Abstract
Nucleotide excision repair (NER) is the main pathway used for the repair of bulky DNA adducts such as those caused by UV light exposure and the chemotherapeutic drug cisplatin. The Xeroderma Pigmentosum group C (XPC)-Rad23B complex is involved in the recognition of these bulky DNA adducts and initiates the global genomic nucleotide excision repair pathway (GG-NER). Photocrosslinking experiments revealed that the human XPC-Rad23B complex makes direct contact with both the cisplatin-damaged DNA strand and the complementary undamaged strand of a duplex DNA substrate. Coupling photocrosslinking with denaturation and immunoprecipitation of protein-DNA complexes, the XPC subunit was identified in complex with damaged DNA. While the interaction of the XPC subunit with DNA was direct, studies revealed that although Rad23B was found in complex with DNA, the Rad23B-DNA interaction was largely indirect via its interaction with XPC. Using site specific crosslinking we determined that XPC-Rad23B is preferentially crosslinked to the damaged DNA when the photoreactive FAP-dCMP (exo-N-{2-[N-(4-azido-2,5-difluoro-3-chloropyridine-6-yl)-3-aminopropionyl]-aminoethyl}-2′-deoxycytidine-5′-monophosphate) analogue is located 5′ of the cisplatin-DNA adduct. When the FAP-dCMP analogue is located 3′ of the adduct, no difference in binding was detected between undamaged and damaged DNA. Collectively, these data suggest a model whereby XPC-DNA interactions drive the damage recognition process contacting both the damaged and undamaged DNA strand. Preferential crosslinking 5′ of the cisplatin-damaged site suggests that XPC-Rad23B displays orientation specific binding to eventually impart directionality to the downstream binding and incision events relative to the site of DNA damage.
Keywords: Nucleotide excision repair (NER), XPC, Rad23B, cisplatin
Lesions, mutations, and bulky adducts caused by external and internal factors, such as UV irradiation and environmental carcinogens interfere with DNA replication and transcription and can lead to cell death, cancer or other diseases (1;2). Nucleotide excision repair (NER) is a versatile DNA repair pathway utilized for the removal of damaged DNA caused by UV irradiation and platinum based chemotherapeutic agents used for the treatment of cancer (1;2). NER is composed of two separate subpathways, transcription coupled-NER (TC-NER) and GG-NER. There are four distinct steps in the NER pathway, including damage recognition, incision of damaged DNA, removal of damage, and resynthesis and ligation of the newly synthesized strand (1;2). The GG-NER and TC-NER pathways differ only in the method of damage recognition. XPC, Rad23B and Centrin 2 (CEN2), form the damage recognition complex and are the first proteins to the site of damage in the GG-NER pathway, whereas the stalling of RNA polymerase during transcription is the method of recognition in the TC-NER pathway (1;3). After recognizing and binding to the damaged DNA, XPC-Rad23B-CEN2 is released and GG-NER progresses through the excision, repair, and resynthesis steps due to the involvement of over 30 additional proteins (2). RPA and XPA have also been shown to possess damage recognition abilities depending on the type of damage in both GG-NER and TC-NER (4). The helicase activity of XPB-TFIIH is responsible for unwinding the damaged DNA followed by 5′ and 3′ incision which is performed by XPF-ERCC1 and XPG, respectively (5). Finally PCNA, RPA, DNA polymerases δ, ε, κ and DNA ligases participate in gap filling and ligation of the newly synthesized and repaired DNA (1;2;4;6;7). More recently, a UV-dependent DNA damage recognition pathway involving DNA-binding protein (DDB) has been shown to recognize and bind to damaged DNA in the absence of XPC (8). DDB has been shown to interact with DNA containing UV-induced CPD lesions, which only moderately distort the DNA, suggesting that DDB may play a role in damage recognition of less distorting lesions difficult for XPC to recognize (9–11).
In vivo, human XPC (125 kDa) forms a heterotrimeric complex with the 58 kDa protein Rad23B along with the 18 kDa protein Centrin 2 (12). In vitro, XPC and Rad23B are sufficient for reconstituting the damage recognition complex responsible for the initiation of NER (13;14) and the XPC-Rad23B complex preferentially binds to cisplatin and UV damaged double strand DNA in the presence of the non-specific competitor poly dI/dC (4;15–18). In addition, XPC-Rad23B recognizes a variety of chemically and structurally diverse DNA adducts and previous data have shown that the more distorting the damage the higher affinity XPC-Rad23B demonstrates for the damaged DNA (19). It has been suggested that XPC-Rad23B interacts with the undamaged strand of a damaged duplex DNA via the single-strand characteristics caused by the bulky UV or cisplatin lesions (16;20). This data is supported by the structural analysis of yeast Rad4, the XPC orthologue, and Rad23B bound to DNA containing a cyclobutane pyrimidine dimer (CPD) lesion where Rad4 contacts were identified on both the undamaged strand of the duplex and base pairs surrounding the CPD lesion (21). The transglutaminase-homology domain (TGD) and the long β-hairpin domain 1 (BHD1) bind to the undamaged strand 3′ of the CPD lesion. β-hairpin domains 2 and 3 (BHD2 and BHD3) bind to the DNA containing the CPD lesion and cause the two base pairs opposite of the lesion to flip outward (21). In comparing the DNA-absent (apo) model to the DNA bound model, BHD2 and 3 move approximately 8–12 Å relative to the position of the DNA. This flexibility is thought to aid in damage recognition and binding of Rad4 and possible XPC to damaged duplex DNA. As Rad4 shares 23% sequence identity with XPC it is unclear, however, if XPC will bind to DNA in the same manner as Rad4. It is also unclear whether other lesions, which result in different structural distortions but are still repaired by NER, will be recognized using the same series of interactions. To address this latter question we examined the interaction of XPC-Rad23B with cisplatin-damaged DNA focusing on specific protein-DNA interactions.
Cisplatin is a common chemotherapeutic agent used for the treatment of several cancers, including testicular, lung and ovarian cancer (22–25). 1,2-d(GpG) cisplatin adducts are the most common lesions and distort the DNA ~13° while 1,3-d(GpXpG) cause a ~27–33° kink in the DNA around the platinum lesion and make up a much smaller portion of the cisplatin intrastrand lesions (26;27). Although XPC-Rad23B recognizes the bulky adduct, it remains unclear if XPC-Rad23B contacts the cisplatin lesion directly. It is also unknown if XPC-Rad23B demonstrates preferential binding either 5′ or 3′ to the site of cisplatin damage or if the protein symmetrically spans the lesion. Determining the general location of XPC-Rad23B in relation to the cisplatin adduct will give directionality to XPC-Rad23B and possibly downstream NER proteins. Protein directionality will provide insight into the mechanism of XPC-Rad23B damage recognition which is versatile and unclear. It may also aid in determining the placement of downstream NER proteins, such as XPF and XPG which are important for excision of the damaged bases and the excision product removed. To further probe the nature of the XPC-Rad23B DNA interactions, we undertook a photocrosslinking approach using a series of cisplatin-damaged DNA substrates. To further increase the efficiency, specificity and resolution of the approach, a photoreactive exo-N-{2-[N-(4-azido-2,5-difluoro-3-chloropyridine-6-yl)-3-aminopropionyl]-aminoethyl}-2′-deoxycytidine-5′-triphosphase (FAP-dCTP) analogue was used to introduce photoreactive residues into specific positions of the DNA substrate. FAP-dCTP is a dCTP analogue which contains a photoreactive FAP group inserted at the 4-C location and aids in crosslinking efficiency of substrates (28). Results demonstrate strand and orientation specific binding of the XPC subunit in complex with Rad23B to DNA containing the cisplatin adduct. These results provide a better understanding of the XPC-DNA interaction and provide details that may allow improvement of platinum based chemotherapeutic treatments possibly through direct inhibition of XPC or development of a platinum based therapeutic which XPC cannot recognize.
Experimental Procedures
Chemicals
Cisplatin was purchased from Sigma-Aldrich (St. Louis, MO) and γ32P-ATP was supplied by Perkin-Elmer (Waltham, MA). Oligonucleotides were purchased from Integrated DNA Technologies (Coralville, IA). Klenow Fragment 3′-5′ exo-, T4 Polynucleotide Kinase (T4 PNK), T4 DNA ligase, and HaeIII were purchased from New England Biolabs (Ipswich, MA). Phosphocellulose (Whatman P-11) was obtained from Fisher Scientific (Houston, TX) along with T4 DNA polymerase. ssDNA-cellulose was obtained from Sigma Chemical Co. (St. Louis, MO). Protein-G agarose, XPC and Rad23B rabbit polyclonal antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Streptavidin MagneSphere Paramagnetic Particles were purchased from Promega (Madison, WI). The photoreactive analog of dCTP exo-N-{2-[N-(4-azido-2,5-difluoro-3-chloropyridine-6-yl)-3-aminopropionyl]-aminoethyl}-2′-deoxycytidine-5′-triphosphate (FAP-dCTP), was synthesized as described (28).
Protein Purification
XPC and Rad23B were purified from SF-9 insect cells to near homogeneity as previously described (16). Briefly SF-9 cells (500 ml) were infected (10 PFU/cell) with recombinant baculovirus expressing XPC-Rad23B passage-2 virus and incubated at 27°C. After 48 hours, a cell free extract was prepared as described by Manley et al. (29). Briefly, cells were lysed in a hypotonic buffer (10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 5 mM DTT) followed by a high salt buffer (50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 2 mM DTT, 25% (w/v) sucrose, 50% (v/v) glycerol). The sample was centrifuged at 30,000 × g for 1 hour and the supernatant was dialyzed overnight in Buffer A (25 mM Tris-Cl (pH 7.5), 1 mM EDTA, 1 mM DTT, 0.001% (v/v) Triton X-100, and 10% (v/v) glycerol) containing 0.3 M NaCl. Following centrifugation, the clarified supernatant was applied to a 10 mL P-cell column equilibrated in Buffer A containing 0.3 M NaCl. The column was washed with 0.3 M NaCl Buffer A and protein was eluted from the column using 1 M NaCl Buffer A. XPC-Rad23B containing fractions were pooled and diluted with Buffer A to achieve a final salt concentration of 0.6 M. The diluted protein pool was applied to a 4 mL ssDNA column equilibrated in 0.6 M NaCl Buffer A. The column was washed and protein was eluted from the column using 1.5 M NaCl Buffer A. XPC-Rad23B containing fractions were pooled and dialyzed overnight in Buffer B (25 mM HEPES (pH 7.8), 0.2 M KCl, 1 mM EDTA, 1 mM DTT, and 50% (v/v) glycerol). Final protein concentration was determined using a Bradford dye-based assay (Bio-Rad) and the dialyzed protein was aliquoted and stored at −80°C. SDS-PAGE followed by coomassie staining or western blot analysis was utilized to determine protein purity. XPC-Rad23B DNA binding activity was assessed using fluorescence polarization on a fluorescein-labeled DNA substrate (56-mer) in 500 μL reaction containing 20 mM HEPES (pH 7.8), 1 mM DTT, 0.01% (v/v) NP-40, 100 mM NaCl (16). XPC-Rad23B was titrated to a final concentration of 200 μM. Activity was also accessed using electrophoretic mobility shift assay (EMSA) on undamaged or cisplatin-damaged substrates (60-mer) (16).
DNA Substrates
All oligonucleotides (22-, 37-, 38-, 60-, 61-mers) (Table 1) were purified by electrophoresis on denaturing polyacrylamide gels, ethanol precipitated, and concentration was determined using a Nanodrop 1000 (Nanodrop Technologies). Damage was induced by incubating the purified oligonucleotides with cisplatin at a drug:molecule ratio of 10:1 in buffer containing 1 mM NaHPO4 (pH 7.5) and 3 mM NaCl for 48 hours. Damaged DNA was quantified using the Nanodrop 1000 (Nanodrop Technologies) following ethanol precipitation (16).
Table 1.
DNA Substrates. Single strand substrates are presented along with the complementary strands. The location of cisplatin damaged is underlined in the substrates and the location of the incorporated photoreactive FAP-dCTP analogue is presented in italics.
| DNA | Sequence (5′-3′) |
|---|---|
| 1,2 d(GpG)a | CCCTTCTTTCTCTTCCCCCTCTCCTTCTTGGCCTCTTCCTTCCCCTTCCCTTTCCTCCCC |
| C-1,2 d(GpG) | GGGGAGGAAAGGGAAGGGGAAGGAAGAGGCCAAGAAGGAGAGGGGGAAGAGAAAGAAGGG |
| 5′FAP1b | CCCTTCTTTCTCTTCCCCCTATC |
| 5′FAP2a | TCTCTTGGCCTCTCTCTTCCCCTTCCCTTTCCTCCCC |
| 3′FAP1a b | CCCTTCTTTCTCTTCCCCCTATCTCTCTTGGCCTCTCTC |
| C-FAP | AGGGGAGGAAAGGGAAGGGGAAGAGAGAGGCCAAGAGAGATAGGGGGAAGAGAAAGAAGGG |
Underlined bases indicate the site for cisplatin damage.
Italic base indicates the incorporated photoreactive FAP-dCTP analogue.
Radio-labeled (5′) DNA was prepared by incubating undamaged or cisplatin-damaged oligonucleotides with γ32P-ATP, T4 PNK and Kinase Buffer at 37°C. After 30 minutes of incubation, 1 mM ATP was added followed by 0.5 M EDTA and then the DNA was purified using a G50-spin column. Counts per minute (CPM) was measured using a scintillation counter (Beckman) and the specific activity and DNA concentrations were calculated. Undamaged and damaged substrates were annealed to the complement strand, C-1,2 d(GpG), by heating to 95°C for 3 minutes in annealing buffer (50 mM Tris (pH 7.5), 10 mM magnesium acetate, and 5 mM DTT) to generate ds-(GpG). Double stranded cisplatin-damaged substrates were subjected to HaeIII digestion and 60-mer products were purified by electrophoresis on native polyacrylamide gels. The site of platination resides within the HaeIII digestion site; therefore, any substrate not platinated is cleaved by HaeIII to a 30-mer; whereas platinated substrates remain 60-mers.
Oligonucleotides possessing the photoreactive analogue of dCMP exo-N-{2-[N-(4-azido-2,5-difluoro-3-chloropyridine-6-yl)-3-aminopropionyl]-aminoethyl}-2′-deoxycytidine-5′-monophosphase (FAP-dCMP) were prepared by extension reactions catalyzed by Klenow Polymerase 3′-5′ exo- and ligation catalyzed by T4 DNA ligase. Duplex DNA substrates were prepared with the FAP-dCMP either 5′ (ds-5′FAP) or 3′ (ds-3′FAP) of the cisplatin adduct (Supplementary Figure 1). 5′FAP1 and 3′FAP1 were 5′ radio-labeled and control (undamaged) and cisplatin damaged 5′FAP2 was cold-phosphorylated and excess ATP removed by Sephadex G50-spin column chromatogrphy. Radiolabeled control or damaged 5′FAP1 and 5′FAP2 were annealed to the complementary strand (C-FAP) and fill-in reactions performed with FAP-dCTP followed by ligation were used to generate control and cisplatin damaged ds-5′FAP. Radio-labeled control or cisplatin-damaged 3′FAP1 was annealed to C-FAP, extended with the FAP-dCTP analogue and purified via Sephadex G50-spin column chromatography. The substrates were incubated with dCTP, dTTP, and Klenow Polymerase 3′-5′ exo- for an additional 30 minutes at 37°C to complete synthesis of the substrate generating control or cisplatin-damaged ds-3′FAP. The free dNTPs were removed using a Sephadex G50-spin column chromatography and cisplatin-damaged substrates were digested with HaeIII (see above), purified by electrophoresis on a preparative denaturing polyacrylamide gel and reannealed.
Electrophoretic Mobility Shift Assay (EMSA)
XPC-Rad23B was bound to 100 fmol of undamaged or cisplatin damaged ds DNA (60-mer) in buffer containing 20 mM HEPES (pH 7.8), 0.001% (v/v) NP-40, 50 mM NaCl, 1 mM DTT, 0.05 mg/ml BSA, and 2 mM MgCl2. Binding was performed for 15 minutes at room temperature in the absence or presence of non-specific competitor poly dI/dC (100 ng). Bound XPC-Rad23B was gluteraldehyde crosslinked to the substrates, by the addition of 0.25% (v/v) gluteraldehyde and incubating the samples at room temperature for 3 minutes. Buffer containing 10 mM EDTA, 10% glycerol, 0.01% bromophenol blue, and 0.01% xylene cyanol was used to stop the reactions and samples were separated on native polyacrylamide gels for 1 hour at 170 volts. Dried gels were imaged using a Storm 820 PhosphorImager (Amersham, Biosciences) and bound protein was quantified using ImageQuant 5.2. Averages and standard deviations of three independent experiments are presented.
Photocrosslinking and SDS-PAGE Analysis
Binding to cisplatin damaged or undamaged/control DNA substrates was performed in the presence or absence of poly dI/dC, as stated above. Samples were then photocrosslinked in eppendorf tubes, on ice, by UV irradiation using General Electric-15 Watt bulbs which emit a wavelength of 254 nm, for the indicated times. Following crosslinking, 6x SDS dye was used to stop the reactions and samples were heated to 95°C for 5 minutes and loaded on SDS-PAGE gels. Gels were imaged and quantified using a Storm 820 PhosphorImager (Amersham, Biosciences) and ImageQuant 5.2. Averages and standard deviations from at least three independent experiments are presented.
Immunoprecipitation (IP)
XPC-Rad23B was bound to the indicated undamaged/control or cisplatin damaged dsDNA (600 fmol) and photocrosslinked for the indicated times. Photocrosslinked samples were incubated for 20 minutes with either XPC or Rad23B rabbit polyclonal antibodies with and without a denaturation step (10 minutes at 95°C) prior to incubation with the antibody. Protein-G agarose was washed three times with PBS, added to each sample along with PBS buffer, and rotated at 4°C for 1.5 hours. Samples processed after IP were washed three times with PBS, resuspended in 1x SDS dye and loaded onto SDS-PAGE gels. Western blot analysis using either a primary XPC or Rad23B antibody (1:1,000) and a goat-anti-rabbit secondary antibody (1:2,000) was used along with chemiluminescense to visualize the results. A Fuji LAS-3000 along with Multi Gauge V3.0 software was used to visualize the results. Radio-labeled substrates were also separated using SDS-PAGE and dried gels were imaged using a Storm 820 PhosphorImager (Amersham, Biosciences).
Results
XPC-Rad23B Covalently Photocrosslinks to Undamaged and Cisplatin-Damaged DNA
We and others have analyzed the interaction of XPC-Rad23B with DNA by a variety of biochemical techniques and assays (15;16;19;30). XPC-Rad23B demonstrates damage specificity only in the presence of the non-specific competitor poly dI/dC, a synthetic double strand oligonucleotide. XPC-Rad23B often requires gluteraldehyde crosslinking to identify the presence of protein-DNA complexes (16). In order to further analyze the specifics of XPC-Rad23B interactions with DNA, we employed DNA containing a single cisplatin lesion in conjunction with site-specific photoreactive dCMP residues. The XPC-Rad23B complex was purified to near homogeneity from insect cells using a recombinant baculovirus construct. Preliminary experiments involved fluorescence polarization and EMSA with gluteraldehyde crosslinking confirmed the DNA binding activity of XPC-Rad23B. XPC-Rad23B was shown to bind to a fluorescein-labeled substrate (56-mer) and gluteraldehyde crosslinking demonstrated damaged specificity in the presence of competitor poly-dI/dC, confirming previously published results (16).
Photocrosslinking of XPC-Rad23B to radio-labeled DNA was conducted by first binding XPC-Rad23B to ds-(GpG) with or without a site-specific 1,2-d(GpG) cisplatin lesion (Table 1). XPC-Rad23B was photocrosslinked to these DNA derivatives for 1 hour and SDS-PAGE gels were utilized to separate the covalent protein-DNA complexes. Preliminary experiments involving UV exposure time course analysis revealed 1 hour to be the optimal for protein stability and crosslinking efficiency (data not shown). PhosphorImager analysis reveals the presence of two bands in reactions containing radio-labeled DNA duplexes and XPC-Rad23B. The lower band migrating at approximately 15 kDa corresponds to unbound DNA and the upper band (170 kDa) corresponds to a covalent protein-DNA complex (Figure 1A). The amount of this covalent protein-DNA complex increased with increasing concentrations of XPC-Rad23B whereas the unbound DNA decreases. In the data presented and quantification of triplicate binding experiments, XPC-Rad23B revealed preferential crosslinking to cisplatin damaged DNA (Figure 1C). However, the results were variable and in some preparations of XPC-Rad23B, there was less of a difference in photocrosslinking of the protein to undamaged and cisplatin damaged DNA. Similar experiments involving photocrosslinking XPC-Rad23B to a damaged ds-DNA substrate (60-mer) containing a site-specific 1,3-d(GpXpG) cisplatin lesion also reveal damage specific crosslinking (Supplementary Figure 2). As stated above, XPC-Rad23B gluteraldehyde crosslinks more efficiently to undamaged DNA in the absence of competitor (data not shown). Therefore the observed increase in photocrosslinking of XPC-Rad23B to cisplatin-damaged DNA suggests there is either a platinum-damage dependent alteration in XPC structure that is more efficiently crosslinked to the DNA bases or, more likely, a direct platinum-protein crosslink. The precedence for UV-dependent direct platinum-protein crosslinks has been established in the analysis of HMGB-1 binding to a single cisplatin-damaged DNA (31). This interpretation would position XPC in direct proximity to the DNA adduct consistent with the recently reported Rad4 crystal structure in conjunction to a CPD lesion (21).
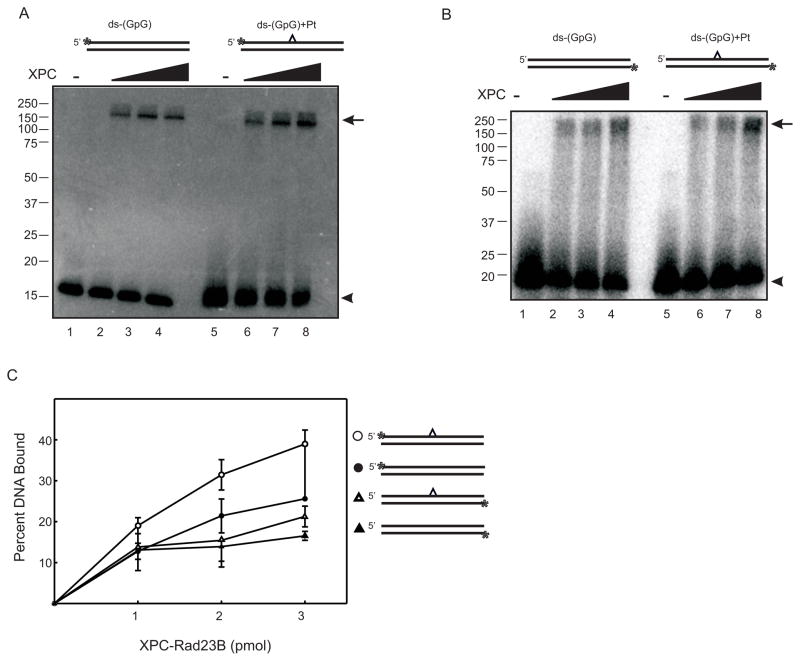
Figure 1.
XPC-Rad23B Photocrosslinks to Undamaged and Cisplatin-Damaged DNA. (A) Increasing amounts of XPC-Rad23B were incubated with undamaged or cisplatin damaged ds-DNA with the 32P-label on the damaged strand and photocrosslinked for 60 minutes. Samples were heat denatured, separated via SDS-PAGE and radioactivity detected by PhosporImager analysis. (B) Similar crosslinking reactions were performed except the 32P-label was positioned on the DNA strand complementary to the damaged DNA strand. A schematic of each DNA substrate is depicted above the gels with the cisplatin indicated by the carat and the position of the 32P-label by the asterisk. The covalent protein-DNA complex is indicated by the arrow and the free DNA by the arrow head. (C) Quantification of XPC-Rad23B photocrosslinked to ds- undamaged or cisplatin-damaged substrates presented in A and B.
XPC-Rad23B Covalently Photocrosslinks to the Undamaged Complementary Strand
Data presented above demonstrates that XPC-Rad23B is able to bind and photocrosslink to cisplatin damaged and undamaged DNA, demonstrating preference for damaged DNA. However, it is not known if, in the context of a duplex DNA containing a single cisplatin lesion, XPC-Rad23B can make contact with the undamaged complementary DNA strand. In order to visualize whether XPC-Rad23B binds to the undamaged complement strand, DNA substrates were prepared with a 5′-32P-label located on the undamaged strand. After binding and photocrosslinking, SDS-PAGE gels were used to separate the crosslinked products and a PhosphorImager was utilized to visualize the results. Again, only covalent interactions will remain intact when separated on SDS-PAGE gels. Therefore, if XPC-Rad23B is not covalently bound to the undamaged complement strand, only free DNA would be expected (15 kDa). However, data reveals the presence of a high molecular weight band (170 kDa) representative of a protein-DNA photo-induced covalent crosslink (Figure 1B). Quantification of the data (Figure 1C) indicates the amount of the crosslinked product observed when the undamaged strand is 32P-labeled is less than that observed in reactions with the damaged strand 32P-labeled. These results suggest that XPC and possibly Rad23B not only make contact with the damaged strand of duplex DNA but also interacts with the complementary undamaged strand of the damaged DNA duplex (Figure 1). Therefore XPC forms direct contacts with both strands of an undamaged DNA duplex (Figure 1A and B). While the photocrosslinking reactions were internally controlled, irradiation with UV light for 60 minutes can impact numerous factors including UV induced DNA damage and decreases in overall protein stability. Control experiments were conducted in which the DNA was UV irradiated and then used in an XPC-Rad23B binding reaction with gluteraldehyde crosslinking. The results demonstrate no significant increase in protein-DNA complex formation versus the control experiment in which the DNA was not UV irradiated prior to gluteraldehyde crosslinking (data not shown). In addition, SDS-PAGE analysis of the protein following UV-irradiation revealed no significant degradation of the XPC or Rad23B subunits (data not shown). Despite the controls which indicated that there was not extensive UV induced damage to the substrate, the crosslinking efficiency is still relatively poor, thus we sought a more efficient methodology with increased resolution to characterize the protein-DNA complexes formed on cisplatin damaged DNA.
XPC-Rad23B Covalently Photocrosslinks to Photoreactive Substrates
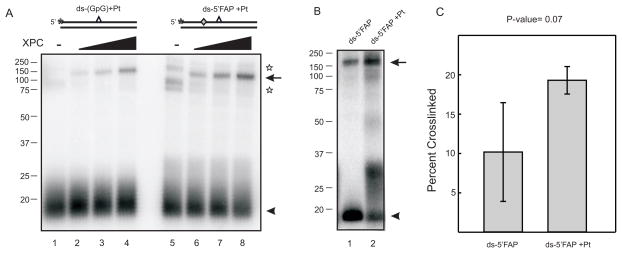
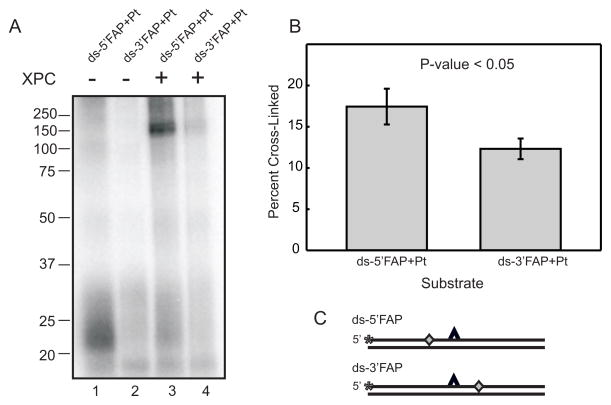
Previously, halogenated bases have been used to improve crosslinking efficiency in experiments involving photoaffinity labeling of proteins. However, only small increases in photocrosslinking efficiency have been shown (32). More recently, photoactivateable crosslinkers have been synthesized as dNTP analogs which allow for their site-specific placement into DNA as a photoreactive dNMP moiety which results in significantly increased crosslinking efficiency (28;33;34). Oligonucleotides possessing a photoreactive FAP-dCMP analogue, 5′ or 3′ of the cisplatin damage site were prepared as described in Experimental Procedures. Photocrosslinking experiments were again conducted with DNA substrates which contained FAP-dCMP moieties and were only exposed to UV light for 10 minutes versus the 60 minute exposure for dCMP containing substrates. The significant decrease in UV exposure reduces the likelihood of UV-induced damage to the DNA substrates and protein which may influence binding and crosslinking. SDS-PAGE and PhosphorImager analysis were utilized for sample separation and visualization of the crosslinking results. Data reveal that XPC-Rad23B photocrosslinks significantly more efficiently to substrates possessing FAP-dCMP than an identical substrate possessing dCMP. The data presented in Figure 2A demonstrate that 60 minute photocrosslinking with the control substrate (dCMP) still resulted in less covalent protein-DNA complex being detected compared to a 10 minute exposure with the FAP-dCMP containing substrate. The FAP-dCMP substrate crosslinks with itself in the absence of XPC-Rad23B; however, this interaction is eliminated in the presence of XPC-Rad23B and doesn’t inhibit a protein-DNA interaction (Figure 2A). In comparing control and cisplatin-damaged substrates containing FAP-dCMP, XPC does not demonstrate damage specific binding when the photoreactive substrate is 5′ or 3′ of the analogue (Figure 2B and C and Supplementary Figure 3A and B). This result suggests that the FAP-dCTP analogue causes some degree of distortion with which XPC-Rad23B can recognize and interact. Data demonstrates that XPC-Rad23B crosslinks slightly more efficiently to substrates containing a 5′ located photoreactive analogue verses a 3′ photoreactive analogue (Figure 3). A comparison of the data presented in Figure 3B is significant to a P-value of <0.05. This data demonstrates that XPC-Rad23B has a preference for interacting 5′ of the site of damage. However, the ability to detect protein-DNA interactions 3′ of the cisplatin lesion suggests the protein spans the damage to enable contacts on both sides of the cisplatin lesion. Overall this data provides a better understanding of where XPC-Rad23B contacts the damaged DNA, more specifically, XPC-Rad23B contacts the DNA both 5′ and 3′ of the cisplatin lesion demonstrating a preference for contacting 5′ of the lesion.
Figure 2.
Damage Specificity of XPC-Rad23B Photocrosslinks revealed by a Photoreactive FAP-dCTP Analogue (A) Increasing amounts of XPC-Rad23B (2, 4 and 6 pmol) were incubated with cisplatin-damaged control or FAP-dCMP modified ds-DNA with the 32P-label on the damaged strand and photocrosslinked for 60 minutes (lanes 1–4) or 10 minutes (lanes 5–8). Samples were heat denatured, separated via SDS-PAGE and radioactivity detected by PhosporImager analysis. The stars indicate a photo-induced DNA crosslink observed in the absence of XPC-Rad23B. (B) Similar crosslinking reactions were performed with the FAP-dCMP modified using either control (lane 1) or cisplatin-damaged DNA (lane 2). The covalent protein-DNA complex is indicated by the arrow and the free DNA by the arrow head. A schematic of each DNA substrate is depicted above the gels with the cisplatin indicated by the carat, the position of the 32P-label by the asterisk and the position of the FAP-dCMP indicated by the diamond. (C) Quantification of three independent crosslinking experiments to compare the efficiency of XPC-Rad23B crosslinking to a cisplatin-damaged or control substrate. A P-value of 0.07 was determined by a paired student T-test analysis.
Figure 3.
Orientation of XPC-Rad23B bound to Cisplatin-Damaged DNA. (A) XPC-Rad23B (6 pmol) was incubated with and photocrosslinked for 10 minutes to cisplatin damaged substrates containing a 5′ (lanes 1 and 3) or 3′ (lanes 2 and 4) photoreactive FAB-dCMP analogue. Products were separated by SDS-PAGE and visualized by PhosphorImager analysis. (B) Quantification of the binding data from three independent crosslinking experiments. A paired T-test was utilized to determine the statistical significance (P-value <0.05). (C) Schematic of DNA substrates with the cisplatin indicated by the carat, the position of the 32P-label by the asterisk and the position of the FAP-dCMP indicated by the diamond.
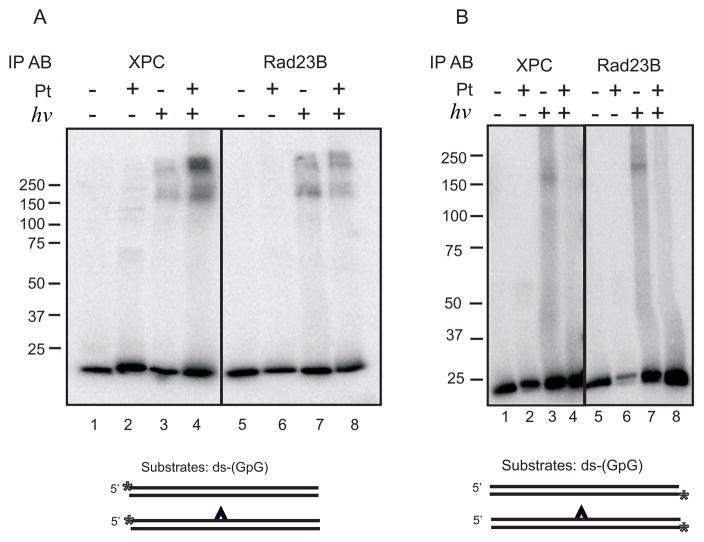
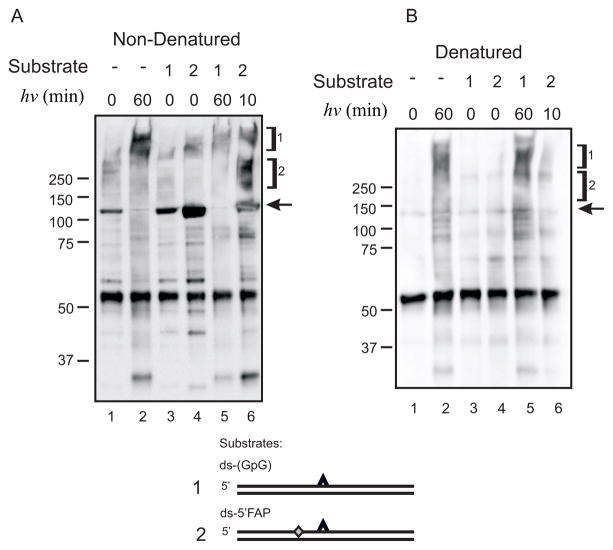
XPC-Rad23B Subunits Covalently Photocrosslink in the Absence and Presence of DNA
The mobility of the covalent protein-DNA complex (170 kDa) presents the possibility that either XPC alone or in complex with Rad23B is present in the crosslinked product. The absence of a product with the mobility ranging between 60–100 kDa suggests that Rad23B alone is not in complex with the DNA. To determine if Rad23B is present in the 170 kDa covalent protein-DNA complex, we extended the crosslinking analysis to include IP after a heat denaturation step used to disrupt the non-covalent protein-protein and protein-DNA complexes. In doing so, only remaining covalent interactions would be pulled-down with IP and detected on the SDS-PAGE. Results demonstrate that both the XPC and Rad23B antibody are capable of pulling-down a photo-induced covalent protein-DNA complex (Figure 4A). Similar analysis performed, in which the undamaged complementary strand of the duplex was radio-labeled, revealed significantly less crosslinking which is apparent in that over-exposure is necessary to detect the complexes (Figure 4B). In addition, quantification of the signal intensity revealed a higher level of binding to the undamaged DNA compared to the cisplatin-damaged DNA (24% bound verses 7%).
Figure 4.
Immunoprecipitation of Photocrosslinked protein-DNA complex with anti-XPC or anti-Rad23B. (A) XPC-Rad23B was bound and photocrosslinked to radio-labeled undamaged or cisplatin damaged double strand substrates containing the 32P-label on the damaged strand. IP was performed with either the XPC (lanes 1–4) or Rad23B antibody (lanes 5–8) and products were imaged using a PhosphorImager. (B) Reactions were performed similar to those in panel A except the 32P-label was on the DNA strand complementary to the damaged strand. A schematic of DNA substrates below each gel is presented with the cisplatin indicated by the carat and the position of the 32P-label by the asterisk.
The ability to pull down a protein-DNA complex with anti-Rad23B suggests that either Rad23B is in direct contact with DNA or was covalently photocrosslinked to XPC. To further study the nature of these interactions, we probed for each component using western blot analysis. In these experiments, samples were UV irradiated for the indicated times, immunoprecipitated with either anti-XPC (data not shown) or anti-Rad23B without a heat denaturation step (Figure 5A) or following heat denaturation (Figure 5B). The control experiments performed without prior heat denaturation demonstrate, as expected, that anti-Rad23B could pull-down XPC in the presence or absence of DNA (Figure 5A lanes 1, 3 and 4). Exposure to UV light reveals a decrease in the intensity of the XPC protein migrating at 125 kDa and the appearance of a smear of products with significantly reduced mobility consistent with covalently interacting XPC-Rad23B complexes (Figure 5A lanes 2, 5, 6). Interestingly, when samples were photocrosslinked and heat denatured prior to pull down with the anti-Rad23B antibody, an XPC-Rad23B complex with reduced mobility was still detected (Figure 5B lanes 2, 5, and 6). These results are consistent with the generation of a photo-induced covalent bond between the XPC and Rad23B subunits in the absence or presence of DNA. Data revealed that the presence of DNA had minimal affect on the photo-induced XPC-Rad23B covalent complexes (Figure 5A and B compare lanes 3 to 5 and 4 to 6). In summary, photocrosslinking induced both protein-protein interactions in the absence and presence of DNA as well as protein-DNA interactions in the presence of DNA.
Figure 5.
Immunoprecipitation and Western Blot Analysis of Photocrosslinked XPC in the Absence or Presence of Damaged DNA. (A) XPC-Rad23B and the indicated DNA substrates were bound, crosslinked for the indicated time, and immunoprecipitated using the anti-Rad23B antibody. The IP was then separated by SDS-PAGE, transferred to PVDF and probed with anti-XPC antibody. Antibody reactivity was visualized by chemiluminescence detection using a HRP-conjugated secondary antibody. Bracket 1 denotes a covalent protein-protein crosslink that occurs between XPC and Rad23B subunits that is independent of DNA. Bracket 2 indicates a covalent protein-DNA crosslink. Free, uncrosslinked XPC is indicated by the arrow. (B) Identical reactions were performed as described in panel A except the samples were heat denatured prior to immunoprecipitation.
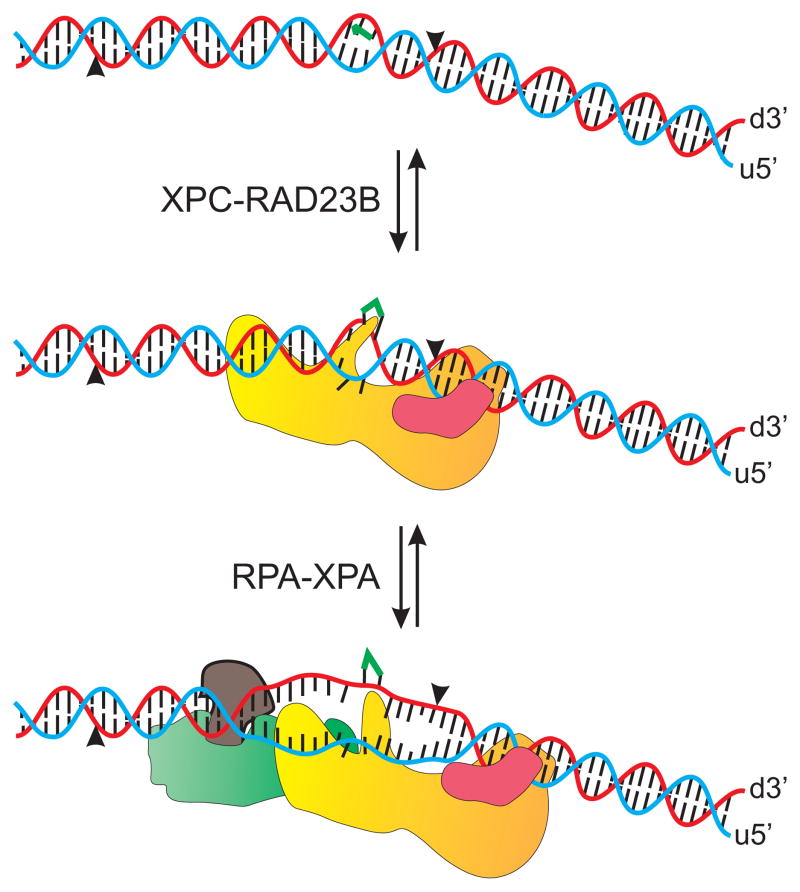
Together these results lead to a model whereby the XPC subunit is responsible for the majority of the interactions with DNA while Rad23B appears to make minimal contact directly with DNA (Figure 6). The XPC-Rad23B complex does contact DNA both 5′ and 3′ of the damaged site though there appears to be a preference for interaction 5′ of the lesion. XPC-Rad23B complex also makes contact with the undamaged strand and in this case the Rad23B subunit contributes to the interaction with DNA (Figure 6). Interestingly, the photocrosslinking procedure resulted in the formation of a covalent bond between the two protein subunits that was not influenced by DNA, suggesting that the interface between these two protein subunits, following DNA binding, is not (or minimally) disrupted.
Figure 6.
Model of XPC-Rad23B Binding to Cisplatin Damaged DNA. XPC is presented as a yellow complex, Rad23B as a mauve complex, XPA as a brown complex and RPA as a green complex. XPC-Rad23B recognizes and binds to cisplatin damaged DNA with placement of the XPC subunit both 5′ and 3′ of the cisplatin lesion. The Rad23B subunit interacts with the undamaged strand of the cisplatin damaged duplex 3′ of the cisplatin lesion. After damage recognition, RPA-XPA binds to the damaged duplex and XPC-Rad23B prepares to dissociate from the damaged DNA duplex.
Discussion
XPC-Rad23B as a DNA binding protein recognizes and interacts with the bases or the sugar-phosphate backbone of DNA. To impart damage specificity in the NER pathway, XPC-Rad23B must display a higher affinity for DNA containing chemical modifications or distortions. These can be caused by exposure to common chemotherapeutic agents like cisplatin or environmental carcinogens such as exposure to UV light. After binding to damaged DNA, XPC-Rad23B recruits downstream NER proteins and then dissociates from the site of damage prior to incision by the XPG and XPF/ERCC1 nucleases (1;2). The binding and dissociation of XPC-Rad23B occurs without any change in the chemical structure of the DNA, although changes in DNA secondary structure are likely to accompany XPC-Rad23B-DNA interactions (21). Based on the crystal structural analysis of Rad4-Rad23B bound to UV damaged DNA, changes in secondary structure of DNA include the displacement of bases opposite the thymine dimer out of the DNA helix and this is accompanied by localized unwinding of the DNA surrounding the lesion, possibly influencing DNA-protein interactions. Similarly, there are changes in the Rad4 protein tertiary structure in which the β-hairpin of BHD2 and BHD3 are inserted between the UV-damaged duplex DNA while in the apo structure these domains are displaced to avoid steric interaction (21). The crosslinking analysis performed in this study using cisplatin damaged DNA revealed XPC-DNA interactions in close proximity to the cisplatin lesion suggesting that while UV and cisplatin cause different DNA structural and chemical distortions, there is a common mechanism of damage recognition involving the BHD2 and BHD3 domains. More specifically BHD3, which shares 31% sequence identity with XPC, may play a major role in stabilizing XPC-DNA interactions since most of the conserved regions are localized in residues with structural or DNA binding properties. The ability to photocrosslink XPC-Rad23B to undamaged DNA is likely through the TGD and BHD1 domains as revealed from the crystal structure modeled with undamaged DNA (21). This suggests that the interaction of XPC-Rad23B to cisplatin-damaged DNA would be similar to that of Rad4-Rad23B with UV-damaged DNA.
With respect to damage and DNA strand orientation, the majority of DNA interactions in the Rad4 structure are 3′ of the DNA damage (21). Interestingly, the crosslinking analysis of XPC-Rad23B demonstrates interactions on both the 5′ and 3′ side of the cisplatin lesion. The potential exists that photo-induced crosslinking of XPC-Rad23B to photoreactive DNA substrates may be influenced by the presence of the photoreactive FAP-dCMP analogue. However, crosslinking analysis, in which the analogue was incorporated into the complementary strand, either 5′ or 3′ of the cisplatin adduct, demonstrates no difference in XPC-Rad23B crosslinking to the DNA substrates (Supplementary Figure 4). The complete C-terminal and N-terminal domains of the Rad4 protein are absent in the crystal structure, however the directionality of the protein domain location is suggestive of an interaction 5′of the lesion. However, further structural analysis is necessary to have a full assessment of the protein-DNA interaction. The crosslinking analysis reveals that XPC interacts with both the damaged and undamaged strands of the DNA duplex. The absence of structural data with the C-terminus of Rad4 makes the identification of amino acids interacting 5′ of the damaged site impossible. However, structural analysis suggests that Rad4 interactions with DNA 3′ of the CPD lesion involve amino acids Val448 to Thr456. Interactions of Rad4 with the undamaged strand of the duplex DNA are likely to occur via amino acids Lys511 through Leu531. The interactions identified by the Rad4 crystal structure were based on DNA containing a CPD lesion; however, it is suggestive that the amino acids in contact with the DNA surrounding an adduct would be the same. In the case of the CPD lesion presented, amino acids Phe599-Val605 insert between the two DNA strands of the duplex and interact with nucleotides directly surrounding the CPD adduct (21). As a cisplatin intrastrand lesion distorts the DNA differently than a CPD lesion, the interaction of XPC-Rad23B with the damaged DNA may in fact be different, with the difference in distortion resulting in the differential positioning of XPC on the damaged duplex. This would then potentially influence the binding and placement of downstream NER proteins and impact the position of incision relative to the adduct.
While previous data has demonstrated that the size of the incised fragment is the same with platinum and UV adducts (35), the position of XPG and XPF catalyzed incision on cisplatin damaged DNA is not known. While initial crosslinking and immunoprecipitation experiments with XPC-Rad23B were suggestive of a direct Rad23B-DNA covalent crosslink, further analysis demonstrates that Rad23B does not interact directly with DNA, the interaction occurs through the XPC subunit. Crosslinking analysis alone does not demonstrate the presence of a Rad23B-DNA interaction because if a covalent interaction was induced by photocrosslinking, a complex between 60–100 kDa would be present. Therefore one must be cautious in the interpretation of data with multi-subunit proteins assessing DNA-protein interactions due to the possibility of protein-protein crosslinks. This also highlights the tremendous utility for the use of photoreactive analogues with high photometric yields for the identification of true protein-DNA covalent crosslinks without induction of UV induced protein-protein crosslinks. Based on the data presented and previous structural data involving Rad4 (21), the model depicts that XPC-Rad23B interacts with both strands of a cisplatin damaged duplex DNA. The XPC subunit interacts both 5′ and 3′ of the cisplatin adduct and contacts both the damaged strand and undamaged strand of the duplex. The Rad23B subunit also interacts with both strands of the duplex DNA and contacts the XPC subunit directly. Following damage recognition and binding of XPC-Rad23B the duplex DNA opens allowing the RPA-XPA complex to bind to the damaged DNA (2). The model depicts an RPA-XPA interaction 5′ of the cisplatin lesion contacting the undamaged DNA strand. The structure of the DNA is further altered by the addition of RPA-XPA and continues to generate a larger single-stranded region with the addition of downstream NER proteins, such as TFIIH, XPF and XPG.
In vitro, the XPC-Rad23B heterodimer is sufficient to reconstitute the NER damage recognition complex and supports dual incision (36). In vivo, Centrin 2 forms a complex with XPC-Rad23B and it remains unknown how Centrin 2 influences these protein DNA interactions in the cell. Similarly, XPC is subject to extensive post-translations modifications such as ubiquitination and sumolyation which can impact DNA binding and protein-protein interactions (37;38). It has been shown that XPC is reversibly ubiquitinated by DDB, however this ubiquitination does not trigger XPC degradation (37) and it remains unknown how, after damage recognition, XPC-Rad23B is released from the site of damage. It is possible that post-translational modifications may influence the XPC-Rad23B DNA-interaction, and cause XPC-Rad23B to dissociate from the damaged site. Taken together, it will also be of interest to determine the role of Centrin 2, DDB and post-translational modifications on XPC-Rad23B-DNA interactions to provide better insight into the mechanisms of damage recognition and dissociation of XPC-Rad23B from the damaged DNA.
Supplementary Material
Acknowledgments
This research was supported by award R01 CA82741 from the NIH, National Cancer Institute (JJT) and the Program on Cellular and Molecular Biology RAS (OL)
We thank the members of the Turchi lab for their insightful scientific discussions and detailed reading of the manuscript.
Abbreviations
- bp
base pair(s)
- BSA
bovine serum albumin
- CPD
cyclobutane pyrimidine dimer
- ds-DNA
double strand DNA
- DTT
dithiothreitol
- EDTA
disodium ethylenediaminetetraacetic acid
- EMSA
electrophoretic mobility shift assay
- GG-NER
global genomic nucleotide excision repair
- IP
immunoprecipitation
- NER
nucleotide excision repair
- Pt
cisplatin
- RPA
replication protein A
- TFIIH
transcription factor II H
- XPA
C,F,G, xeroderma pigmentosum group A,C,F,G proteins
Footnotes
TMN was supported by the NIH, NRSA T32 CA 111198 Cancer Biology Training Program
Supporting Information Available: Supporting data, including a flow chart for substrate preparation, photocrosslinking analysis involving a 1,3-cisplatin lesion, photocrosslinking analysis of in which the photoreactive analogue is located 3′ of the cisplatin lesion or in the complementary strand has been included. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Nouspikel T. DNA repair in mammalian cells: Nucleotide excision repair: variations on versatility. Cell Mol Life Sci. 2009;66:994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shuck SC, Short EA, Turchi JJ. Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology. Cell Res. 2008;18:64–72. doi: 10.1038/cr.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugasawa K, Ng JMY, Masutani C, Iwai S, vanderSpek PJ, Eker APM, Hanaoka F, Bootsma D, Hoeijmakers JHJ. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 4.Wakasugi M, Sancar A. Order of assembly of human DNA repair excision nuclease. J Biol Chem. 1999;274:18759–18768. doi: 10.1074/jbc.274.26.18759. [DOI] [PubMed] [Google Scholar]
- 5.Araujo SJ, Tirode F, Coin F, Pospiech H, Syvaoja JE, Stucki M, Hubscher U, Egly JM, Wood RD. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes & Development. 2000;14:349–359. [PMC free article] [PubMed] [Google Scholar]
- 6.Lehmann AR. New functions for Y family polymerases. Mol Cell. 2006;24:493–495. doi: 10.1016/j.molcel.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 7.Tapias A, Auriol J, Forget D, Enzlin JH, Scharer OD, Coin F, Coulombe B, Egly JM. Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors. J Biol Chem. 2004;279:19074–19083. doi: 10.1074/jbc.M312611200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishi R, Alekseev S, Dinant C, Hoogstraten D, Houtsmuller AB, Hoeijmakers JH, Vermeulen W, Hanaoka F, Sugasawa K. UV-DDB-dependent regulation of nucleotide excision repair kinetics in living cells. DNA Repair (Amst) 2009;8:767–776. doi: 10.1016/j.dnarep.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Sugasawa K. UV-DDB: A molecular machine linking DNA repair with ubiquitination. DNA Repair (Amst) 2009;8:969–972. doi: 10.1016/j.dnarep.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Wakasugi M, Kawashima A, Morioka H, Linn S, Sancar A, Mori T, Nikaido O, Matsunaga T. DDB Accumulates at DNA Damage Sites Immediately after UV Irradiation and Directly Stimulates Nucleotide Excision Repair. J Biol Chem. 2002;277:1637–1640. doi: 10.1074/jbc.C100610200. [DOI] [PubMed] [Google Scholar]
- 11.El-Mahdy MA, Zhu Q, Wang QE, Wani G, Praetorius-Ibba M, Wani AA. Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J Biol Chem. 2006;281:13404–13411. doi: 10.1074/jbc.M511834200. [DOI] [PubMed] [Google Scholar]
- 12.Araki M, Masutani C, Takemura M, Uchida A, Sugasawa K, Kondoh J, Ohkuma Y, Hanaoka F. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J Biol Chem. 2001;276:18665–18672. doi: 10.1074/jbc.M100855200. [DOI] [PubMed] [Google Scholar]
- 13.Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hubscher U, Egly JM, Wood RD. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- 14.Park CH, Sancar A. Reconstitution of mammalian excision repair activity with mutant cell-free extracts and XPAC and ERCC1 proteins expressed in Escherichia coli. Nucleic Acids Res. 1993;21:5110–5116. doi: 10.1093/nar/21.22.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krasikova YS, Rechkunova NI, Maltseva EA, Petruseva IO, Silnikov VN, Zatsepin TS, Oretskaya TS, Scharer OD, Lavrik OI. Interaction of nucleotide excision repair factors XPC-HR23B, XPA, and RPA with damaged DNA. Biochemistry (Mosc ) 2008;73:886–896. doi: 10.1134/s0006297908080063. [DOI] [PubMed] [Google Scholar]
- 16.Trego KS, Turchi JJ. Pre-steady state binding of damaged DNA by XPC-hHR23B reveals a kinetic mechanism for damage discrimination. Biochemistry. 2006;45:1961–1969. doi: 10.1021/bi05196t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes & Development. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.You JS, Wang M, Lee SH. Biochemical analysis of the damage recognition process in nucleotide excision repair. J Biol Chem. 2003;278:7476–7485. doi: 10.1074/jbc.M210603200. [DOI] [PubMed] [Google Scholar]
- 19.Roche Y, Zhang D, Segers-Nolten GM, Vermeulen W, Wyman C, Sugasawa K, Hoeijmakers J, Otto C. Fluorescence correlation spectroscopy of the binding of nucleotide excision repair protein XPC-hHr23B with DNA substrates. J Fluoresc. 2008;18:987–995. doi: 10.1007/s10895-008-0390-6. [DOI] [PubMed] [Google Scholar]
- 20.Maillard O, Solyom S, Naegeli H. An aromatic sensor with aversion to damaged strands confers versatility to DNA repair. PLoS Biol. 2007;5:e79. doi: 10.1371/journal.pbio.0050079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 22.Rigas JR, Kelly K. Current treatment paradigms for locally advanced non-small cell lung cancer. J Thorac Oncol. 2007;2(Suppl 2):S77–S85. doi: 10.1097/01.JTO.0000269735.21209.bc. [DOI] [PubMed] [Google Scholar]
- 23.Juergens RA, Brahmer JR. Adjuvant therapy for resected non-small-cell lung cancer: past, present, and future. Curr Oncol Rep. 2005;7:248–254. doi: 10.1007/s11912-005-0046-5. [DOI] [PubMed] [Google Scholar]
- 24.Einhorn LH. Curing metastatic testicular cancer. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4592–4595. doi: 10.1073/pnas.072067999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armstrong DK, Bundy B, Wenzel L, Huang HQ, Baergen R, Lele S, Copeland LJ, Walker JL, Burger RA. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. New England Journal of Medicine. 2006;354:34–43. doi: 10.1056/NEJMoa052985. [DOI] [PubMed] [Google Scholar]
- 26.Teuben JM, Bauer C, Wang AHJ, Reedijk J. Solution structure of a DNA duplex containing a cis- diammineplatinum(II) 1,3-d(GTG) intrastrand cross-link, a major adduct in cells treated with the anticancer drug carboplatin. Biochemistry. 1999;38:12305–12312. doi: 10.1021/bi9904757. [DOI] [PubMed] [Google Scholar]
- 27.Liedert B, Pluim D, Schellens J, Thomale J. Adduct-specific monoclonal antibodies for the measurement of cisplatin-induced DNA lesions in individual cell nuclei. Nucleic Acids Res. 2006;34:e47. doi: 10.1093/nar/gkl051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dezhurov SV, Khodyreva SN, Plekhanova ES, Lavrik OI. A new highly efficient photoreactive analogue of dCTP. Synthesis, characterization, and application in photoaffinity modification of DNA binding proteins. Bioconjugate Chemistry. 2005;16:215–222. doi: 10.1021/bc0497867. [DOI] [PubMed] [Google Scholar]
- 29.Manley JL, Fire A, Cano A, Sharp P, Gefter ML. DNA-Dependent transcription of adenovirus genes is a soluble whole cell extract. Proc Natl Acad Sci U S A. 1980;77:3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park CJ, Choi BS. The protein shuffle. Sequential interactions among components of the human nucleotide excision repair pathway. FEBS J. 2006;273:1600–1608. doi: 10.1111/j.1742-4658.2006.05189.x. [DOI] [PubMed] [Google Scholar]
- 31.Lippard SJ, Bond PJ, WUKC, Bauer WR. Stereochemical requirements for intercalation of platinum complexes into double-stranded DNA’s. Science. 1976;194:726–728. doi: 10.1126/science.982037. [DOI] [PubMed] [Google Scholar]
- 32.Hermanson-Miller IL, Turchi JJ. Strand-specific binding of RPA and XPA to damaged duplex DNA. Biochemistry. 2002;41:2402–2408. doi: 10.1021/bi0112863. [DOI] [PubMed] [Google Scholar]
- 33.Maltseva EA, Rechkunova NI, Gillet LC, Petruseva IO, Scharer OD, Lavrik OI. Crosslinking of the NER damage recognition proteins XPC-HR23B, XPA and RPA to photoreactive probes that mimic DNA damages. Biochim Biophys Acta. 2007;1770:781–789. doi: 10.1016/j.bbagen.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 34.Khodyreva SN, Lavrik OI. Photoaffinity labeling technique for studying DNA replication and DNA repair. Current Medicinal Chemistry. 2005;12:641–655. doi: 10.2174/0929867053202179. [DOI] [PubMed] [Google Scholar]
- 35.Zamble DB, Mu D, Reardon JT, Sancar A, Lippard SJ. Repair of cisplatin--DNA adducts by the mammalian excision nuclease. Biochemistry. 1996;35:10004–10013. doi: 10.1021/bi960453+. [DOI] [PubMed] [Google Scholar]
- 36.Mu D, Park CH, Matsunaga T, Hsu DS, Reardon JT, Sancar A. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem. 1995;270:2415–2418. doi: 10.1074/jbc.270.6.2415. [DOI] [PubMed] [Google Scholar]
- 37.Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Tanaka K, Hanaoka F. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 38.Wang QE, Zhu QZ, Wani G, El-Mahdy MA, Li JY, Wani AA. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 2005;33:4023–4034. doi: 10.1093/nar/gki684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.