Abstract
The structural and functional role of conserved residue G86 in HIV-1 protease (PR) was investigated by NMR and crystallographic analyses of substitution mutations of glycine to alanine and serine (PRG86A and PRG86S). While PRG86S had undetectable catalytic activity, PRG86A exhibited ~6000-fold lower catalytic activity than PR. 1H-15N NMR correlation spectra revealed that PRG86A and PRG86S are dimeric, exhibiting dimer dissociation constants (Kd) of ~0.5 and ~3.2 μM, respectively, which are significantly lower than seen for PR with R87K mutation (Kd > 1 mM). Thus, the G86 mutants, despite being partially dimeric under the assay conditions, are defective in catalyzing substrate hydrolysis. NMR spectra revealed no changes in the chemical shifts even in the presence of excess substrate, indicating very poor binding of the substrate. Both NMR chemical shift data and crystal structures of PRG86A and PRG86S in the presence of active-site inhibitors indicated high structural similarity to previously described PR/inhibitor complexes, except for specific perturbations within the active site loop and around the mutation site. The crystal structures in the presence of the inhibitor showed that the region around residue 86 was connected to the active site by a conserved network of hydrogen bonds, and the two regions moved further apart in the mutants. Overall, in contrast to the role of R87 in contributing significantly to the dimer stability of PR, G86 is likely to play an important role in maintaining the correct geometry of the active site loop in the PR dimer for substrate binding and hydrolysis.
Keywords: NMR, X-ray, crystal, AIDS, retrovirus, protease, HIV-1
Introduction
Mature human immunodeficiency virus type 1 (HIV-1) protease (PR) is an aspartic protease that is responsible for processing the Gag and Gag-Pol polyproteins to yield mature proteins required for viral maturation.1 Due to its critical role, the protease has been a valuable target for structure-based drug-design.2–4 Although several PR inhibitors are in clinical use, selection of drug-resistant variants hampers the long-term effectiveness of these inhibitors.5 Thus, detailed structural analyses of HIV-1 PR mutants have been conducted in the presence of inhibitors or substrates and their analogues to understand how the resistant mutants lose susceptibility to inhibitor while maintaining their catalytic activity. 3,6–11 These structural observations, together with other experimental results, such as thermodynamic studies12,13, have successfully uncovered several molecular mechanisms contributing to drug resistance.
Mutational and structural studies of essential conserved residues have contributed, directly or indirectly, to the structure-based design of HIV-1 PR inhibitors. Amino acid residues that are essential for the PR structure and/or function are unlikely to be selected for drug-resistant mutations. Therefore, inhibitors designed to interact with such essential residues are expected to be more effective under drug pressure. Sequence alignment of retroviral proteases reveals three conserved regions in HIV-1 PR: the active site region, the flap region, and the region encompassing the single α-helix.14,15 The active site triad, D25-T/S26-G27, is shared by all aspartic acid proteases, whereas the second conserved triad G86-R87-D/N88 in the α-helix is found only in retroviral proteases. 14 The G86-R87-D/N88 triad is close to the active site loop and dimer interface but does not directly contact the substrate (Figure 1). Loeb and colleagues previously showed that mutation at N88 does not completely abolish the enzymatic activity of the PR16. Similarly, this mutation does not significantly alter the structure.6 The importance of the other conserved residues, such as those in the flaps and active site, has been described in terms of enzymatic activity,16,17 although the mechanisms are not well understood.
Figure 1.
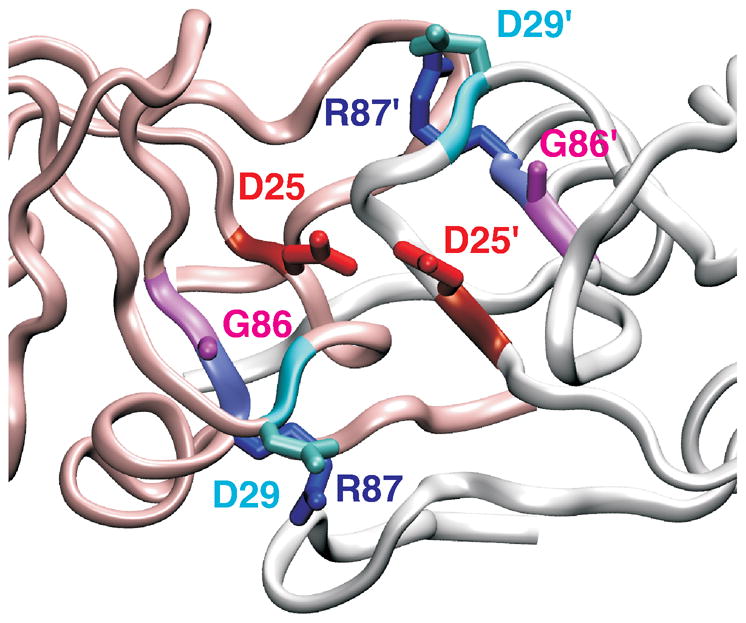
Ribbon structure of HIV-1 protease active site region. In the HIV-1 protease, active site D25 residues (red color) from the two identical subunits (white and pink ribbons) interact with each other. Residues R87 (blue) and G86 (pink) are conserved in retroviral proteases. Interaction of R87 with D29 (cyan) is important for dimer formation.18,19. PDB 1A8K41 was used to generate the figure.
We previously demonstrated that the R87 side chain is critical for the monomer-dimer equilibrium of the mature PR, and even a conservative mutation, R87K, drastically alters dimer formation.18,19 In the PR structures, the guanidiniumgroup of R87 forms a salt-bridge with the carboxylate side chain of D29 within one subunit, and hydrogen bonds with L5′ and W6′ in the other subunit.20,21 In solution, the intra-subunit salt-bridge between R87 and D29 was shown to be crucial for dimer stability of the mature PR; both the D29N and R87K mutants increased the dimer dissociation constant.19 Increased dimer dissociation is expected to diminish the catalytic activity, when activity is measured in the concentration range where enzyme dissociates, because the active site is formed by D25 and D25′, from each subunit of the dimer (Figure 1). Thus, the mutation of R87K increases dissociation of the dimer resulting in drastically reduced catalytic activity.
In this study, we investigated the role of the highly conserved G86 residue in PR structure and function (Figure 1). Substitutions G86A and G86S were introduced into PR to construct mutants, termed PRG86A and PRG86S. These mutants exhibited very low catalytic activity. Here, we show that NMR 1H-15N heteronuclear correlation spectra of PRG86A and PRG86S are very similar to that of PR, demonstrating that these mutants exist predominantly as dimers in solution with low μM Kds. Chemical shift analysis of these mutants in the presence of the inhibitor DMP323 indicates small but significant conformational perturbation in residues in the active site loop and the single α-helix that includes residue 86. Moreover, crystal structures of PRG86A and PRG86S in complex with darunavir (DRV) demonstrate high similarity to the corresponding PR complexes with rmsd differences for Cα atom positions of less than 0.5 Å. The largest structural changes were observed in PRG86S/DRV at residues 66 through 69, correlating with significant side chain rearrangements in the α-helix. These observations for the G86 residue are in contrast to those for the adjacent R87 residue, which contributes primarily to the monomer-dimer equilibrium of the PR.
Materials and Methods
Sample preparation
An HIV-1 protease construct containing five mutations that retard autoproteolysis and avoid cysteine-thiol oxidation (Q7K, L33I, L63I, C67A and C95A), termed PR, was used. The G86A and G86S mutations were introduced into the PR template to generate PRG86A and PRG86S, respectively, using the QuikChange mutagenesis protocol (Stratagene, La Jolla, CA). The PR mutants were expressed in Escherichia coli BL21(DE3) at 37°C in LB medium to yield unlabeled protein for enzymatic assays and crystallization studies, or in minimal medium containing 15NH4Cl or 15NH4Cl and D-(13C6)glucose for NMR studies. Proteins were isolated from inclusion bodies, purified by size-exclusion and reversed-phase high-pressure liquid chromatography, and folded as described previously.21
Protease Assay
Proteins were folded in the same manner as prepared for NMR experiments. Enzyme assays were conducted using only freshly prepared samples, at a maximum concentration of 3 μM, and with 460 μM chromogenic substrate IV, Lys-Ala-Arg-Val-Nle-(4-nitrophenylalanine)-Glu-Ala-Nle-NH2 (California Peptide Research, Napa, CA) in 50 mM sodium acetatebuffer, pH 5, at 25 °C, as described previously.18,19
NMR spectroscopy
NMR experiments were performed in 15 mM acetate buffer at pH 4.5 in 95% H2O/5%D2O and a sample volume of ~ 300 μl in a 5-mm Shigemi tube (Shigemi, Inc., Allison Park, PA) with protein concentrations of 4 μM to 0.3 mM for 15N-1H HSQC experiments and ca. 0.3 mM for the HNCA experiment (in dimer concentration). Experiments were conducted on Bruker DMX 500, Avance 600 MHz, and Avance 700 MHz spectrometers equipped with cryoprobes (Bruker Instruments, Billerica, MA).
15N-1H HSQC spectra of PRG86S and PRG86A were recorded in the absence and presence of excess DMP323 at 20 °C. Each HSQC spectrum was acquired with 16–80 scans, 400–512 complex at t1 increments and 1024 complex at t2 increments with ~2000 Hz F1 and ~8000 Hz F2 spectral widths, respectively. Kd was calculated using the volume ratios of the signals. Backbone signals of PRG86A and PRG86S in the presence of DMP323 were assigned by HNCA at 25 °C. NMR data were processed and analyzed using the nmrPipe 22 and Sparky (Goddard and Kneller, Univ. California, SF). Chemical shifts were compared with those of PR/DMP323 determined at pH 5.8.23 Differences between the chemical shifts were estimated to be significant only when they were larger than 0.05 ppm for 1H, 0.3 ppm for 13C, and 0.5 ppm for 15N, respectively.24
15N-1H HSQC spectra of the freshly prepared PRG86A were also recorded in the presence of substrate IV in 15 mM acetate buffer at pH 4.5. The spectra were recorded twice at 0.1 mM and 15 μM concentrations in the presence of 0.5 mM (5 fold excess) and 0.3 mM (20 fold excess) substrate, respectively.
Crystallographic analysis
Crystals were grown by the hanging drop vapor diffusion method. The protein sample was concentrated to about 5 mg/ml and the inhibitor, DRV 25 or DMP323,26 was dissolved in dimethyl sulfoxide (DMSO). Protein and inhibitor were preincubated on ice for one hour at a molar ratio of 1:5. Crystallization drops contained 1 μl protein and 1 μl of the reservoir solution of 10% sodium chloride and 0.1 M MES buffer at pH 6.5. Crystals of the minimal size required for synchrotron data collection (0.1 ×0.05 × 0.03 mm3) were obtained at 4 °C after 3 weeks.
X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beam line at the Advanced Photon Source, Argonne National Laboratory. Data were processed in the space group P21212 by HKL 2000.27 The structures were solved by molecular replacement with AmoRe 28 and refined by SHELX97,29 where the electron density map was refitted by using the computer graphic program O 8.0.30 Alternative conformations were modeled for the inhibitor and protease residues when they were observed in the electron density maps. The solvent was modeled with ~100 water molecules, some with partial occupancy. The PRG86A structure in complex with DRV was refined with a sodium cation, two chloride anions, and 113 water molecules, including partial occupancy sites. The PRG86S structure in complex with DRV was refined with a sodium cation, a chloride anion, and 132 water molecules, including partial occupancy sites. The PRG86A structure in complex with DMP323 was refined with 85 water molecules, including partial occupancy sites. The mutant structures of PRG86S and PRG86A in complex with DRV were compared with PR/DRV structure (PDB 2IEN). The structure of PRG86A in complex with DMP323 was not used for comparison since no structure was available of the equivalent PR (with five mutations) complex with DMP323. Structural figures were made using Bobscript and Weblab viewer (Molecular Simulations Inc.).
Results
Enzymatic activity of G86A/S mutants
Neither mutant protein sustained a stable fold for long periods in the absence of inhibitor at 20 °C, as indicated by the loss of backbone 1H-15N signals within ~12 h and ~24 h for PRG86A and PRG86S, respectively. In addition, as described below, consistent growth of crystals was obtained only with freshly folded samples in the presence of inhibitors. Thus, enzyme assays were conducted only using freshly folded samples and at a maximum concentration of 3 μM protein. Under these conditions, PRG86S showed no detectable activity, whereas PRG86A exhibited at least 6000-fold reduced activity relative to PR (Table 1). The loss of activity of PR bearing these mutations was more substantial than described previously for PRR87K and PRD29N mutants that showed reductions of ~4600 fold and 920 fold, respectively18,19. It was impossible, however, to determine the detailed kinetic parameters due to the extremely low activity of the G86 mutants.
Table 1.
Approximate dimer dissociation constants (Kd) and enzymatic activity of G86 mutants of PR.
Dimer of the PRG86A mutant in solution
Since mutations at residues D29 or R87 significantly increased the dimer dissociation constant (Kd) of the mature PR,18,19 it is conceivable that substitution mutations of residue 86 also perturb the monomer-dimer equilibrium. Thus, NMR experiments were performed to elucidate the role of the conserved G86 residue on dimer formation and to characterize the conformation in solution.
We previously demonstrated that the 1H-15N HSQC spectrum of the mature PR exhibits characteristic changes in chemical shifts upon dimer dissociation.18,31 Particularly, signals at the dimer interface, such as residues I3, I93, and A95, change their chemical shifts significantly. At 20 μM protein concentration, 1H-15N HSQC spectrum of the freshly prepared PRG86A in the absence of inhibitors showed peaks in positions characteristic for the PR dimer32,33 (Figure 2A). Since PRG86A in the absence of inhibitor precipitated within a day, experiments to assign signals could not be performed for the free protein.
Figure 2.
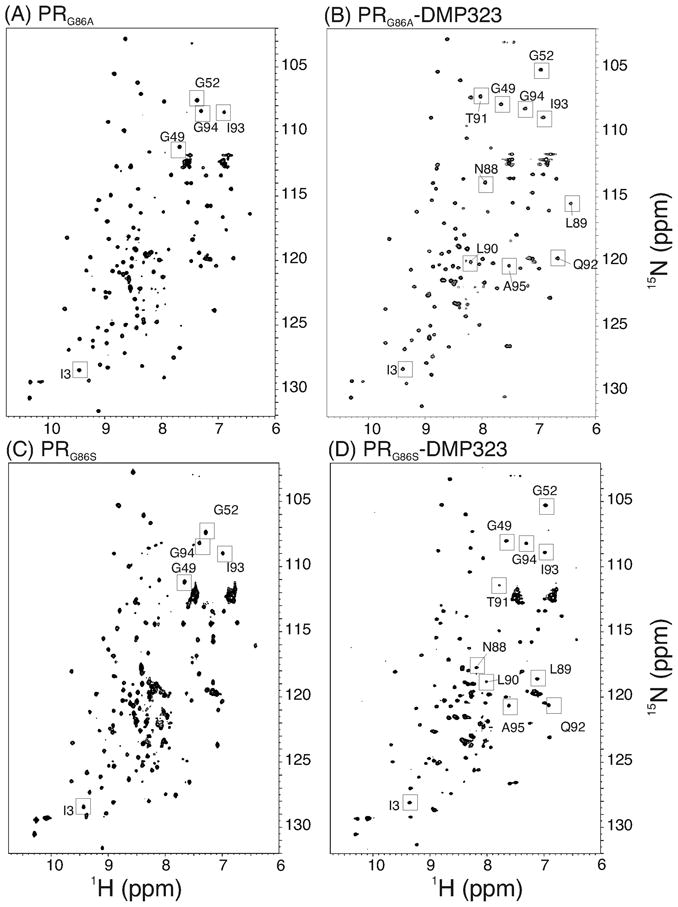
15N-1H HSQC spectra of PRG86A (A) in the absence and (B) in the presence of DMP323, and spectra of PRG86S (C) in the absence and (D) in the presence of DMP323. Signal assignments were conducted in (B) and (D). Labeling signals are based on similarity of peak positions in (A) and (C). Characteristic peaks for dimers are shown by squares.
An overview of the PRG86A HSQC spectrum qualitatively indicated that the protein is mostly folded as a dimer in solution. Based on the protein concentration used in the HSQC experiment (20 μM as dimer) and on the signal-to-noise ratio of the spectrum, the upper limit Kd was estimated to be < 1 μM. This value was further confirmed by recording the HSQC spectrum at 4 and 7 μM concentrations, at which small peaks consistent with the monomer signals in PR mutants were observed (supplementary material Figure S1).19,34 Based on the ratios of the signal intensities, Kd was semi-quantitatively estimated to be ~0.5 μM, which is close to the value for PRD25N (Table 1). Addition of DMP323 (Figure 2B) resulted in changes in chemical shifts of only a small set of signals. These chemical shift changes were similar to those observed for PR dimer with DMP32335, demonstrating that the PRG86A in the absence of the inhibitor is predominantly a dimer.
To understand the low catalytic activity of PRG86A, an 1H-15N HSQC spectrum of the PRG86A was recorded in the presence of excess substrate IV, and compared with the substrate free spectrum in Figure 2A. Experiments repeated twice at two different protein concentrations with 5–20 fold excess substrate showed no significant changes in the NMR spectra (Supplemental material). This result indicated that the substrate-bound enzyme fraction was less than noise-level even when the complex was present, with presumably a mM substrate dissociation constant.
NMR studies reveal local structural perturbations caused by G86A mutation
To gain more insight into chemical shift perturbation, backbone chemical shifts (Cα, N, and HN) of PRG86A in the presence of DMP323 were assigned and compared with those of PR.18,35 Due to the limited stability and sensitivity of the NMR sample, DRV could not be used because, as an asymmetric inhibitor, it splits the degenerated dimer signals and, therefore, reduces the apparent intensity of the signal by half. For the same reason, NMR experiments to characterize protein backbone dynamics could not be performed. As shown in Figure 3A, differences in backbone chemical shifts of PRG86A/DMP323 relative to those of PR/DMP323 were small except for residues from 28 to 32 and R87, which is adjacent to the mutation site. There was no significant change in chemical shifts in the flap region between PRG86A and PR in the presence of DMP323 (Figure 3A), indicating that flaps of the PRG86A/DMP323 were closed and similar to those of PR/DMP323. Chemical shift changes of the terminal residues, 1 to 4 and 96 to 99, were insignificant, indicating that PRG86A in the presence of DMP323 forms an inter-subunit 4-stranded β-sheet similar to that of the PR/DMP323 complex. The insignificant changes in chemical shifts in the terminal regions were distinct from chemical shift differences between PR and PR monomer mutants studied previously.18
Figure 3.
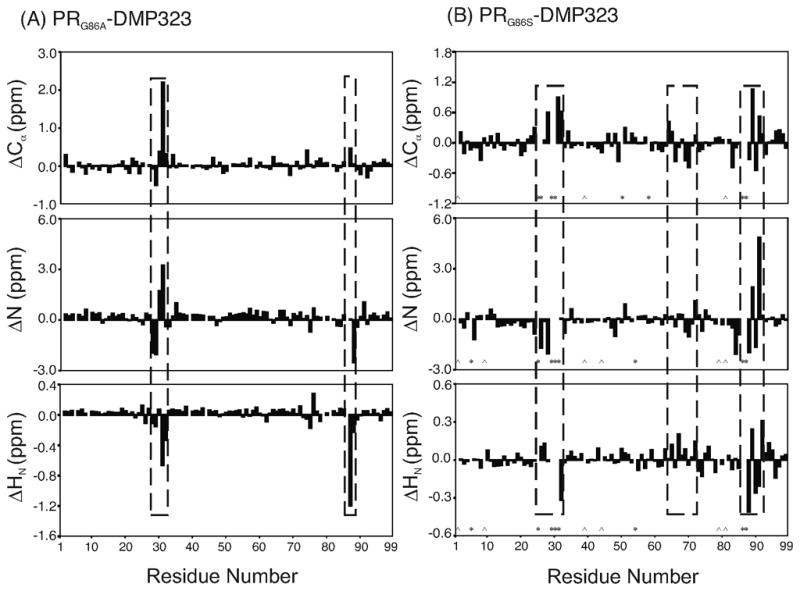
Differences in backbone Cα, N, and HN chemical shifts (A) between PR and PRG86A and (B) PR and PRG86S in the presence of DMP323. In (B), the residues whose backbone signals were not assigned are marked by *, and proline residues positions are marked by ^. In both mutants, chemical shift perturbation is observed in the active site loop as well as around residue 86.
We postulate that local conformational changes transmitted from residue 86 to the catalytic D25 affect substrate binding in PRG86A. Among the local changes, up-field shift of more than 1 ppm was observed for the amide proton of the R87 residue in the PRG86A HSQC spectrum compared to that of PR (Figure 3A). Typically, mutations cause changes in the magnetic environment of adjacent residues. However, mutation of glycine to alanine does not introduce changes in electrostatic charge, therefore, such a significant shift (>1 ppm) is likely due to changes in the secondary structure or hydrogen bonding.36 Of the two possible interpretations, secondary structure changes of the mutation site are unlikely because neighboring residues in the active site loop exhibit no significant chemical shift perturbation. Instead, a change in a salt bridge or hydrogen bond may explain this observation, which is consistent with distance changes observed in the crystal structures as described below. It is plausible that the large change in R87 backbone amide chemical shift is caused by loss of the salt bridge, and as a result, the active site is more flexible or open in the G86A mutant in complex with DMP323.
G86S mutation causes more pronounced effects than G86A mutation in solution
In contrast to the PRG86A spectrum, the HSQC spectrum of PRG86S in the absence of an inhibitor showed distinct differences from that of PR (squared regions in Figure 2C). Since the protein is not stable over extended periods, backbone chemical shifts of the free PRG86S could not be assigned. In the HSQC spectrum, peaks for residues in the α-helix (Q92, I93, and G94) in the dimer 18,19 were not observed in the same positions as those in the free form of PRG86S, suggesting that the G86 mutation may induce long-range structural changes. Nevertheless, since the PRG86S formed a homodimer in the presence of DMP323 (described below) with changes in only a small number of signal positions, PRG86S most likely exists as a dimer even in the absence of inhibitor (Figure 2D). Consistent with these observations, the PRG86S HSQC spectrum recorded at a lower concentration, 4 μM (as dimer concentration), showed additional peaks that are similar to those appearing in the low concentration spectrum of the PRG86A (Supplemental material). Assuming that the additional signals are due to the monomer fraction, the Kd of PRG86S was semi-quantitatively estimated to be 1.7 μM based on the ratio of signal intensities of the spectrum (Table 1).
To understand the chemical shift perturbation caused by G86S mutation in more detail, backbone chemical shifts of PRG86S in the presence of DMP323 were assigned, and compared with those of PR (Figure 3B). Many residues in these spectra showed almost identical chemical shifts. In particular, there were no significant changes in chemical shifts for the residues in the N- and C-termini that contribute to the dimer interface, demonstrating that the PRG86S forms a homodimer in the presence of DMP323. Perturbations of the chemical shifts were observed in the entire α-helical region (residues 86 to 93) and in the region from residues 64 to 72 in PRG86S/DMP323 as well as around the mutation site, indicating more pronounced conformational changes with the G86S mutation compared to that of G86A.
The PRG86S/DMP323 spectrum showed another difference from the PRG86A/DMP323; several signals could not be observed in the HSQC spectrum for D25, D29, T31, S86, and R87, most likely due to chemical exchange broadening. Chemical exchange broadening is observed when a site undergoes exchange of two (or more) chemical shifts in milli-microsecond time scale, presumably due to (1) conformational exchange or (2) exchange of chemical shifts between inhibitor free and the bound forms. The flap region exhibited the largest chemical shift changes upon DMP323 binding to PR (the amide 15N and 1H signals change 0 – 4 ppm and 0 – 1.7 ppm, respectively 32,35). Since no signal broadening was observed in the flap region, diminished signals are not likely to be due to exchange of chemical shifts between inhibitor free and bound forms, but rather to conformational flexibility. Although further NMR relaxation experiments will help to answer this question, it was impossible to obtain quantitative data for the conformational flexibility of the G86 mutants due to the short life-time of the sample.
Crystallographic studies of the inhibitor complexes of G86S and G86S mutants
The crystal structures of PRG86A and PRG86S in complex with DRV (Figure 4A), and PRG86A in complex with DMP323 were determined at high resolution. No diffraction quality crystals were obtained for PRG86S in complex with DMP323. As the folded G86 mutant proteins tend to aggregate rapidly, the crystallization studies were conducted only using freshly refolded protein. The three complexes were crystallized in the same space group (P21212), and diffracted to 1.6–1.8 Å resolution. Crystallographic statistics are summarized in Table 2. The electron density for inhibitor and protein is clear, as shown for the mutated Ser86 side chain in PRG86S/DRV (Figure 4B). The protein formed a dimer in the crystal structure with inhibitor bound in the active site cavity. In the PRG86A/DRV and PRG86S/DRV complexes, the asymmetric inhibitor was observed in two alternate conformations in the active site cavity. DMP323 is a symmetric inhibitor and only a single conformation was observed in the dimer structure. The structures were refined with anisotropic B-factors giving final R-factors in the range of 18.6–22%. These R-factors are slightly larger than values for other PR mutant structures determined previously, probably due to the small size and weak diffraction of the crystals.37,38 Two alternate conformations were observed for the side chains of 13 residues in the PRG86S/DRV structure, 19 residues in PRG86A/DRV and 6 residues in PRG86A/DMP. The average atomic B-factors were 19.1–23.4 Å2 for the main chain and 23.4–31.5 Å2 for the side chain atoms.
Figure 4.
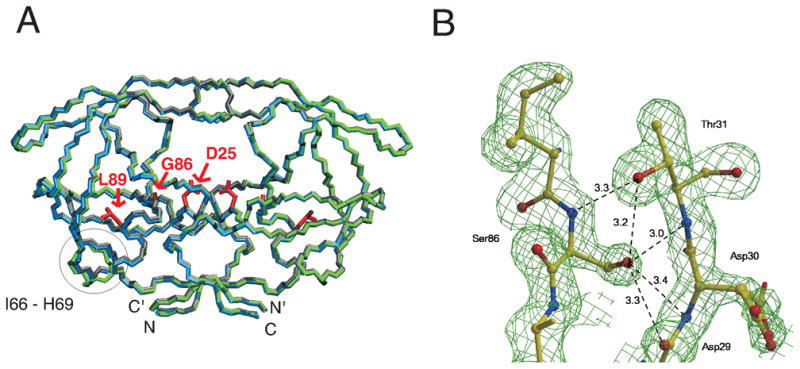
(A) Comparison of backbone traces of the crystal structures of PR (PDB 2IEN 40, gray with atoms colored by type), PRG86A (blue), and PRG86S (green) in complexes with DRV and (B) Electron density map for region around S86 in crystal structure of PRG86S/DRV. In (A), side chains of residue 25, 86, and 89 are shown in red, and a circle indicates a region (residues 66 to 69) that exhibits the largest Cα rmsd for mutants and PR. In (B), the map is contoured at a level of 2.0 σ, showing side chain conformation of S86 clearly. The hydrogen bond interactions of S86 are indicated by broken lines with distances in Å.
Table 2.
Crystallographic Data Collection and Refinement Statistics
| Complexes | G86A-DMP | G86A-DRV | G86S-DRV |
|---|---|---|---|
| Space group | P21212 | P21212 | P21212 |
| Unit cell dimensions: (Å) | |||
| a | 58.07 | 58.33 | 58.36 |
| b | 86.36 | 86.43 | 86.15 |
| c | 45.59 | 46.20 | 46.37 |
| Resolution range (Å) | 50-1.8 | 50-1.6 | 50-1.8 |
| Unique reflections | 22278 | 31701 | 22187 |
| Rmerge (%) overall (final shell) | 9.5 (25.4) | 13.2 (47.4) | 11.8 (35.2) |
| I/σ overall (final shell) | 14.0 (3) | 10.7 (3) | 9.6 (3) |
| Completeness (%) overall (final shell) | 89.5 (53.2) | 95.4 (87.3) | 98.4 (94.8) |
| Data range for refinement (Å) | 10-1.8 | 10-1.6 | 10-1.8 |
| R (%) | 22.0 | 18.6 | 22.3 |
| Rfree (%) | 28.9 | 25.6 | 27.2 |
| No. of solvent atoms (total occupancies) | 68 | 102.8 | 130 |
| RMS deviation from ideality | |||
| Bonds (Å) | 0.006 | 0.009 | 0.005 |
| Angle distance (Å) | 0.023 | 0.029 | 0.023 |
| Average B-factors (Å2) | |||
| Main-chain atoms | 23.4 | 20.5 | 19.1 |
| Side-chain atoms | 31.5 | 27.2 | 23.4 |
| Inhibitor | 25.5 | 19.5 | 17.4 |
| Solvent | 29.9 | 33.0 | 29.0 |
| Occupancies for alternate conformations of darunavir (%) | 100 | 67/33 | 60/40 |
The overall backbone structures of PRG86S/DRV and PRG86A/DRV were almost identical to that of PR/DRV with rmsd (Cα) values of 0.3 Å and 0.5 Å, respectively. The biggest shifts of 1.7 Å and 1.5 Å occurred at residues 67 and 68, respectively. Earlier comparison of 73 HIV-1 protease crystal structures showed that the most flexible part of the protein is residues 37–42 (rmsd >1 Å) and the other two regions with larger variation are residues 79–83 and region 65–72 (rmsd about 0.75 Å).39 Much of this variation of surface residues arises from crystal structures in different space groups with different intermolecular packing. Here, the G86 mutant crystal structures are in the same space group as PR/DRV, so changes of 1.5–1.7 Å at residues 67–68 are significant and exceed the normal variation observed in protease structures in a range of space groups.
Interactions with DRV
DRV was found to bind in two orientations with occupancy of 0.7/0.3 and 0.6/0.4 to PRG86A and PRG86S, respectively. Similar to the PR/DRV complex, the inhibitor bound in subsites S2 to S2′of PRG86A and PRG86S, and formed van der Waals interactions with protease residues L23, D25/25′, G27/27′, A28/28′, D29/29′, D30/30′, V32, G48/48′, G49/49′, I50′, P81, V82/82′, and I84/84′. (The residues in the two subunits of the PR dimer are labeled 1 to 99 and 1′ to 99′). The two oxygen atoms of the bis-THF group of DRV formed hydrogen bond interactions with the main chain amides of D29′ and D30′ and with the carboxylate oxygen of D29′, similar to those in the wild type PR/DRV structure.40 However, the interaction of the aniline nitrogen of DRV with the side chain of D30 was lost in one subunit of the PRG86A complex due to rotation of the D30 side chain away from the inhibitor, while the bis-THF ring gained extra hydrogen bonds with one conformation of the side chain of D30′ in the other subunit.
The catalytic residues D25 and D25′ showed small changes in their interactions consistent with the shift in position of residues 29–31 (Figure 5A). The relative positions of the Cα atom in residue 86 and the D25 Cα atom in PRG86A/DRV and PRG86S/DRV became slightly shorter (0.2 Å) than that in PR/DRV. Also, the distance between the two OD1 inner carboxylate oxygen atoms of the catalytic D25 and D25′ was shortened to 2.5–2.6 Å in the PRG86A/DRV and PRG86S/DRV structures, instead of 2.9 Å in the wild type complex (Figure 5A). Therefore, the structural changes due to the mutations propagate to the catalytic residues.
Figure 5.
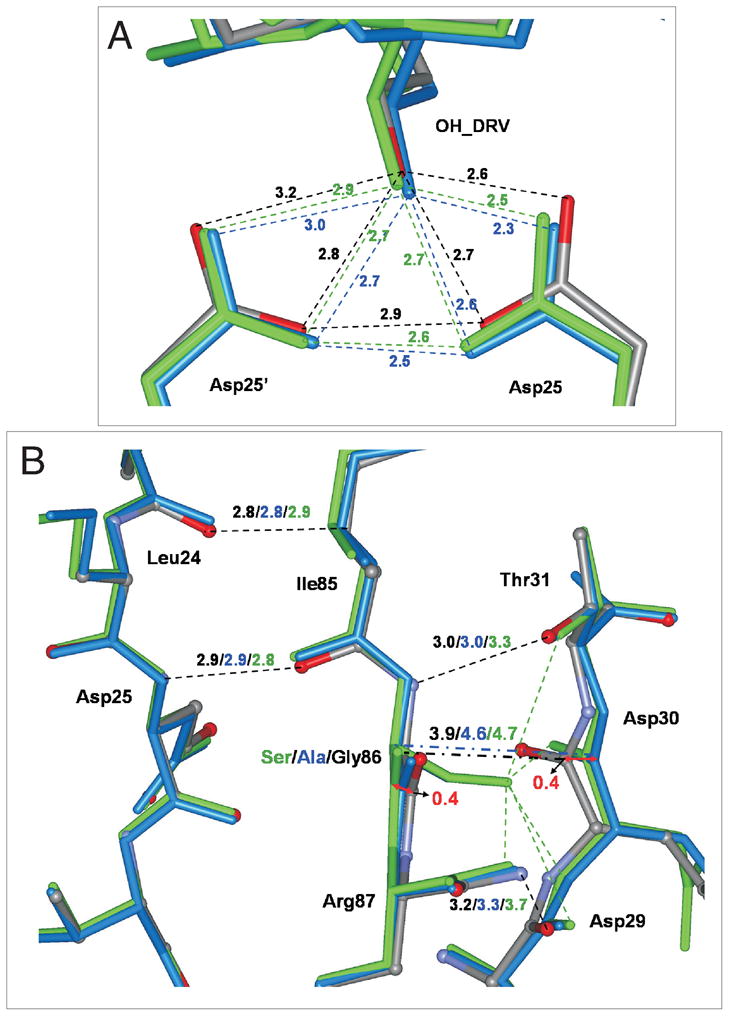
(A) The catalytic sites and (B) interactions of residue 86 with the active site loop in crystal structures of PR/DRV(gray with atoms colored by type), PRG86A/DRV (blue), and PRG86S/DRV (green). Hydrogen bonds are indicated by dashed lines with the distances in Å in the same colors as the structures. The hydrogen bond distances in the catalytic site of PRG86S/A are shorter than those of PR. A network of hydrogen bond interactions connects the main chains of L24, D25, D29, D30 and I85-R87. The main chain atoms of 86 and D30 are 0.8 Å further apart in the PRG86S/A relative to PR structure, as indicated by broken lines between the alpha carbon atom of 86 and the carbonyl oxygen of D30.
Residue 86 is connected to the active site and dimer interface
The region around residue 86 forms a conserved network of connections with the active site loop comprising residues 25–32, as shown in Figure 5B. The amide of residue 86 forms a hydrogen bond with the hydroxyl side chain of T31, while the adjacent main chain carbonyl oxygen of I85 interacts with the amide of D25 in both subunits of the wild type and mutant complexes. The main chain-main chain interactions of I85 with D25 and L24 are maintained in all the complexes. The adjacent R87 residue also forms several intersubunit interactions (Figure 6A): the guanidiniumgroup of R87 forms intersubunit hydrogen bonds with the main chain carbonyl oxygens of L5′ and W6′ on one side, while on the other side R87 forms a complex salt bridge with the side chains of D29 and R8′.20 The R8′ and R8 side chains were observed in alternate conformations, although one conformation always formed the intersubunit salt bridge with D29.
Figure 6.
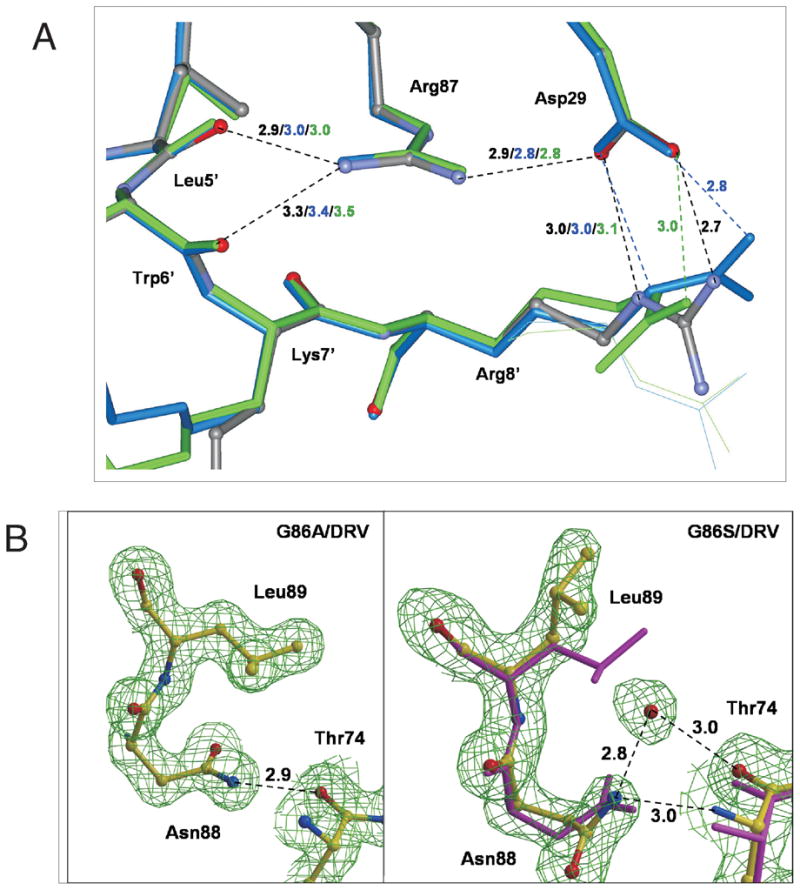
(A) Intersubunit interactions around R87 in the PR/DRV (gray with atoms colored by type), PRG86A/DRV (blue), and PRG86S/DRV (green) structures, and (B) side chain conformation in PRG86A/DRV (left) and PRG86S/DRV (right). In (A), minor conformations of side chains are shown as thin lines, and hydrogen bonds are indicated by broken lines with the distances in Å in the same colors as the structures. In (B), the atoms are shown with yellow bonds for both of PRG86S/DRV and PRG86A/DRV, and with magenta for PR/DRV. In PRG86S/DRV, a water molecule is observed next to the L89 side chain and interacting with N88 and T74. The maps are contoured at a level of 1.8 σ.
While the overall interaction was well maintained, the interactions of residues 86–87 with active site loop residues 29–31 showed small differences in the mutants. In the G86 mutants the distance between the Cα atoms of 86 and D30 increased to 4.6 Å from 3.9 Å in the wild type structure (Figure 5B). The G86S mutation produced larger structural changes: the β-sheet-like interaction of the amide of N88 with the carbonyl oxygen of D29 was greatly weakened by elongation to 3.7 Å, and a slight elongation of 0.3 Å appeared for the hydrogen bond between the amide of S86 and hydroxyl side chain of T31. Consistent with these observations, the NMR data described above showed the chemical shift perturbation of the active site loop by the G86 mutations (Figure 3).
Figure 5B clearly indicates that the entire helix was slightly pushed away from the active site loop region. Cα - Cα distances between G86 - D29 and G86′ - D29′ were observed to be 5.3 Å and 5.2 Å, respectively in PR/DRV. In contrast, these distances increased to 5.5 Å and 5.6 Å in PRG86A/DRV, and 5.7 Å in PRG86S/DRV. Although these distance changes (< 0.5 Å) are relatively small, the slight shift of the helix position is not surprising because the glycine side chain of residue 86 was changed to a more bulky side chain in PRG86A/DRV and PRG86S/DRV.
Overall, small but consistent changes of about 0.8 Å were observed in main chain inter-atom distances between the active site loop around residues D29–D30 and the α-helix of the PRG86A and PRG86S mutant relative to PR (Figure 5). This movement pushes the α-helix away from the active site region, which is accompanied by smaller changes of ~0.3 Å that reduce the separation of the catalytic D25/25′ side chains.
Significant side chain rearrangements observed in crystal structure PRG86S/DRV
In addition to the small changes in active site conformation, the β-turn from residue 66 to 69 exhibited large changes in Cα positions in PRG86S/DRV (rmsd > 1 Å, Figure 4A) compared to PR/DRV, with a cascade of rearrangements in the side chain conformations as clearly seen in the electron density map (Figure 6B). The bulky S86 side chain shifted the position of the N88 side chain, which was accompanied by a rotation of about 180 degree of the χ1 angle of L89 maintaining the hydrophobic interaction between L89 and I66. This rotation of the side chain of L89 pushed the I66 side chain towards the surface of the molecule such that the I66 Cδ1 position was shifted with 123.4 °degree χ1 angle and3.1 Å. As a result, the β-turn between residues 66 to 69 was twisted towards the solvent. An oxygen atom of a water molecule was observed at a position that is 2.7 Å from N88 Nδ2 atom, 1.8 Å from L89 Cγ atom, and 3.1 Å from T74 Oγ atom.
These significant changes in side chain orientations at the α-helix (residues 86 to 93) and the β-turn region (that includes residues 66–69) were observed only in one of the two subunits of PRG86S/DRV, although DRV is bound in two alternative conformations. Therefore, the side chain rearrangements are not related to the relative orientation of DRV, but instead more likely due to structural flexibility or structural heterogeneity caused by the mutation. Such rearrangements were not observed in PRG86A/DRV, suggesting that more drastic structural perturbations in PRG86S/DRV compared to PRG86A/DRV are due to the larger size of the S86 side chain and the formation of new hydrogen bonds of the S86 hydroxyl group (Figure 4B). As described above, NMR data also showed more pronounced changes in the conformation and possible flexibility of PRG86S than seen in PRG86A in the presence of DMP323. Both NMR and crystal studies suggest increased flexibility of the β-turn region at residues 66–69 in PRG86S in the presence of the active site inhibitors.
Discussion
The aim of this study was to understand the role of the G86 residue in the highly conserved triad G86-R87-N/D88 of the mature HIV-1 PR in dimer formation and catalytic activity. Although mutations of G86 were known to significantly impair catalytic activity 16, the structural basis for this loss in activity was not elucidated. In fact, the very low catalytic activity of the G86 mutants does not permit measuring either the detailed kinetic parameters or the Kd of the mutant by monitoring enzymatic activity as a function of protein concentration. Moreover, structural studies have been limited by the short life-time observed for G86 mutants: only one HSQC spectrum could be recorded for each freshly prepared NMR sample of the G86 mutants in the absence of inhibitor, and small crystals grew only from freshly prepared protein. Therefore, our structural studies of the PRG86A and PRG86S mutants applied the complementary methodologies of crystallography as well as NMR solution studies in the presence and the absence of the active-site inhibitors.
NMR was used to semi-quantitatively measure the Kds of G86 mutants, which were found to be more than 102 fold higher than that of the wild-type PR,22 but similar to the Kd determined for PRD25N (Table 1). The D25N mutant is inactive, being the catalytic residue, and is predominantly a dimer at 20 μM concentration as seen by NMR. This active-site mutant D25N has been used extensively to study the protease-substrate complex by solution NMR and crystallography.7,32,34 In contrast to the G86 mutants, substitution of the adjacent conserved Arg residue (PRR87K) results in ~105 fold higher Kd than that of the wild-type PR and reduced catalytic activity mainly because of dimer dissociation.18,19 Thus, although both PRR87K and the G86 mutants share extremely low catalytic activities, the considerable differences in Kd values indicate that the reduced catalytic activity of G86 mutants occurs through a mechanism other than increased dimer dissociation.
Although the G86 mutants exhibit significant dimerization at 0.5 – 3 μM concentrations, they exhibit very poor catalytic activity similar to the PRR87K mutant, which is mainly monomeric under similar assay conditions. NMR HSQC spectra analyzed at concentrations where the G86 mutants are mostly dimeric also showed no evidence of substrate interaction. The absence of changes in the HSQC spectrum contrasts with our previous NMR studies of PRD25N in which significant chemical shift changes were observed in the presence of substrate IV with a substrate dissociation constant, KS, of 0.27 mM.33 Overall, the current NMR data suggest that the Km for substrate binding to PRG86A dimer is high, explaining the poor catalytic activity.
Key features to understand the conformational differences between these G86 mutants and PR were obtained by comparing inhibitor bound forms by NMR and X-ray crystallography. Overall, the crystal structures for PRG86A and PRG86S in complex with DRV were almost identical to the wild type complex with rmsd (Cα) values of 0.3 Å and 0.5 Å (Figure 4A), in agreement with mostly similar NMR chemical shifts for the mutants and PR in the presence of DMP323 (Figure 3). Despite the overall similarities, small but consistent shifts were observed in main chain inter-atom distances between the active site residues 24–31 and the adjacent α-helix in the crystal structures of PRG86A and PRG86S mutants relative to PR with DRV (Figure 5). These shifts reached a maximum of ~0.8 Å between residue 86 and D29–D30. This movement pushes the α-helix away from the active site region, and is accompanied by smaller changes of ~0.3 Å reducing the separation of the catalytic D25/25′ side chains, which will tend to reduce the size of the active site cavity.
Interestingly, changes in the NMR chemical shifts (more than 0.5 ppm shifts for Cα) in the G86 mutants in the presence of DMP323 were observed in the same regions that showed the differences in the crystal structures (Figure 3). In the previous study comparing the chemical shifts of PRR87K to PR in the presence of DMP323, such specific changes of chemical shifts in the active site residues and the α-helix region were not observed.19 Current NMR data demonstrate more pronounced differences in the interaction of the G86 mutants with active-site inhibitors than seen for the R87K mutant. Overall, both the methodologies of NMR and X-ray crystallography in different sample conditions consistently demonstrate that the mutants have conformational changes connecting residue 86 to the active site region.
In contrast to the R87K mutation that reduces the catalytic activity by increasing the dimer dissociation constant18, the G86 mutation has a different mechanism to reduce the catalytic activity without severely increasing Kd; mutation of G86 causes a decrease in the dimer interaction with substrate. In conclusion, based on our observation that the G86 mutants have correlated conformational changes in the G86 site and the active site region when bound to active-site inhibitors, we propose that the altered conformation or dynamics of the active site region influence the substrate binding.
Supplementary Material
Acknowledgments
This study was supported by University of Pittsburgh, in part by the Intramural Research program of the NIDDK, NIH, and by the Georgia State University Molecular Basis of Disease Program, the Georgia Research Alliance, the Georgia Cancer Coalition and the National Institutes of Health grant GM062920. We also thank the staff at the SER-CAT beamline at the Advanced Photon Source, Argonne National Laboratory, for assistance during X-ray data collection. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38.
Abbreviations
- PR
protease construct containing the five other mutations (Q7K, L33I, L63I, C67A and C95A)
- DRV
darunavir
- HSQC
heteronuclear single quantum coherence
- rmsd
root-mean-square deviation
- Kd
dimer dissociation constant
References
- 1.Oroszlan S, Luftig RB. Retroviral Proteinases. Curr Topics in Microbial and Immu. 1990;157:153–185. doi: 10.1007/978-3-642-75218-6_6. [DOI] [PubMed] [Google Scholar]
- 2.Erickson JW, Burt SK. Structural mechanisms of HIV drug resistance. Ann Rev Pharm and Toxic. 1996;36:545–571. doi: 10.1146/annurev.pa.36.040196.002553. [DOI] [PubMed] [Google Scholar]
- 3.Vondrasek J, Buskirk CP, Wlodawer A. Database of three-dimensional structures of HIV proteinases. Nat Struct Biol. 1997;4:8. doi: 10.1038/nsb0197-8. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn P, Wilson K, Patch MG, Stevens RC. The genesis of high-throughput structure-based drug discovery using protein crystallography. Curr Opin Chem Biol. 2002;6:704–710. doi: 10.1016/s1367-5931(02)00361-7. [DOI] [PubMed] [Google Scholar]
- 5.Condra JH. Resistance to HIV protease inhibitors. Haemophilia. 1998;4:610–615. doi: 10.1046/j.1365-2516.1998.440610.x. [DOI] [PubMed] [Google Scholar]
- 6.Mahalingam B, Louis JM, Hung J, Harrison RW, Weber IT. Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes. Proteins. 2001;43:455–464. doi: 10.1002/prot.1057. [DOI] [PubMed] [Google Scholar]
- 7.Prabu-Jeyabalan M, Nalivaika E, Schiffer CA. How does a symmetric dimer recognize an asymmetric substrate? A substrate complex of HIV-1 protease. J Mol Biol. 2000;301:1207–1220. doi: 10.1006/jmbi.2000.4018. [DOI] [PubMed] [Google Scholar]
- 8.Tie Y, Boross PI, Wang YF, Gaddis L, Liu F, Chen X, Tozser J, Harrison RW, Weber IT. Molecular basis for substrate recognition and drug resistance from 1.1 to to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs. FEBS J. 2005;272:5265–5277. doi: 10.1111/j.1742-4658.2005.04923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prasanna MD, Vondrasek J, Wlodawer A, Bhat TN. Application of InChI to curate, index, and query 3-D structures. Proteins. 2005;60:1–4. doi: 10.1002/prot.20469. [DOI] [PubMed] [Google Scholar]
- 10.Kumar M, Prashar V, Mahale S, Hosur MV. Observation of a Tetrahedral Reaction Intermediate in the HIV-1 Protease-Substrate Complex. Biochem J. 2005;389:365–371. doi: 10.1042/BJ20041804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovalevsky AY, Chumanevich AA, Liu F, Weber IT. Caught in the Act: 1.5 Å Resolution Crystal Structures of the HIV-1 Protease and the I54V Mutant Reveal a Tetrahedral Reaction Intermediate. Biochemistry. 2007;46:14854–14864. doi: 10.1021/bi700822g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velazquez-Campoy A, Vega S, Freire E. Amplification of the effects of drug resistance mutations by background polymorphisms in HIV-1 protease from African subtypes. Biochemistry. 2002;41:8613–8619. doi: 10.1021/bi020160i. [DOI] [PubMed] [Google Scholar]
- 13.Ohtaka H, Freire E. Adaptive inhibitors of the HIV-1 protease. Prog Biophys Mole Biol. 2005;88:193–208. doi: 10.1016/j.pbiomolbio.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Pearl LH, Taylor WR. A structural model for the retroviral proteases. Nature. 1987;329:351–354. doi: 10.1038/329351a0. [DOI] [PubMed] [Google Scholar]
- 15.Rao JKM, Erickson JW, Wlodawer A. Stuctural and Evolutionary Relationships between Retroviral an Eucaryotic Aspartic Proteinases. Biochemistry. 1991;30:4663–4671. doi: 10.1021/bi00233a005. [DOI] [PubMed] [Google Scholar]
- 16.Loeb DD, Swanstrom R, Everitt L, Manchester M, Stamper SE, Hutchison CA., 3rd Complete mutagenesis of the HIV-1 protease. Nature. 1989;340:397–400. doi: 10.1038/340397a0. [DOI] [PubMed] [Google Scholar]
- 17.Shao W, Everitt L, Manchester M, Loeb DD, Hutchison CA, 3rd, Swanstrom R. Sequence requirements of the HIV-1 protease flap region determined by saturation mutagenesis and kinetic analysis of flap mutants. Proc Natl Acad Sci U S A. 1997;94:2243–2248. doi: 10.1073/pnas.94.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishima R, Ghirlando R, Tozser J, Gronenborn AM, Torchia DA, Louis JM. Folded monomer of HIV-1 protease. J Biol Chem. 2001;276:49110–49116. doi: 10.1074/jbc.M108136200. [DOI] [PubMed] [Google Scholar]
- 19.Louis JM, Ishima R, Nesheiwat I, Pannell LK, Lynch SM, Torchia DA, Gronenborn AM. Revisiting monomeric HIV-1 protease. Characterization and redesign for improved properties. J Biol Chem. 2003;278:6085–6092. doi: 10.1074/jbc.M209726200. [DOI] [PubMed] [Google Scholar]
- 20.Weber IT. Comparison of the crystal structures and intersubunit interactions of human immunodeficiency and Rous sarcoma virus proteases. J Biol Chem. 1990;265:10492–10496. [PubMed] [Google Scholar]
- 21.Ishima R, Torchia DA, Louis JM. Mutational and structural studies aimed at characterizing the monomer of HIV-1 protease and its precursor. J Biol Chem. 2007;282:17190–17199. doi: 10.1074/jbc.M701304200. [DOI] [PubMed] [Google Scholar]
- 22.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 23.Ishima R, Louis JM, Torchia DA. Characterization of two hydrophobic methyl clusters in HIV-1 protease by NMR spin relaxation in solution. J Mol Biol. 2001;305:515–521. doi: 10.1006/jmbi.2000.4321. [DOI] [PubMed] [Google Scholar]
- 24.Ishima R, Louis JM. A diverse view of protein dynamics from NMR studies of HIV-1 protease flaps. Proteins. 2008;70:1408–1415. doi: 10.1002/prot.21632. [DOI] [PubMed] [Google Scholar]
- 25.Koh Y, Nakata H, Maeda K, Ogata H, Bilcer G, Devasamudram T, Kincaid JF, Boross P, Wang YF, Tie Y, Volarath P, Gaddis L, Harrison RW, Weber IT, Ghosh AK, Mitsuya H. Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob Agents Chemother. 2003;47:3123–3129. doi: 10.1128/AAC.47.10.3123-3129.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam PYS, Ru Y, Jadhav PK, Aldrich PE, DeLucca GV, Eyermann CJ, Chang CH, Emmett G, Holler ER, Daneker WF, Li LZ, Confalone PN, McHugh RJ, Han Q, Li RH, Markwalder JA, Seitz SP, Sharpe TR, Bacheler LT, Rayner MM, Klabe RM, Shum LY, Winslow DL, Kornhauser DM, Jackson DA, EricksonViitanen S, Hodge CN. Cyclic HIV protease inhibitors: Synthesis, conformational analysis, P2/P2′ structure-activity relationship, and molecular recognition of cyclic ureas. J Med Chem. 1996;39:3514–3525. doi: 10.1021/jm9602571. [DOI] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:306–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Navaza J. AMoRe: An automated package for molecular replacement. Acta Cryst. 1994;A50:157–163. [Google Scholar]
- 29.Sheldrick GM, Schneider TR. SHELXL: High-resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 30.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 31.Sayer JM, Liu F, Ishima R, Weber IT, Louis JM. Effect of the active site D25N mutation on the structure, stability, and ligand binding of the mature HIV-1 protease. J Biol Chem. 2008;283:13459–13470. doi: 10.1074/jbc.M708506200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freedberg DI, Ishima R, Jacob J, Wang YX, Kustanovich I, Louis JM, Torchia DA. Rapid structural fluctuations of the free HIV protease flaps in solution: relationship to crystal structures and comparison with predictions of dynamics calculations. Protein Sci. 2002;11:221–232. doi: 10.1110/ps.33202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh E, Louis JM, Yamazaki T, Gronenborn AM, Torchia DA, Ishima R. A solution NMR study of the binding kinetics and the internal dynamics of an HIV-1 protease-substrate complex. Protein Sci. 2003;12:1376–1385. doi: 10.1110/ps.0300703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishima R, Torchia DA, Lynch SM, Gronenborn AM, Louis JM. Solution structure of the mature HIV-1 protease monomer: insight into the tertiary fold and stability of a precursor. J Biol Chem. 2003;278:43311–43319. doi: 10.1074/jbc.M307549200. [DOI] [PubMed] [Google Scholar]
- 35.Yamazaki T, Hinck AP, Wang YX, Nicholson LK, Torchia DA, Wingfield P, Stahl SJ, Kaufman JD, Chang CH, Domaille PJ, Lam PY. Three-dimensional solution structure of the HIV-1 protease complexed with DMP323, a novel cyclic urea-type inhibitor, determined by nuclear magnetic resonance spectroscopy. Protein Sci. 1996;5:495–506. doi: 10.1002/pro.5560050311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wishart DS, Sykes BD. Chemical shifts as a tool for structure determination. Methods Enzymol. 1994;239:363–392. doi: 10.1016/s0076-6879(94)39014-2. [DOI] [PubMed] [Google Scholar]
- 37.Kovalevsky AY, Tie Y, Liu F, Boross PI, Wang YF, Leshchenko S, Ghosh AK, Harrison RW, Weber IT. Effectiveness of nonpeptide clinical inhibitor TMC-114 on HIV-1 protease with highly drug resistant mutations D30N, I50V, and L90M. J Med Chem. 2006;49:1379–1387. doi: 10.1021/jm050943c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu F, Kovalevsky AY, Tie Y, Ghosh AK, Harrison RW, Weber IT. Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir. J Mol Biol. 2008;381:102–115. doi: 10.1016/j.jmb.2008.05.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zoete V, Michielin O, Karplus M. Relation between sequence and structure of HIV-1 protease inhibitor complexes: a model system for the analysis of protein flexibility. J Mol Biol. 2002;315:21–52. doi: 10.1006/jmbi.2001.5173. [DOI] [PubMed] [Google Scholar]
- 40.Tie Y, Boross PI, Wang YF, Gaddis L, Hussain AK, Leshchenko S, Ghosh AK, Louis JM, Harrison RW, Weber IT. High resolution crystal structures of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) active against multi-drug-resistant clinical strains. J Mol Biol. 2004;338:341–352. doi: 10.1016/j.jmb.2004.02.052. [DOI] [PubMed] [Google Scholar]
- 41.Weber IT, Wu J, Adomat J, Harrison RW, Kimmel AR, Wondrak EM, Louis JM. Crystallographic analysis of human immunodeficiency virus 1 protease with an analog of the conserved CA-p2 substrate -- interactions with frequently occurring glutamic acid residue at P2′ position of substrates. Eur J Biochem. 1997;249:523–530. doi: 10.1111/j.1432-1033.1997.00523.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


