated and recognized by c-Cbl and ultimately disposed of by lysosomes. Interestingly, these investigators saw significant increases in the levels of Notch1-ICD after application of two lysosome inhibitors (cholorquine and NH4Cl), but failed to detect changes in protein levels with proteasome inhibitors. Unlike earlier studies, Jehn et al. focused on endogenous Notch1 protein expressed in C2C12 cells. In addition, Itch/AIP4 has now been shown to mediate polyubiquitylation-dependent targeting of retrovirally expressed Notch1 ectodomain to lysosomes in the absence of ligand (Chastagner et al., 2008). In aggregate, prior work suggest that proteolytic clearance of Notch proteins could utilize both lysosomal and proteasomal pathways.
In Drosophila, considerable evidence has also suggested that lysosomal degradation plays an important role in regulated Notch activity (Wilkin et al., 2008). However, much less work has been performed on degradation of other mammalian Notch isoforms (Notch 2–4). N4 degradation was investigated by (Wu et al., 2001), who showed that Notch4 is targeted by ubiquitin ligase SEL-10; ubiquitylation and/or SEL-10 binding of C-terminal constructs distal to the ankryn repeats of Notch4 targeted the protein for proteasome mediated destruction. However, unlike Notch1 ICD, the intact Notch4 ICD was relatively unaffected by a dominant negative SEL-10 and was not affected by proteasome inhibition, revealing a complex mode of degradation regulation apparently distinct from Notch1. Other work suggests that Notch isoforms in vivo may indeed differ in mechanisms of degradation. In vivo studies of mouse knockouts of the E3 ubiquitin ligase Fbw7/Sel-10/Cdc4 (Tsunematsu et al., 2004) demonstrate profound defects in angiogenesis that were associated with preferential increases in Notch4 ICD, without alterations in Notch1–3.
Limited work has been performed on Notch3 degradation. Oberg et al. (Oberg et al., 2001) showed that E3 ligases colocalize with Notch3 in overexpression studies in the nucleus and also form direct complexes with the Notch3 ICD via protein interactions; although they described ubiquitylation of Notch1 ICD and upregulation of Notch1 ICD levels with proteasome inhibitors, they did not report similar results for Notch3 ICD. The mechanisms of Notch3 turnover are thus still undefined, prompting us to test whether proteasomes or lysosomes were responsible for Notch3 degradation.
2. Materials and methods
2.1 Constructs
The Notch3 intracellular domain construct (Dang et al., 2006) and the full length human cDNAs (Zhang et al., 2007) were constructed in pCMV-Sport6 (Invitrogen). TM-N3-IC was generated by PCR from the predicted S2 site of Notch3 to the C-terminus and cloned into pSecTag, which includes a canonical secretion tag from IgG for direction into the secretory pathway (Zhang et al., 2007). The C49Y Notch3 CADASIL mutant was generated by PCR mutagenesis of the full length wild type Notch3 clone.
2.2 Cell lines and cell culture
Human embryonic kidney (HEK) 293, mouse myoblast C2C12, human H460 lung and human cervical adenocarcinoma HeLa cells were originally purchased from ATCC and propagated in conventional FBS supplemented media. HEK293-Notch3 cell lines were constructed by stable transfection of 293 cells with wild type or mutant Notch3. Stable clones were selected by cotransfection with a resistance plasmid followed by puromycin selection for one week.
2.3 Notch3 accumulation assay
To evaluate the accumulation of intercellular domain (ICD) and the extracellular domain (ECD) of Notch3 (N3-ICD and N3-ECD, respectively), we cultured cells in the presence of lysosome inhibitors (chloroquine at 25 µM or NH4Cl at 25 mM) or proteasome inhibitors (MG132 at 10 µM and, in some experiments, lactacystin at 10 µM). At 0h, 12h, 24h and 36h post treatment, cells were collected and cell lysates were prepared for immunoblotting of N3-ICD or N3-ECD. Western blotting for tubulin (DSHB, University of Iowa) was used as a protein loading control.
2.4 N3-ICD degradation assay
Cells cultured in 12-well plates were treated with lysosome inhibitor chloroquine at 25 µM for 24h to induce accumulation of N3-ICD. Then the cells were treated with the protein synthesis inhibitor cycloheximide (CHX; 100µg/mL) plus chloroquine (25 µM) or CHX (100µg/mL) plus vehicle to following protein degradation over time. At 0h, 12h, 24h and 36h post treatment, cells were collected and cell lysates were prepared for immunoblotting of N3-ICD, with tubulin as a loading control.
2.5 Immunoblotting
Western blots was performed as previously described (Meng et al., 2009; Zhang et al., 2007) using antibodies against N3-ICD (M-134, Santa Cruz Biotechnologies) from rabbit and wild type N3-ECD (R&D) from sheep. In some experiments, we used a custom antipeptide rabbit antibody directed against N3-ECD-C49Y. IRDye800-labeled secondary antibodies (Rockland) were used for analyzing and quantitating the probed membranes with an Odyssey imaging system (LI-COR Biosciences).
2.6 Notch3 signaling activity assay
The activity of Notch3 signaling was evaluated using the HES-luciferase reporter described (Meng et al., 2009; Zhang et al., 2007). Briefly, H460 and C2C12 cells were transfected with a total of 1 µg of DNA (200 ng HES-luciferase, and 100 ng Renilla luciferase control plasmid (pRL-TK; Promega), adjusted to 1 µg with empty vector). Twenty four hours after transfection, cells were treated with chloroquine at 25 µor vehicle for 24 hours. Firefly and renilla luciferase activities were determined using the Dual Luciferase Assay Kit (Promega) and relative luciferase units (firefly to renilla luciferase ratio) were presented.
3. Results
3.1 N3-ICD accumulates after inhibition of lysosomal degradation
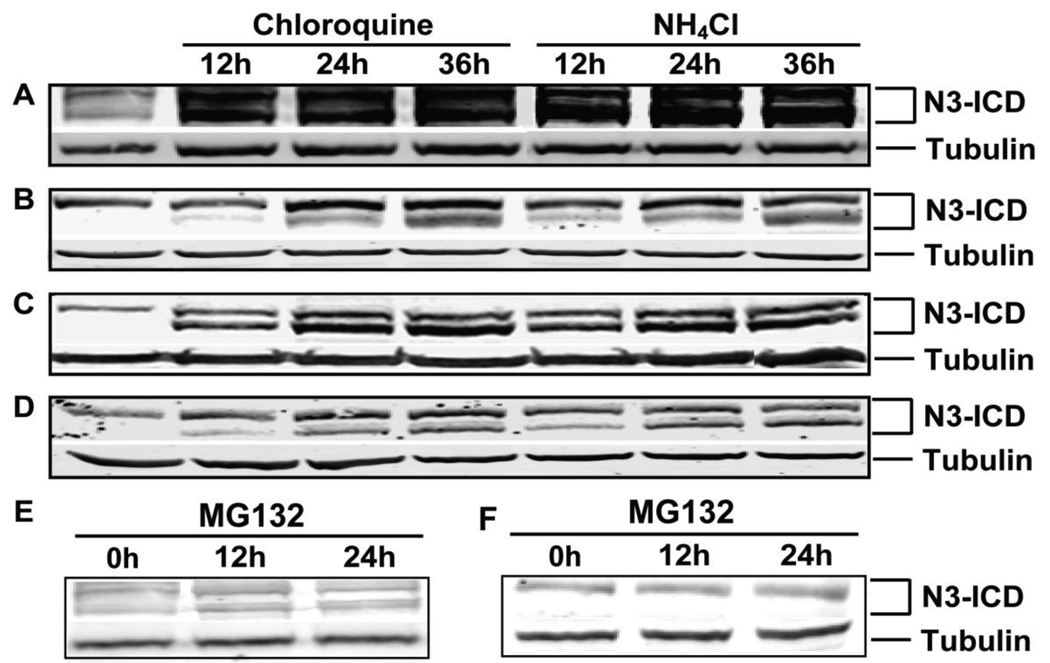
To determine whether proteasomal or lysosomal degradation systems regulate N3-ICD, we determined the response of N3-ICD to lysosome or proteasome inhibition by immunobloting against the C-terminus of Notch3. As shown in Figure1A, N3-ICD accumulated in wild type Notch3-expressing 293 cells (293-Notch3) after application of the lysosome inhibitors chloroquine or NH4Cl (Fig. 1A). Importantly, similar increases in endogenous N3-ICD were seen in mouse myoblast C2C12 cells (Fig. 1B), human H460 lung carcinoma cells (Fig. 1C), as well as human HeLa cervical adenocarcinoma cells (Fig. 1D) after inhibition of lysosomes with either chloroquine or NH4Cl. Two bands were observed in all cell lines at approximately 80 and 85 kDa. Both bands were enhanced by lysosome inhibition in all four cell lines, with the lower band showing more marked changes in expression. The proteasome inhibitor MG132 had no effect on N3-ICD levels in any of the cell lines examined (Fig. 1E and 1F).
Fig 1. Lysosome inhibitor treatment and N3-ICD accumulation in cells.
Cells were treated with lysosome inhibitors chloroquine or NH4Cl. After 0h, 12h, 24h and 36h post treatment, N3-ICD was detected by immunoblotting using tubulin as loading control. (A) HEK 293-Notch3 cells; (B) mouse myoblast C2C12 cells; (C) H460 human lung carcinoma cells and (D) HeLa human cervical adenocarcinoma cells. Proteasome inhibitor MG132 had no effect on the levels of N3-ICD when applied to HEK 293-Notch3 cells (E) and H460 cells (F).
3.2 Delay of N3-ICD degradation by lysosome inhibition
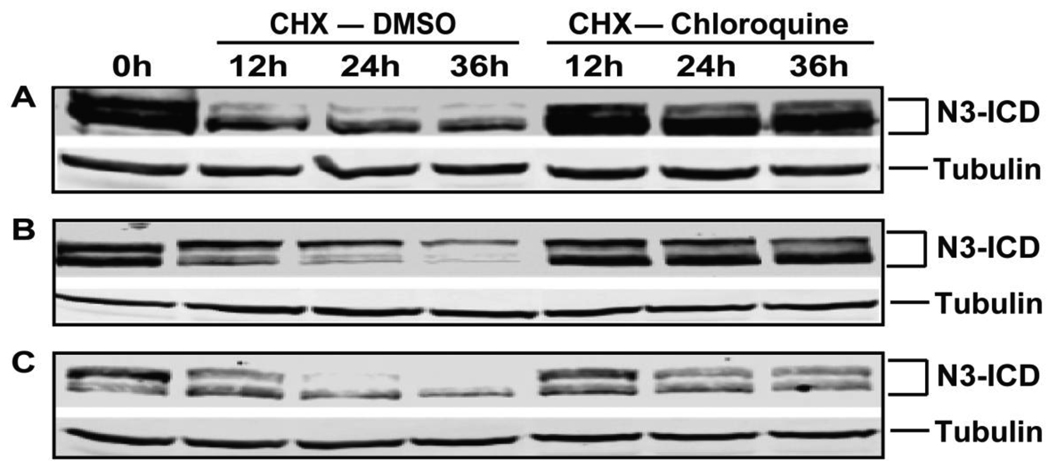
We next determined whether lysosome inhibition delays the degradation of N3-ICD proteins. To test this, cells were firstly treated with chloroquine for 24 hours leading to the accumulation of N3-ICD (the first lane of Figure 2), followed by periods during which we inhibited further protein synthesis by applying cycloheximide (CHX). In the presence of CHX alone, N3-ICD consistently degraded over the 36 hour period of observation. In contrast, when CHX and chloroquine were applied together, N3-ICD levels were virtually unchanged over 36 hours. N3-ICD degradation was significantly inhibited by lysosome inhibition in all cell lines tested. The proteasome inhibitor MG132 failed to alter the degradation kinetics of N3-ICD (not shown). The accumulation of N3-ICD (Figure 1) and the delay of its degradation (Figure 2) with lysosome inhibition clearly demonstrated that N3-ICD is regulated by the lysosomal degradation system and not by the ubiquitin proteasome system.
Fig 2. Lysosome inhibitor treatment and the degradation of N3-ICD.
Cells were treated with chloroquine for 24 hours resulting in the accumulation of N3-ICD (the first lane, 0 hour). Cells were then treated with cycloheximide (CHX) together with vehicle or chloroquine for 0h, 12h, 24h and 36h. Protein lysates were analyzed for N3-ICD and tubulin. (A) HEK 293-Notch3 cells; (B) H460 human lung carcinoma cells; and (C) HeLa human cervical adenocarcinoma cells.
3.3 Identification of N3-ICD fragments
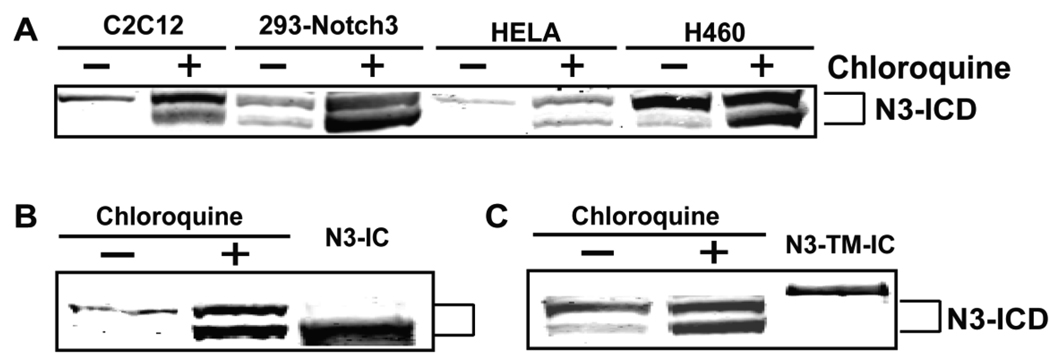
As shown in Figure 1 and Figure 2, two fragments are recognized by the N3-ICD antibody with molecular weights about 80 and 85 kDa, which both accumulate after lysosome inhibition. Both fragments are absent in untransfected 293 cells, demonstrating that they are generated from the Notch3 protein. Proteins from multiple cell lines (treated for 24 hours with chloroquine) run on the same gel confirmed that the two accumulated fragments are conserved in size between mouse and human cells of multiple origins. Constructs expressing the intracellular domain of Notch3 (N3-ICD) or membrane-associated Notch3 (N3-TM-IC which is identical in size to the S2-cut Notch3 protein including a transmembrane domain and the intracellular domain of Notch3) were expressed by transfection and compared by immunoblotting of proteins which accumulated after lysosome inhibitor treatment. As shown in Figure 3B, N3-ICD comigrates with the lower band induced by chloroquine treatment, whereas N3-TM-IC was significantly larger in size compared to both accumulated fragments. The results suggested that the fragment with lower MW accumulated upon lysosome inhibition is the active N3-ICD; the other induced band could represent a novel digestion product of Notch3 or a new posttranslationally modified intracellular product.
Fig 3. Conserved apparent molecular weights of N3-ICD fragments.
(A) Multiple cell lines were treated with media or chloroquine for 24 hours. Two fragments with apparent molecular weights of 80 and 85 kDa were detected using an antibody against the C-terminus of Notch3. The sizes of the two enhanced fragments were compared to transfected N3-ICD (B) and to N3-TM-IC, which corresponds to the S2 cut Notch3 fragment.
3.4 N3-ICD accumulation after lysosome inhibition does not enhance Notch3 signaling
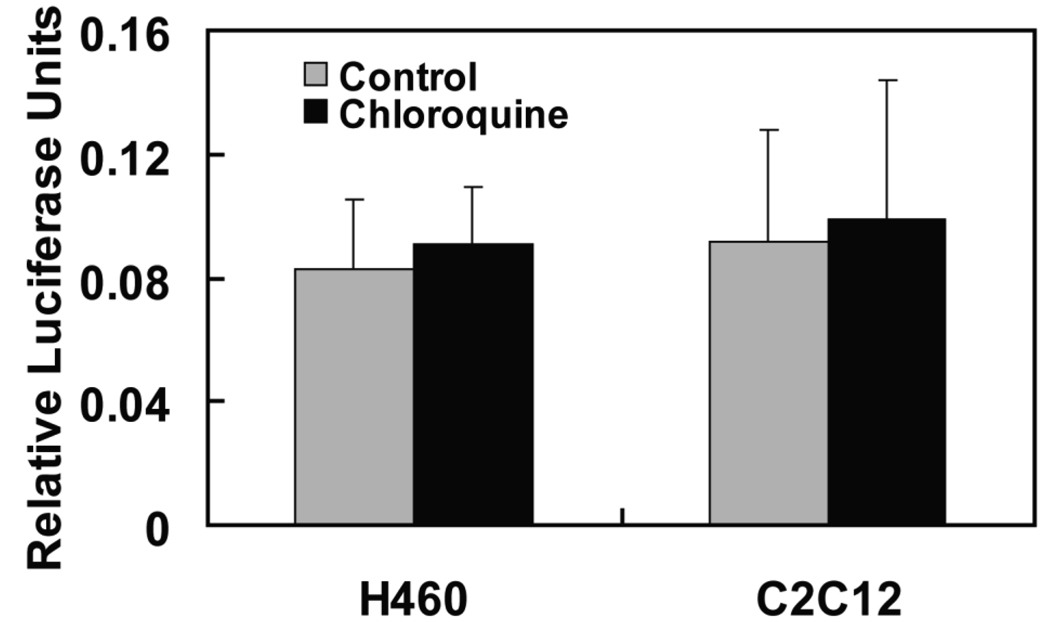
Notch signaling regulates many physiological and pathological processes and is strictly controlled by degradation of Notch-ICD. Since we observed strong stabilztion of N3-ICD with lysosome inhibitors, we tested whether lysosome inhibition regulates Notch3 signaling. Cotransfection of HES-luciferase, an established measure of the efficiency of canonical Notch signaling, was used to quantify Notch3 activity. The ectopic expression of N3-ICD by transient transfection significantly upregulated the reporter activity (data not shown), confirming the responsiveness of this reporter to Notch3. However, no significant enhancement of Notch3 signaling was seen after treatment of both H460 and C2C12 cells with chloroquine (Figure 4), although the treatment consistently led to the accumulation of N3-ICD (Figure 1 and Figure 2). The results suggested that N3-ICD accumulation after lysosome inhibition does not functionally activate Notch3 signaling.
Fig 4. N3-ICD accumulation did not enhance Notch3 signaling.
H460 and C2C12 cells were transfected with a HES-luciferase reporter and a renilla luciferase plasmid to normalize transfection efficiencies between wells. Cell were then treated with chloroquine or vehicle for 24 hours. Luciferase activities were determined to measure the levels of Notch signaling activity, as described in the Materials and Methods.
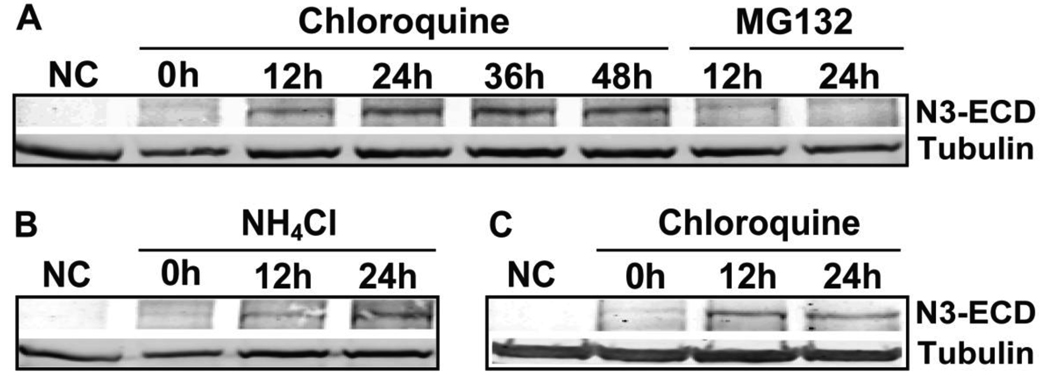
3.5 N3-ECD accumulation upon the inhibition of lysosome, but not proteasome
The extracellular domain of Notch3 (N3-ECD) are rarely detected in normal tissues; however, it accumulates in smooth muscle cells of CADASIL patients (Joutel et al., 2000), which suggests relevance of ectodomain degradation to disease progression. As shown in Figure 5A, N3-ECD expressed in cell lysates accumulates after treatment of 293-Notch3 cells with the lysosome inhibitor chloroquine, but not the proteasome inhibitor MG132, suggesting that N3-ECD is also regulated by a lysosomal pathway. The cellular accumulation of N3-ECD was also observed in the cells treated with NH4Cl (Figure 5B). Similar results were obtained in 293 cell lines expressing a mutated Notch3 (C49Y) (Figure 5C). In summary, these findings demonstrated that N3-ECD is regulated by lysosome, but not proteasome pathways.
Fig 5. Lysosome inhibition and N3-ECD accumulation in cells.
(A) Chloroquine, but not MG132, treatment led to the accumulation of N3-ECD in HEK 293-Notch3 cells, detected in cell lysates with an antibody against the extracellular domain of Notch3. Tubulin was used as a loading control. (B) NH4Cl treatment led to a similar accumulation of N3-ECD in 293-Notch3 cells. (C) Mutant N3-ECD accumulated after treatment with chloroquine. The protein was detected in lysates from mutant Notch3 cell lines using an antibody that specifically recognizes N3-C49Y. NC: negative control wild type 293 cells that lack Notch3.
4. Discussion
Normal cellular function is controlled by the balance of protein synthesis and degradation. Proteasomal and lysosomal mechanisms are two classical protein degradation systems. Prior work has implicated both of these systems in Notch degradation. In this study, we investigate the degradation of both intracellular domain and extracellular domain of Notch3 in multiple cells lines; our experiments implicate lysosomal degradation as the principle route of Notch3 clearance.
Degradation of Notch3 appears to be distinct from that of Notch1, the best characterized mammalian isoform. Prior studies have suggested a dual role for UPS and lysosomes in Notch1 degradation with consistent identification of ubiquitylated Notch1 ICD that may target the protein to either UPS or lysosomal pathways. However, we have not found evidence of ubiquitylation for Notch3; cells overexpressing Notch3 and treated with MG132 did not contain ubiquitylated Notch3, unlike Notch1 and Notch4. In addition, in sharp contrast to Notch1 studies, we have never observed increases in Notch3 ICD with any cell line using two independent proteasome inhibitors. In contrast, two lysosome inhibitors caused robust increases in Notch3 protein levels, leading us to conclude that the principle mechanism of Notch3 clearance requires the lysosomal system. Our ability to confirm all of our findings consistently in cell lines expressing endogenous Notch3 gives us confidence that our conclusions were not affected by protein overexpression, which has been a necessary, but perhaps confounding factor in studies of other Notch isoforms. Of note, the single study which has implicated lysosomal degradation in the processing of Notch1 ICD (Jehn et al., 2002) was performed on endogenous Notch1 in C2C12 mouse cells. The studies by our group and Jehn et al. indicated that further investigation should be performed on the potential processing of Notch1, 2, and 4 by lysosomes.
Our experiments hint at additional complexities of Notch3 signaling. Although protein levels increased robustly after lysosomal inhibition, we were not able to detect differences in transcriptional activation. It is unlikely that Notch3 ICD is trapped in lysosomes after treatment with lysosome inhibitors, since overexpression studies demonstrate uniform expression in the nucleus (Oberg et al., 2001). An analogous paradox was found identified by Oberg et al. (Oberg et al., 2001) who noted that proteasome inhibitors increased the levels of Notch1 protein, but failed to increase Notch1 signaling efficiency. A possible explanation that links our findings with other observations is that Notch ICD could be irreversibly tagged for destruction prior to degradation by proteasomes (for Notch1) and lysosomes (for Notch3); modification of Notch proteins may simultaneously mark the proteins for destruction while inactivating their transcriptional potency. We note that nuclear hormone receptors are also ubiquitylated prior to degradation and removal from transcriptional targets; degradation is in fact required for full transcriptional activation of the nuclear hormone targets. This and other potential mechanisms that prevent the over-activation of Notch signaling by accumulated Notch-ICD remain to be tested.
Another complexity is the unexpected molecular weight of Notch3 which accumulates after lysosome inhibition. The larger of the doublet of proteins does not correspond to known products of Notch3 proteolysis. Dephosphorylation with phosphatases did not reveal a change in molecular weight of either form, suggesting that phosphorylation is not a cause of the molecular weight shift (unpublished results). Failure to observe either of the bands after transfection of TM-N3-IC suggests that the large band is not the result of S2 cleavage and implies that processing of Notch3 to the larger ICD requires the expression of the full length protein. This again differs from what is known about Notch1, whose S2 fragment is constitutively processed by the gamma secretase complex. Alternative posttranslational modifications that could account for a relatively large shift in molecular weight observed between the two bands (eg. glycosylation, SUMOylation) or novel proteolytic processing are most likely responsible for this finding, but must be directly addressed in more extensive studies.
Until now, few studies have been performed to determine the molecular fate of Notch3 ECD. In Drosophila and mammalian Notch1, increasing evidence favors a role of transendocytosis in clearance of Notch ectodomain (Klueg and Muskavitch, 1999; Morel et al., 2003; Nichols et al., 2007; Parks et al., 2000). Interestingly, signal transduction is coupled to ectodomain clearance and requires endocytosis in the signal sending (ligand expressing) cell. Constititutive recycling of Notch3 ECD by endosomal sorting with lysosomal disposal is certainly feasible given recent demonstration of this mechanism for Notch1 (Chastagner et al., 2008). Our findings further support a role of lysosomal disposal of Notch3 ECD for both wild type and CADASIL mutant N3-ECD, suggesting that enhancement of lysosomal function could lower even mutant N3 ECD levels in CADASIL. Potentially, emerging knowledge from studies of molecular mechanisms of Notch transendocytosis and endosomal sorting can be applied to devise strategies for ameliorating CADASIL, a disease hallmarked by the pathological accumulation of Notch3 ectodomain (Joutel et al., 2000).
Acknowledgements
We thank members of the Wang lab for constructive discussions and suggestions and Drs. Yi Sun and Dan Michele for cell lines used in this study. This work was funded by NIH grants NS054724 and NS052681.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Artavanis-Tsakonas SRM, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Blaumueller CM, Qi H, Zagouras P, Artavanis-Tsakonas S. Intracellular cleavage of Notch leads to a heterodimeric receptor on the plasma membrane. Cell. 1997;90:281–291. doi: 10.1016/s0092-8674(00)80336-0. [DOI] [PubMed] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- Chastagner P, Israel A, Brou C. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS ONE. 2008;3:e2735. doi: 10.1371/journal.pone.0002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, Fan X, Chaudhry A, Wang M, Gaiano N, Eberhart CG. Notch3 signaling initiates choroid plexus tumor formation. Oncogene. 2006;25:487–491. doi: 10.1038/sj.onc.1209074. [DOI] [PubMed] [Google Scholar]
- Dang TP, Gazdar AF, Virmani AK, Sepetavec T, Hande KR, Minna JD, Roberts JR, Carbone DP. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J Natl Cancer Inst. 2000;92:1355–1357. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, Ciechanover A, Israel A. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- Iso T, Hamamori Y, Kedes L. Notch signaling in vascular development. Arterioscler Thromb Vasc Biol. 2003;23:543–553. doi: 10.1161/01.ATV.0000060892.81529.8F. [DOI] [PubMed] [Google Scholar]
- Jehn BM, Dittert I, Beyer S, von der Mark K, Bielke W. c-Cbl binding and ubiquitin-dependent lysosomal degradation of membrane-associated Notch1. J Biol Chem. 2002;277:8033–8040. doi: 10.1074/jbc.M108552200. [DOI] [PubMed] [Google Scholar]
- Joutel A, Andreux F, Gaulis S, Domenga V, Cecillon M, Battail N, Piga N, Chapon F, Godfrain C, Tournier-Lasserve E. The ectodomain of the Notch3 receptor accumulates within the cerebrovasculature of CADASIL patients. J Clin Invest. 2000;105:597–605. doi: 10.1172/JCI8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, Alamowitch S, Domenga V, Cecillion M, Marechal E, et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–710. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- Klueg KM, Muskavitch MA. Ligand-receptor interactions and trans-endocytosis of Delta, Serrate and Notch: members of the Notch signalling pathway in Drosophila. J Cell Sci. 1999;112(Pt 19):3289–3297. doi: 10.1242/jcs.112.19.3289. [DOI] [PubMed] [Google Scholar]
- Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, Israel A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci U S A. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MA, McGlade CJ. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem. 2003;278:23196–23203. doi: 10.1074/jbc.M302827200. [DOI] [PubMed] [Google Scholar]
- Meng H, Zhang X, Hankenson KD, Wang MM. Thrombospondin 2 potentiates notch3/jagged1 signaling. J Biol Chem. 2009;284:7866–7874. doi: 10.1074/jbc.M803650200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel V, Le Borgne R, Schweisguth F. Snail is required for Delta endocytosis and Notch-dependent activation of single-minded expression. Dev Genes Evol. 2003;213:65–72. doi: 10.1007/s00427-003-0296-x. [DOI] [PubMed] [Google Scholar]
- Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- Nichols JT, Miyamoto A, Olsen SL, D'Souza B, Yao C, Weinmaster G. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J Cell Biol. 2007;176:445–458. doi: 10.1083/jcb.200609014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J Biol Chem. 2001;276:35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- Okochi M, Steiner H, Fukumori A, Tanii H, Tomita T, Tanaka T, Iwatsubo T, Kudo T, Takeda M, Haass C. Presenilins mediate a dual intramembranous gamma-secretase cleavage of Notch-1. Embo J. 2002;21:5408–5416. doi: 10.1093/emboj/cdf541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JT, Li M, Nakayama K, Mao TL, Davidson B, Zhang Z, Kurman RJ, Eberhart CG, Shih Ie M, Wang TL. Notch3 gene amplification in ovarian cancer. Cancer Res. 2006;66:6312–6318. doi: 10.1158/0008-5472.CAN-05-3610. [DOI] [PubMed] [Google Scholar]
- Parks AL, Klueg KM, Stout JR, Muskavitch MA. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development. 2000;127:1373–1385. doi: 10.1242/dev.127.7.1373. [DOI] [PubMed] [Google Scholar]
- Qiu L, Joazeiro C, Fang N, Wang HY, Elly C, Altman Y, Fang D, Hunter T, Liu YC. Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J Biol Chem. 2000;275:35734–35737. doi: 10.1074/jbc.M007300200. [DOI] [PubMed] [Google Scholar]
- Saxena MT, Schroeter EH, Mumm JS, Kopan R. Murine notch homologs (N1–4) undergo presenilin-dependent proteolysis. J Biol Chem. 2001;276:40268–40273. doi: 10.1074/jbc.M107234200. [DOI] [PubMed] [Google Scholar]
- Schouwey K, Delmas V, Larue L, Zimber-Strobl U, Strobl LJ, Radtke F, Beermann F. Notch1 and Notch2 receptors influence progressive hair graying in a dose-dependent manner. Dev Dyn. 2007;236:282–289. doi: 10.1002/dvdy.21000. [DOI] [PubMed] [Google Scholar]
- Tsunematsu R, Nakayama K, Oike Y, Nishiyama M, Ishida N, Hatakeyama S, Bessho Y, Kageyama R, Suda T, Nakayama KI. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J Biol Chem. 2004;279:9417–9423. doi: 10.1074/jbc.M312337200. [DOI] [PubMed] [Google Scholar]
- Wilkin M, Tongngok P, Gensch N, Clemence S, Motoki M, Yamada K, Hori K, Taniguchi-Kanai M, Franklin E, Matsuno K, Baron M. Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of notch in the endosomal trafficking pathway. Dev Cell. 2008;15:762–772. doi: 10.1016/j.devcel.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Wu G, Lyapina S, Das I, Li J, Gurney M, Pauley A, Chui I, Deshaies RJ, Kitajewski J. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol Cell Biol. 2001;21:7403–7415. doi: 10.1128/MCB.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi N, Oyama T, Ito E, Satoh H, Azuma S, Hayashi M, Shimizu K, Honma R, Yanagisawa Y, Nishikawa A, et al. NOTCH3 signaling pathway plays crucial roles in the proliferation of ErbB2-negative human breast cancer cells. Cancer Res. 2008;68:1881–1888. doi: 10.1158/0008-5472.CAN-07-1597. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Jia L, Lee SJ, Wang MM. Conserved signal peptide of Notch3 inhibits interaction with proteasome. Biochem Biophys Res Commun. 2007;355:245–251. doi: 10.1016/j.bbrc.2007.01.151. [DOI] [PMC free article] [PubMed] [Google Scholar]