Abstract
The voltage-gated potassium channel modulatory membrane protein KCNE3 was overexpressed and purified into both micelles and bicelles. Remarkably, microinjection of KCNE3 in bicelles into Xenopus oocytes resulted in functional co-assembly with the human KCNQ1 channel expressed therein. Microinjection of LMPC micelles containing KCNE3 did not result in channel modulation, indicating that bicelles sometimes succeed at delivering a membrane protein into a cellular membrane when classical micelles fail. Backbone NMR resonance assignments were completed for KCNE3 in both bicelles and LMPC, indicating that the secondary structure distribution in KCNE3’s N-terminus and transmembrane domains exhibits only modest differences from that of KCNE1, even though these KCNE family members have very different affects on KCNQ1 channel function.
The KCNE(1–5) family of small single span membrane proteins modulate certain voltage-gated potassium channels in a manner that is critically linked to the physiological functions of these channels(1). For example, association of KCNE1 with human KCNQ1 (KV7.1, KvLQT1) causes delayed channel activation and enhanced conductance, resulting in the IKs current, a critical component of the cardiac action potential(2). On the other hand, association of KCNE3 with KCNQ1 results in a constitutively open channel with enhanced conductance, as may be critical for acid secretion by parietal cells of the stomach epithelium and salt transport by colonic or trachael epithelial cells(3–5). Mutations in KCNE3 are linked to long QT syndrome(6), atrial fibrillation(7), periodic paralysis(8), and Brugada syndrome(9). KCNE3 may also play a role in the etiology of Alzheimer’s disease(10). Although the structural basis for how different KCNE proteins can modulate channel function with very distinct effects is not well understood, the recent determination of the 3D-structure of KCNE1 combined with previous structure-function data led to a testable working model for how this protein modulates KCNQ1(11).
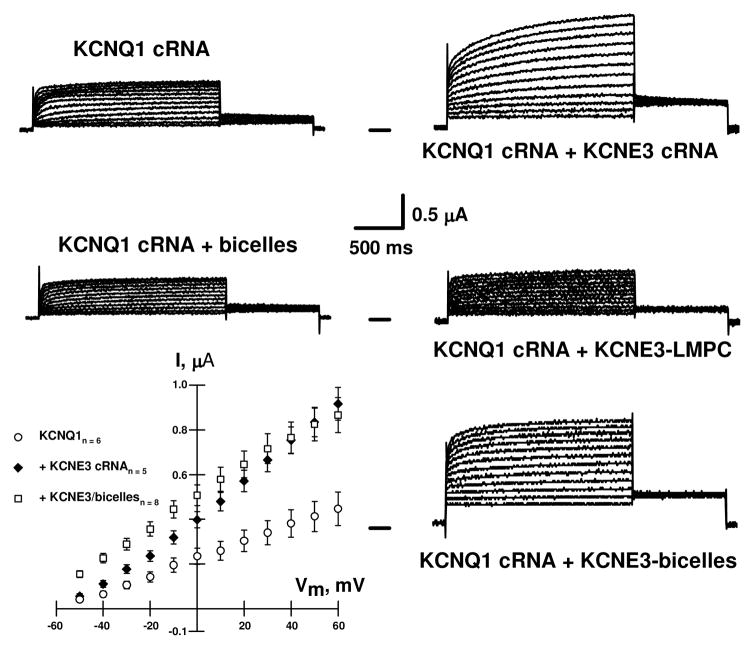
N-Terminal hexaHis-tagged KCNE3 was overexpressed in E. coli and purified into non-denaturing LMPC detergent micelles (Figure S1). LMPC was used rather than LMPG (which was employed in the previous studies of KCNE1) because KCNE3 was observed to be aggregation-prone in LMPG. In an effort to validate the suitability of LMPC micelles for structural studies of KCNE3, solutions of the purified protein were microinjected into Xenopus oocytes expressing human KCNQ1 channels. As shown in Figure 1, microinjection of LMPC micelles containing recombinant KCNE3 into KCNQ1-expressing oocytes did not affect channel gating kinetics or current amplitudes when compared to oocytes injected only with KCNQ1 cRNA. This result indicates that LMPC micelles were not able to successfully deliver KCNE3 to the oocyte membranes in a way that supported functional assembly with KCNQ1 channel. This is surprising since microinjection of KCNE1 in lyso-phospholipid micelles had previously been shown to modulate KCNQ1 channels in a native-like manner(12).
Figure 1.
Representative whole-cell current traces recorded from oocytes injected with KCNQ1 and KCNE3 cRNAs, and from oocytes expressing KCNQ1 channels and then injected with either purified His6-KCNE3 in LMPC micelles, His6-KCNE3 in DHPC-DMPG (q= 0.33) bicelles, or empty bicelles. These results show that injection of either KCNE3 cRNA or His6-KCNE3 in DHPC-DMPG bicelles increased the KCNQ1-induced current magnitude and caused the channel to be open at all potentials tested without slowing the rate of channel activation. Similar results were previously observed after co-injection of KCNQ1 and KCNE3 cRNAs into Xenopus oocytes(5;30). Injection of either His6-KCNE3 in LMPC micelles or empty micelles did not alter KCNQ1-induced currents. Xenopus oocytes were injected with 25 nl of solutions containing 6 ng of KCNQ1 cRNA and 3 ng of KCNE3 cRNA; or with 6 ng of KCNQ1 cRNA and then, 48 h later, with 10 nl of 1 mg/ml purified KCNE3 in LMPC micelles. Whole-cell currents were recorded 72 h after injection of cRNAs for KCNQ1 and KCNE3 or 18 after micelle or bicelle injections.
The fact that micelles composed of LMPC, normally considered to be a very mild detergent, were unable to successfully deliver KCNE3 to oocyte membranes led us to purify KCNE3 into a model membrane medium that is considered to be more native-like than even the most optimal micelles: bicelles(13;14). KCNE3 was purified into 3:1 (mol:mol) DHPC/DMPG bicelles (see Supporting Information) and then microinjected into KCNQ1-expressing oocytes. As shown in Figure 1 His6-tagged KCNE3 in bicelles modulated KCNQ1 channels to yield whole-cell currents that are very similar to those recorded when KCNE3 and KCNQ1 are co-expressed from cRNA. This suggests that His6-tagged KCNE3 in bicelles can assemble with KCNQ1 channels into functional complexes. In addition, the results in Figure 1 strongly suggest that bicelles alone do not perturb the plasma membrane properties. Injection of protein-free bicellar solutions did not affect KCNQ1-induced currents. To our knowledge, this is the first use of bicelles as a membrane protein delivery reagent. The degree to which other integral membrane proteins can be delivered from bicelles into pre-formed membranes is unclear2, but the results for KCNE3 suggest this issue is worthy of further investigation.
Why bicelles led to functional assembly of the KCNE3/KCNQ1 complex2 but lyso-phospholipid micelles did not is unclear, but may be related to the higher morphological and compositional similarity of bicelles to native bilayers(13–15). Why KCNE1 but not KCNE3 could be successfully delivered into oocyte membranes from lyso-phospholipids micelles is not clear, may be related to structural differences between KCNE1 and KCNE3, comparison of which follows.
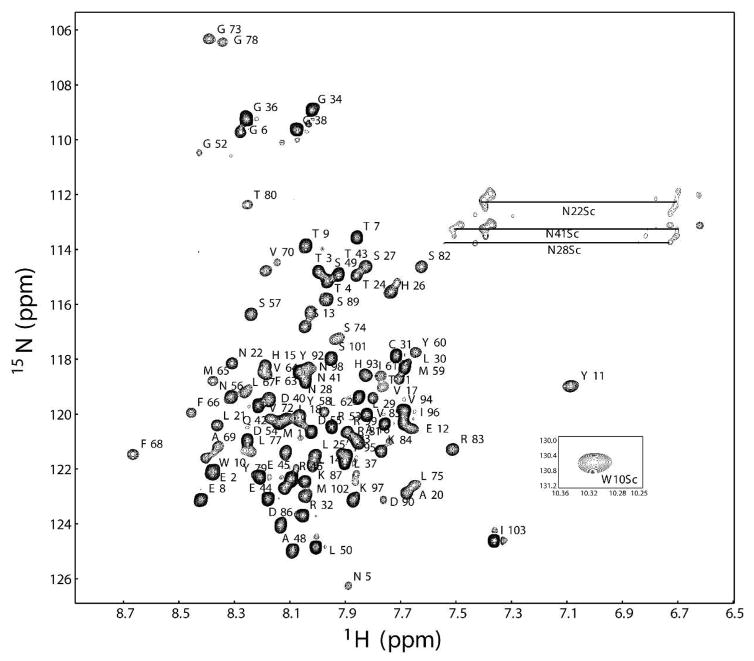
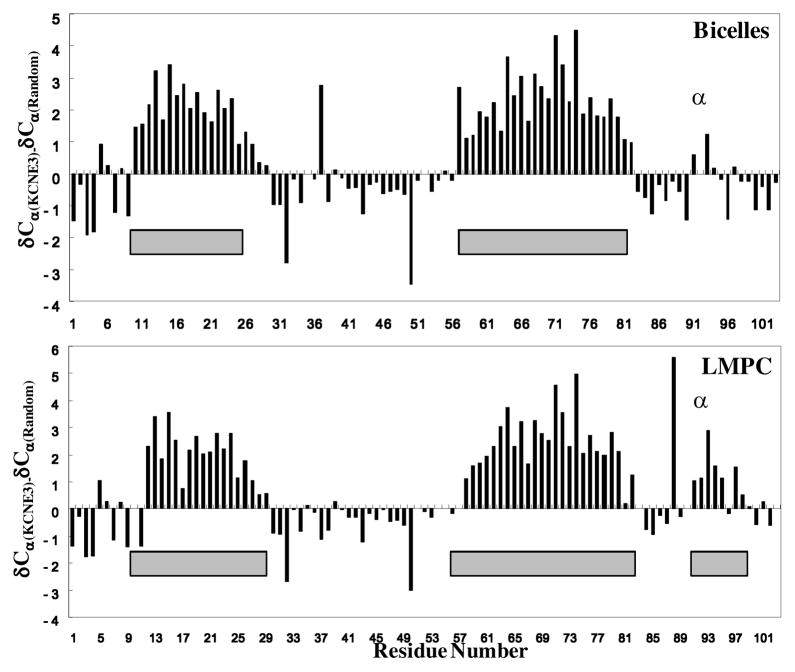
KCNE3 in bicelles and in LMPC micelles yields high quality NMR spectra (Figures 2 and S2), which were assigned using 3-D TROSY-based experiments (c.f. Figures S3–S4). The chemical shifts associated with these assignments were analyzed using chemical shift index analysis(16) and the program TALOS+(17) to gain insight into KCNE3’s secondary structure in bicelles. Results are summarized in Figure 3, which extend the recent observation(18) that KCNE3 is a largely helical protein.
Figure 2.
Assigned 1H-15N TROSY-HSQC NMR spectrum of uniformly-2H,13C,15N-labeled KCNE3 in isotropic bicelles (buffer: 250 mM imidazole, 2 mM DTT, pH 6.5). The sample contained 22% (w/v) bicelles with DHPC:DMPG = 3:1 (q = 0.33). NMR experiments were carried out at 40 °C on a Bruker 800 MHz spectrometer equipped with a cryogenic probe as described in the Supporting Information. Backbone resonance assignments are tabulated in BioMagResBank entry 16621.
Figure 3.
Summary of chemical shift index(16) and TALOS+(17) analysis of KCNE3’s secondary structure in bicelles (top panel) and LMPC micelles (lower panel). The differences between the observed residue-specific 13Cα chemical shifts for KCNE3 and the corresponding random coil chemical shift values are plotted in the bar graph. The gray bars indicate segments that are confidently determined to have dihedral angles in the α-helical range based on backbone/13Cβ chemical shift analysis using TALOS+.
The transmembrane domain of KCNE3 is predicted to span residues 58–80 and is seen to be α-helical for KCNE3 in LMPC and in bicelles. In this regard, KCNE3 resembles KCNE1, although in the case of KCNE1 the helix is known to be highly curved with a possible break in the middle(11;12). The transmembrane domain of KCNE3 does not appear to exhibit a break, but the chemical shift data alone cannot establish whether the helix is bent or curved—additional measurements such as residual dipolar couplings will be required further illuminate its structure. These future studies are critical as there is considerable evidence that the primary differences for how KCNE1 and KCNE3 modulate KCNQ1 reside in this domain(19;20).
The extracellular N-terminus domain of KCNE3 is about 13 residues longer than that of KCNE1 and there is little or no sequence homology between these two domains. Nevertheless, KCNE3 resembles KCNE1(11) in that both proteins exhibit amphipathic α-helices spanning at least residues 11–24. While for neither protein does the N-terminus appear to be critical for dictating the channel-modulatory properties, the N-terminus is known to affect the pH sensitivity and pharmacological properties of the assembled KCNE-KCNQ1 complex(21;22). The N-terminus likely(11) plays a role in defining the known 2:4 subunit stoichiometry of the KCNE1:KCNQ1 complex(23;24), a role for the N-terminus that likely also pertains to the KCNE3-KCNQ1 channel. This domain likely contributes to overall affinity for KCNQ1. For both KCNE1 and KCNE3 the sequence between the N-terminal amphipathic helix and the transmembrane domain does not appear to have regular secondary structure.
KCNE3’s 23 residue C-terminus appears to be unstructured in bicelles but exhibits an α-helix spanning residues 91–99 in LMPC micelles. This segment corresponds to the juxtamembrane region (residues 72–91) of KCNE1’s much longer C-terminus, which is thought to contribute to KCNE1’s affinity for the channel and to its channel modulatory function(25–27). In the case of KCNE1 this segment was previously observed to be unstructured in LMPG micelles; however, there is evidence that this segment adopts a helical structure when KCNE1 binds to the KCNQ1 channel(28). While KCNE3’s C-terminal domain does not appear to be essential for its modulation of KCNQ1(29), its propensity to form an α-helix may reflect its conformation when KCNE3 is associated with the channel. That this domain is seen to be disordered in DMPG-based bicelles, but contains a helix in neutral LMPC micelles suggests that this helix, which contains three positively charged residues (but is not amphipathic), is destabilized by the anionic bicelle surface.
In summary, our results demonstrate that not only are bicelles a suitable medium for NMR structural studies of KCNE3, they are can also be used to deliver this membrane protein to the membranes of Xenopus oocytes, where it can co-assemble with the KCNQ1 channel and modulate its function. Our preliminary structural studies of KCNE3 in both micelles and bicelles show considerable similarity in overall structure between this protein and KCNE1. Evidently, the basis for the very differing effects of KCNE1 and KCNE3 on KCNQ1 channel function cannot be explained on the basis of gross structural differences between these KCNE family members, but will require a higher level of structural detail for KCNE3 than is provided by this preliminary structural study and/or by additional data that illuminate the details of KCNE3-KCNQ1 interactions.
Supplementary Material
Acknowledgments
We thank Dr. Masayoshi Sakakura for helping with NMR experiments and Prof. Alfred George for useful discussion.
Abbreviations
- DHPC
dihexanoylphosphatidylcholine
- DMPG
dimyristoylphosphatidylglycerol
- LMPC
lyso-myristoylphosphatidylcholine
- LMPG
lyso-myristoylphosphatidylglycerol
- NMR
nuclear magnetic resonance
- TROSY
transverse relaxation-optimized NMR spectroscopy
Footnotes
NMR resonance assignments of KCNE3 in isotropic bicelles and in LMPC micelles have been deposited in the BioMagResBank with accession codes 16621 and 16602.
A preliminary test indicated that KCNE1 in bicelles also can be successfully microinjected into KCNQ1-expressing oocytes to generate an IKs-like channel.
SUPPORTING INFORMATION PARAGRAPH
Methods and Figures S1–S4. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by RO1 DC007416 (to CRS) and by NIH training grant T32 NS007491 (to WDVH).
References
- 1.McCrossan ZA, Abbott GW. Neuropharmacology. 2004;47:787–821. doi: 10.1016/j.neuropharm.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 2.Jespersen T, Grunnet M, Olesen SP. Physiology (Bethesda ) 2005;20:408–416. doi: 10.1152/physiol.00031.2005. [DOI] [PubMed] [Google Scholar]
- 3.Grahammer F, Warth R, Barhanin J, Bleich M, Hug MJ. J Biol Chem. 2001;276:42268–42275. doi: 10.1074/jbc.M105014200. [DOI] [PubMed] [Google Scholar]
- 4.Grahammer F, Herling AW, Lang HJ, Schmitt-Graff A, Wittekindt OH, Nitschke R, Bleich M, Barhanin J, Warth R. Gastroenterology. 2001;120:1363–1371. doi: 10.1053/gast.2001.24053. [DOI] [PubMed] [Google Scholar]
- 5.Schroeder BC, Waldegger S, Fehr S, Bleich M, Warth R, Greger R, Jentsch TJ. Nature. 2000;403:196–199. doi: 10.1038/35003200. [DOI] [PubMed] [Google Scholar]
- 6.Ohno S, Toyoda F, Zankov DP, Yoshida H, Makiyama T, Tsuji K, Honda T, Obayashi K, Ueyama H, Shimizu W, Miyamoto Y, Kamakura S, Matsuura H, Kita T, Horie M. Hum Mutat. 2009;30:557–563. doi: 10.1002/humu.20834. [DOI] [PubMed] [Google Scholar]
- 7.Lundby A, Ravn LS, Svendsen JH, Hauns S, Olesen SP, Schmitt N. Cell Physiol Biochem. 2008;21:47–54. doi: 10.1159/000113746. [DOI] [PubMed] [Google Scholar]
- 8.Abbott GW, Butler MH, Goldstein SA. FASEB J. 2006;20:293–301. doi: 10.1096/fj.05-5070com. [DOI] [PubMed] [Google Scholar]
- 9.Delpon E, Cordeiro JM, Nunez L, Thomsen PE, Guerchicoff A, Pollevick GD, Wu Y, Kanters JK, Larsen CT, Burashnikov E, Christiansen M, Antzelevitch C. Circ Arrhythm Electrophysiol. 2008;1:209–218. doi: 10.1161/CIRCEP.107.748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi E, Abbott GW. Mol Pharmacol. 2007;72:499–501. doi: 10.1124/mol.107.039206. [DOI] [PubMed] [Google Scholar]
- 11.Kang C, Tian C, Sonnichsen FD, Smith JA, Meiler J, George AL, Jr, Vanoye CG, Kim HJ, Sanders CR. Biochemistry. 2008;47:7999–8006. doi: 10.1021/bi800875q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian C, Vanoye CG, Kang C, Welch RC, Kim HJ, George AL, Jr, Sanders CR. Biochemistry. 2007;46:11459–11472. doi: 10.1021/bi700705j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanders CR, Prosser RS. Structure. 1998;6:1227–1234. doi: 10.1016/s0969-2126(98)00123-3. [DOI] [PubMed] [Google Scholar]
- 14.Sanders CR, Kuhn HA, Gray DN, Keyes MH, Ellis CD. Chembiochem. 2004;5:423–426. doi: 10.1002/cbic.200300830. [DOI] [PubMed] [Google Scholar]
- 15.Prosser RS, Evanics F, Kitevski JL, Al-Abdul-Wahid MS. Biochemistry. 2006;45:8453–8465. doi: 10.1021/bi060615u. [DOI] [PubMed] [Google Scholar]
- 16.Wishart DS, Sykes BD. Nuclear Magnetic Resonance, Pt C. 1994;239:363–392. [Google Scholar]
- 17.Shen Y, Delaglio F, Cornilescu G, Bax A. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goncharuk SA, Shulga AA, Ermolyuk YS, Kuzmichev PK, Sobol VA, Bocharov EV, Chupin VV, Arseniev AS, Kirpichnikov MP. Biochemistry (Mosc ) 2009;74:1344–1349. doi: 10.1134/s0006297909120074. [DOI] [PubMed] [Google Scholar]
- 19.Melman YF, Domenech A, de la LS, McDonald TV. J Biol Chem. 2001;276:6439–6444. doi: 10.1074/jbc.M010713200. [DOI] [PubMed] [Google Scholar]
- 20.Melman YF, Krumerman A, McDonald TV. J Biol Chem. 2002;277:25187–25194. doi: 10.1074/jbc.M200564200. [DOI] [PubMed] [Google Scholar]
- 21.Heitzmann D, Koren V, Wagner M, Sterner C, Reichold M, Tegtmeier I, Volk T, Warth R. Cell Physiol Biochem. 2007;19:21–32. doi: 10.1159/000099189. [DOI] [PubMed] [Google Scholar]
- 22.Peretz A, Schottelndreier H, haron-Shamgar LB, Attali B. J Physiol. 2002;545:751–766. doi: 10.1113/jphysiol.2002.028381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H, Kim LA, Rajan S, Xu S, Goldstein SA. Neuron. 2003;40:15–23. doi: 10.1016/s0896-6273(03)00570-1. [DOI] [PubMed] [Google Scholar]
- 24.Morin TJ, Kobertz WR. Proc Natl Acad Sci U S A. 2008;105:1478–1482. doi: 10.1073/pnas.0710366105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Zheng R, Melman YF, McDonald TV. PLoS One. 2009;4:e5143. doi: 10.1371/journal.pone.0005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Splawski I, Tristani-Firouzi M, Lehmann MH, Sanguinetti MC, Keating MT. Nat Genet. 1997;17:338–340. doi: 10.1038/ng1197-338. [DOI] [PubMed] [Google Scholar]
- 27.Tapper AR, George AL., Jr J Gen Physiol. 2000;116:379–390. doi: 10.1085/jgp.116.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rocheleau JM, Gage SD, Kobertz WR. J Gen Physiol. 2006;128:721–729. doi: 10.1085/jgp.200609657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gage SD, Kobertz WR. KCNE3 J Gen Physiol. 2004;124:759–771. doi: 10.1085/jgp.200409114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anantharam A, Lewis A, Panaghie G, Gordon E, McCrossan ZA, Lerner DJ, Abbott GW. Journal of Biological Chemistry. 2003;278:11739–11745. doi: 10.1074/jbc.M212751200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.