Abstract
The protein from Arabidopsis thaliana gene locus At1g79260.1 is comprised of 166-residues and is of previously unknown function. Initial structural studies by the Center for Eukaryotic Structural Genomics (CESG) suggested that this protein might bind heme, and consequently, the crystal structures of apo and heme-bound forms were solved to near atomic resolution of 1.32 Å and 1.36 Å respectively. The rate of hemin loss from the protein was measured to be 3.6 × 10−5 s−1, demonstrating that it binds heme specifically and with high affinity. The protein forms a compact ten-stranded β-barrel that is structurally similar to the lipocalins and fatty acid binding proteins (FABPs). One group of lipocalins, the nitrophorins (NP), are heme proteins involved in nitric oxide (NO) transport and show both sequence and structural similarity to the protein from At1g79260.1 and two human homologues, all of which contain a proximal histidine capable of coordinating a heme iron. Rapid-mixing and laser photolysis techniques were used to determine the rate constants for carbon monoxide (CO) binding to the ferrous form of the protein (k'CO = 0.23 µM−1s−1, kCO = 0.050 s−1) and NO binding to the ferric form ( k'NO = 1.2 µM−1 s−1, kNO = 73 s−1). On the basis of both structural and functional similarity to the nitrophorins, we have named the protein nitrobindin and hypothesized that it plays a role in NO transport. However, one of the two human homolog of nitrobindin contains a THAP domain, implying a possible role in apoptosis.
Keywords: nitric oxide, carbon monoxide, structural genomics, nitrobindin, lipocalins
Introduction
Heme-containing proteins are broadly-utilized across all domains of life and have evolved to perform a wide range of essential biological functions including: electron transfer (e.g., cytochrome c) [1], substrate oxidation (e.g., cytochrome P450s) [2], ligand sensing (e.g., HemAT) [3, 4], and oxygen binding and storage (e.g., myoglobin) [5]. This diversity of function stems from the heme redox chemistry and reactivity with diatomic gases and from its ability to bind to a variety of distinct protein folds.
Respiration [6], O2 transport and storage, signaling [7], and heme degradation [8] are all essential physiologically functions that involve the interaction of diatomic ligands with the heme iron. Although these functions are performed by a number of distinct heme-protein families, each with a unique set of biophysical and structural properties, the underlying principles of diatomic ligand binding are similar and were developed using large libraries of mammalian myoglobin (Mb) variants as a model system [5, 9, 10]. Thus, an extensive mechanistic framework is available to interpret the properties of any newly discovered heme protein that is involved in the binding of diatomic ligands.
The gene At1g79260.1 of A. thaliana encodes a 166-residue protein with a molecular mass of 18.5 kDa. This gene product was selected as a structural target by CESG as part of the NIH-funded Protein Structure Initiative due to its low sequence similarity to any protein in the Protein Data Bank at the time of its selection. Initial preparations of the purified protein, which were utilized for crystallization trials, exhibited a slight reddish color and a UV-Vis spectrum that had a weak absorbance at 407 nm, indicative of a heme bound at low occupancy. The heme precursor delta-aminolevulonic acid was supplemented into the growth media to increase the rate of heme biosynthesis and facilitate its incorporation into the protein. Although, the resulting purified protein had a higher percentage of heme incorporation, the bound heme was not present at full occupancy. The addition of a concentrated stock hemin solution to purified protein resulted in full occupancy and a stable, bright red holoprotein preparation. The crystal structure of the heme-bound protein has a well-defined heme binding site with 100% occupancy of the porphyrin ring and iron atom. We determined the crystal structures of both the apo and heme-bound forms at 1.32 Å and 1.36 Å resolution, respectively.
Several biophysical techniques were utilized to probe the heme and ligand binding properties of the protein. The results are compared to those for mammalian myoglobins (Mbs), human hemoglobin (Hb), and insect nitrophorins (NPs). The CO and NO association rates were determined by laser flash photolysis, while the CO and NO dissociation rates were measured by stopped-flow, rapid mixing spectrophotometry. The FTIR spectrum of the protein-CO complex was measured to probe the electrostatic environment of the heme pocket. The rate of hemin loss from the protein was estimated by mixing the holoprotein with excess of the apoform of the "green" H64Y/V68F Mb mutant as a heme scavenging reagent [11]. The structural data combined with these hemin and ligand binding studies provide the first characterization of this previously unknown A. thaliana protein, which we have named nitrobindin.
Sequence and three-dimensional structural homologues of nitrobindin are found in a range of species from bacteria to humans. The majority have not been functionally characterized and identified. Nitrobindin is structurally similar to proteins from two families, the fatty acid binding proteins (FABP) and the lipocalins. FABPs belong to the superfamily of lipid-binding proteins and are involved in the intracellular transport of hydrophobic ligands [12]. Lipocalins, including the heme-binding NPs, form a compact β-barrel and have been shown to be involved in ligand transport [13]. The NPs are one of the most well-studied group of lipocalins and are found in the salivary glands of the blood-feeding insect Rhodnius prolixus, which carries the parasite that causes Chagas’ disease [14]. The insect NPs utilize heme iron to store, transport and release NO upon entering the host, which facilitates feeding by causing vasodilation [15, 16]. NPs are capable of reversibly binding NO in a pH dependent manner [17], and based on the structural similarity between nitrobindin and the insect NPs, we hypothesize that nitrobindin could be involved in NO transport in plants.
To better understand the role of heme in nitrobindin, we characterized its ligand binding properties, rapid autooxidation in the presence of O2, slow autoreduction in the presence of excess NO, and resistance to hemin loss. These properties were compared to those observed for Mb and Hb models and insect NPs. Nitrobindin exhibits functional behavior which is very similar to that of the NPs and quite distinct from the globin family of heme proteins, implying a functional role in NO transport or metabolism.
Material and Methods
Protein Purification
The gene from locus At1g79260.1 was cloned from an A. thaliana cDNA library, and the selenomethionine labeled protein was purified following the standard CESG pipeline protocols for cloning, protein expression, protein purification, and overall information management [18–21]. The cDNA encoding the protein was cloned into the pVP16 plasmid containing a tobacco etch protease (TEV)-cleavable His8-maltose-binding protein tag. The pvP16–At1g79260 construct was expressed in Eschericha coli B834p(Rare2). The cells were lysed by sonication, and the supernate was purified using immobilized nickel affinity chromatography. TEV protease was used to cleave the affinity/solubility tag, which was subsequently captured by subtractive nickel affinity chromatography.
Following gel filtration the protein was concentrated to 10 mg•ml−1 and dialyzed against 5 mM 2-(N-morpholino)ethanesulfonic acid (MES–HCl) buffer at pH 6.0 containing NaCl (50 mM), NaN3 (3 mM), and tris(2-carboxyethyl)phosphine (TCEP) (0.3 mM). Heme-bound nitrobindin was purified in a similar manner as apo nitrobindin with the following modifications. Unlike the apo form, heme-bound nitrobindin was expressed without selenomethionine. Prior to gel filtration, 1 mL aliquots of a freshly-prepared hemin solution (5 mM hemin in 50 mM NaOH) were slowly diluted into a solution of purified apo nitrobindin until a 2:1 molar ratio of hemin : protein was attained. The hemin-protein mixture was then loaded onto a prep grade Superdex 200 26/60 gel filtration column (GE Health Care, Piscataway, NJ) which had been preequlibrated with 5 mM MES–HCl buffer at pH 6.0 containing NaCl (50 mM) and TCEP (0.3 mM). Fractions containing the heme-bound protein were then concentrated to 16.5 mg•ml−1.
Crystallization
Crystals of apo nitrobindin were grown by hanging drop vapor diffusion by mixing 1 µL of protein solution described above with an equal volume of 100 mM tris(hydroxymethyl)-aminomethane (TRIS) buffer at pH 8.0 containing polyethylene glycol 2000 (18%) at 277 K. The heme-bound crystals were grown by hanging drop vapor diffusion by mixing 2 µL of protein solution described above with an equal volume of 50 mM 1,3-bis(tris(hydroxymethyl)methylamino)propane (BTP) buffer at pH 7.0 containing polyethylene glycol 4000 (30%) at 277 K.
Diffraction Data Collection and Structure Solution
X-ray diffraction data used for phasing and refinement of apo nitrobindin were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID-D beamline at the Advanced Photon Source (APS), Argone National Laboratory. Diffraction images for both apo and heme-bound crystals were indexed, integrated, and scaled using HKL2000 [22]. The apo protein crystal selenium substructure was determined using Hyss [23]. The results from Hyss indicated there were two heavy atom sites corresponding to the two selenomethionine in the protein. The protein was phased to 1.4 Å using a two wavelength phasing dataset with autoSharp [24]. The experimental electron density map was of excellent quality and the automated tracing procedures of ARP/warp produced an initial model containing 84% of all possible residues [25]. The structure was completed with alternate cycles of model building in Xfit [26] and refinement in Refmac5 [27]. The B-factors were refined anisotropically in the final stages of refinement. All refinement steps were monitored using an Rfree value based on 5.1% of the independent reflections. The diffraction data for the heme-bound protein were collected at the Life Sciences Collaborative Access Team (LS-CAT) 21-ID-G beamline at the APS, Argone National Laboratory. Molecular replacement was carried out with MOLREP using the apo form (PDB ID 2A13) as an initial model [28]. The heme-bound structure was completed in a similar manner as apo nitrobindin with the exception that Coot was used for model building [29], aniosotropic B-factors were only utilized to model atomic displacement of the heme moiety, and the final rounds of refinement included four TLS groups [30].
The final model of apo nitrobindin was refined to a resolution of 1.32 Å with an Rcryst of 0.156 and an Rfree of 0.186, the heme-bound form was refined to a resolution of 1.36 Å with an Rcryst of 0.171 and an Rfree of 0.198. All pertinent information on data collection, refinement, phasing, and model statistics of apo and heme-bound forms are summarized in Table I. Model quality was assessed using PROCHECK and MolProbity [31, 32]. All figures were generated using PyMOL [33].
Table I.
Summary of crystal parameters, data collection, structure phasing and refinement statistics. Values in parentheses are for the highest resolution shell.
| nitrobindin Refinement |
nitrobindin Phasing-peak |
nitrobindin Phasing-remote |
nitrobindin Heme |
|
|---|---|---|---|---|
| Crystal parameters | ||||
| Space group | P21212 | P21212 | P21212 | P21212 |
| Unit-cell | a = 60.1, b = 80.0, c = 36.9 |
a = 58.9, b = 78.5, c = 36.2 |
a = 59.6, b = 79.5, c = 36.6 |
a = 59.7, b = 79.7, c = 37.0 |
| parameters (Å, °) | α = β = γ = 90 | α = β = γ = 90 | α = β = γ = 90 | α = β = γ = 90 |
| Data collection statistics | ||||
| Wavelength (Å) | 0.97166 | 0.97934 | 0.96863 | 0.97856 |
| Resolution range | 29.26–1.31 | 32.86–1.46 | 33.26–1.46 | 50.00–1.36 |
| (Å) | (1.34–1.31) | (1.49–1.46) | (1.49–1.46) | (1.41–1.36) |
| No. of reflections (measured / unique) |
306132 / 41019 | 414533 / 29610 | 285069 / 28976 | 177708 / 37822 |
| Completeness (%) | 96.3 (93.1) | 95.3 (40.8) | 93.2 (86.3) | 97.2 (97.6) |
| Rmerge* | 0.066 (0.240) | 0.070 (0.144) | 0.059 (0.140) | 0.041 (0.236) |
| Redundancy | 7.5 (6.8) | 14.0 (12.1) | 9.8 (8.7) | 4.7 (3.7) |
| Mean I / sigma (I) | 14.91 (6.18) | 22.08 (13.00) | 17.38 (9.14) | 22.94 (5.41) |
| Phasing statistics ‡ | ||||
| Mean FOM (centris / acentric) | 0.48/0.17 | |||
| Phasing power (anomalous / isomorphous) |
1.65 | 0.81 / 0.59 | ||
| Cullis R-factor (anomalous / isomorphous) |
0.68 | 0.85 / 0.92 | ||
| Refinement and model statistics | ||||
| Resolution range | ||||
| (Å) | 48.06–1.32 | 47.84–1.36 | ||
| No. of reflections (work / test) |
38582 / 2068 | 35935 / 1864 | ||
| Rcryst§ | 0.156 (0.160) | 0.171 | ||
| Rfree¶ | 0.186 (0.210) | 0.198 | ||
| RMSD bonds (Å) | 0.011 | 0.010 | ||
| RMSD angles () | 1.311 | 1.354 | ||
| ESU from Rfree | 0.053 | 0.054 | ||
| B factor (protein / solvent) (Å2) |
12.5 / 34.8 | 18.8 / 36.9 | ||
| No. of protein atoms |
1237 | 1237 | ||
| No. of waters | 329 | 289 | ||
| No. of auxiliary molecules |
0 | 1 Hemin, 1 Ethylene Glycol |
||
| Ramachandran plot (%) | ||||
| Most favorable region |
90.6 | 90.6 | ||
| Additional allowed region |
9.4 | 9.4 | ||
| Generously allowed region |
0.0 | 0.0 | ||
| Disallowed region | 0.0 | 0.0 | ||
| PDB code | 2A13 | 3EMM | ||
Rmerge = ∑h ∑i | Ii (h) – <I(h)>| / ∑h∑i Ii(h), where Ii(h) is the intensity of an individual measurement of the reflection and <I(h)> is the mean intensity of the reflection.
Phasing in autoSharp
Rcryst = ∑h ‖Fobs| – |Fcalc‖ / ∑h |Fobs|, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes, respectively.
Rfree was calculated as Rcryst using 5.1 % of randomly selected unique reflections that were omitted from the structure refinement. Heme-bound nitrobindin Rcryst was calculated using the same set of Rfree reflections as the apo structure.
Metal Analysis
The total metal content of purified nitrobindin was determined by inductively coupled plasma-MS at the Environmental Health Division of the Wisconsin State Laboratory of Hygeine (Madison, WI).
Dynamic Light Scattering
All dynamic light scattering measurements were done utilizing a Protein Solutions DynaPro-99 (Wyatt Technology Corp., Santa Barbara, CA).
Knockout Plant Growth and Genotyping
Six seed lines (SALK_144412, SALK_088124, SALK_140278, CS100050, SALK_151606, and SALK_094969C) containing a T-DNA insert were purchased from the Arabidopsis Biological Resource Center (Columbus, OH). The knockout seeds were imbibed in water for 48 hours at 2° C before planting. All plants were grown at 22° C under a cool-white fluorescent light source. Long-day conditions consisted of a 16-hour period of light followed by an 8-hour period of darkness. Short-day conditions consisted of an 8-hour period of light followed by a 16-hour period of darkness.
Genomic DNA was extracted from the leaves of plants grown from the SALK_144412 and the SALK_151606 seed lines as previously described [34]. Genotyping of the SALK_144412 and the SALK_151606 varieties was conducted using the method described on the SIGnAL website (http://signal.salk.edu/tdnaprimers.html). Genomic DNA from the SALK_144412 line was amplified with an LP primer (5'- TGTCAACACCACATAAAAAGCC-3') and an RP primer (5'- GAAGTCGTCATTGCTCAGAGC-3'), while the SALK_151606 line was amplified with an LP primer (5'- TGAAATGAAGAAAGCAGGTGG-3') and an RP primer (5'- GAGCTCCTGATTCCAATTTCC-3'). To test for T-DNA insertion the genomic DNA from both the SALK_144412 and the SALK_151606 lines were amplified with the RP primers described above and the LBb1 primers described on the SIGnAL website.
CO Association and Dissociation Rate Constants
Data collection and analysis were carried out as previously described with only minor differences due to the rapid autooxidation of the protein [35]. To measure the overall CO association rate, two syringes of 100 mM potassium phosphate buffer at pH 7 containing EDTA (1 mM) were purged with either 100% CO(g) to produce a solution of 1000 µM CO or N2(g) for 20 minutes to remove any residual oxygen. The purged buffers and purified protein were drawn into a 1-ml Hamilton syringe to produce a series of solutions containing ~30 µM protein and 750 µM, 500 µM, and 250 µM CO. Each solution was used to fill a N2(g)-purged 1 mm path length cuvette, to which a few grains of sodium dithionite had been added. Protein reduction by dithionite and CO binding were verified by measuring the UV-Vis absorbance spectrum in a Varian 50 spectrophotometer.
Flash photolysis of liganded CO was initiated by a 300-ns pulse from a Phase-R 2100B dye laser (λ = 575 nm, Phase-R Co., New Durham, NH). The absorbance changes for bimolecular rebinding were monitored spectrophotometrically at 436 nm. Four to eight traces were collected at each CO concentration, averaged, and fitted to a single exponential expression. The overall association rate (k`CO) was obtained from the slope of a plot of the observed pseudo first order rate constants (kobs) versus [CO].
CO dissociation rates were determined by measuring time courses for the replacement of bound CO by NO. Because nitrobindin was found to be highly sensitive to autoxidation by trace levels of O2, ~ 50 µM sodium dithionite was added to a solution containing nitrobindin in 100 µM CO before it was loaded into the stopped flow spectrometer and mixed with 2,000 µM NO. Because the rate of NO binding is much faster than that for CO (see Table III) and [NO] > [CO], the observed rate for CO displacement is equal to the kCO, the first order rate constant for CO dissociation.
Table III.
Kinetic parameters for the binding of NO and CO to nitrobindin compared to sperm whale myoglobin, leghemoglobin, and nitrophorins 1–4.
| Protein | k’CO(µM−1 s−1) | kCO(s−1) | KCO(µM−1) |
|---|---|---|---|
| Nitrobindin (Fe2+) A | 0.23 | 0.050 | 4.5 |
| SWMb WT(Fe2+) B | 0.53 | 0.019 | 28 |
| SWMb H64Q(Fe2+) B | 0.94 | 0.012 | 78 |
| SWMb H64L (Fe2+) B | 26 | 0.024 | 1,100 |
| Soybean Lba(Fe2+) C | 17 | 0.0078 | 2,200 |
| Nitrophorin 4 pH 8.0 D | 7.9 | n.d. | n.d. |
| Protein | k’NO(µM−1 s−1) | kNO(s−1) | KNO(µM–1) |
| Nitrobindin (Fe2+) A | 81 | ~0.08 | ~1,000 |
| SWMb WT (Fe2+) B | 22 | 0.000098 | 220,000 |
| SWMb H64Q (Fe2+) B | 43 | 0.00011 | 370,000 |
| SWMb H64L (Fe2+) B | 190 | 0.00013 | 1,500,000 |
| Soybean Lb a (Fe2+) C | 170 | 0.00002 | 9,000,000 |
| Nitrobindin (Fe3+) A | 1.2 | 73 | 0.016 |
| metSWMb WT (Fe3+) B | 0.080 | 12 | 0.0067 |
| SW metMb H64Q (Fe3+) B | 11 | 59 | 0.19 |
| SW metMb H64L (Fe3+) B | 44 | 80 | 0.55 |
| Soybean metLba(Fe3+) E | 0.14 | 3.0 | 0.047 |
| Nitrophorin 1 pH 8.0 F | 1.5 | 4.3 | 0.34 |
| Nitrophorin 2 pH 8.0 F | 33 | 32 | 1.0 |
| Nitrophorin 3 pH 8.0 F | 6.7 | 30 | 0.22 |
| Nitrophorin 4 pH 8.0 F | 2.5 | 15 | 0.17 |
Rate constants that were determined in this study were measured in 100 mM potassium phosphate buffer at pH 7 containing EDTA (1 mM) at 20 °C.
The rates were taken from Rohlfs et al. [35], Eich et al. [76], Thorsteinsson et al. [55], Foley [36].
Taken from Hargrove et al. [75].
Rates taken from Maes et al. [48]
Taken from Herold et al. [77]
NO Association Rates
The overall rate of NO association with the ferric form of the protein was measured by laser flash photolysis as described above for CO. To measure the association rate constant NO binding to the ferrous protein, the samples of the ferric NO complex were allowed to incubate anaerobically overnight at 4 °C in the presence of excess NO. Under these conditions, the ferric NO heme will slowly autoreduce to the ferrous NO form [36, 37]. These samples were examined by UV-Vis spectroscopy to verify their conversion to Fe2+NO, photolyzed by our 300 ns dye laser, and bimolecular rebinding traces were obtained at 436 nm to determine the NO association rate constant for the ferrous state. Unlike most globin ferrous-NO complexes, the reduced nitrobindin-NO complex has a high quantum yield, and large absorbance changes associated with bimolecular rebinding were observed.
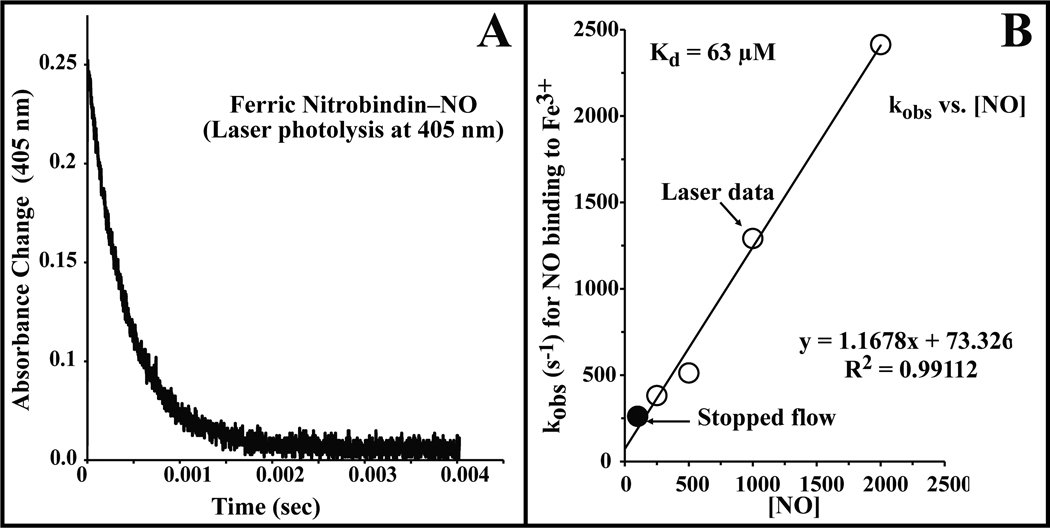
The NO concentrations of the Fe2+NO samples were adjusted to account for the NO consumed during the reduction of the ferric heme. The reported Fe2+NO association rates may be underestimated due to the possibility of trace O2 leaching into the vials and consuming some of the free NO during the overnight incubation. The overall association rate constants (k`NO) for NO binding to the ferric and ferrous forms of nitrobindin were obtained from the slope of a plot of the pseudo first order observed rates (kobs) versus [NO] (see Fig. 5B).
Figure 5.
Time course for NO rebinding to ferric nitrobindin after flash photolysis. Absorbance changes were measured at 405nm. The NO concentration for this trace was 1 atmosphere (2000 µM) of NO, and the protein concentration was approximately 100 µM. For all NO association experiments nitrobindin was in 100 mM potassium phosphate buffer at pH 7 containing EDTA (1 mM) at 20 °C. The overall association rate (k`NO) is taken from the slope of a linear fit of the rate of absorbance change versus the NO concentration.
Autoreduction of the Ferric Protein by NO
Autoreduction of nitrobindin in the presence of excess NO and absence of oxygen was measured as described previously for metMb [36]. A septum-stoppered, 1-cm path length cuvette was prepared with 100 mM potassium phosphate buffer at pH 7.0 containing EDTA (1 mM) equilibrated with 1 atmosphere (2000 µM) NO. To monitor the reduction rate, ferric protein was added to yield a final concentration of 15 µM and a series of UV-Vis spectra were collected at 20°C.
Hemin Loss
Hemin loss from nitrobindin was measured using the protocol developed by Hargrove et al. [11]. Purified ferric nitrobindin was diluted to a concentration of 50 µM in 200 mM potassium phosphate buffer at pH 7.0 containing sucrose (600 mM) to inhibit precipitation of the resultant apoprotein and excess (150 µM) hemin loss reagent, apo-H64Y/V68F sperm whale myoglobin (SWMb). As nitrobindin loses hemin, holo-H64Y/V68F SWMb is formed, which results in a green color due to a large increase in absorbance at 600 nm. The reaction was monitored using a spectrophotometer (Cary 50 Bio), and the time courses were fit to single exponential expressions.
FTIR Spectroscopy
The CO complexes were analyzed at room temperature by FTIR spectroscopy as described previously [38]. 20 µL of 1–2 mM protein was equilibrated with 1 atmosphere CO, reduced with 1 µL of 200 mM sodium dithionite and loaded into a 40-µm path length IR cell. The FTIR spectrometer was set to average sixty interferograms giving a final resolution of 1 cm−1 for each spectrum.
Results
Overall Structure and Structure Quality
In both the apo and heme-bound nitrobindin structures well-defined electron density was observed for residues 14–166. Electron density was lacking for the N-terminal residues 1–13, presumably resulting from intrinsic disorder. Both structures contain one molecule per asymmetric unit and belong to the P21212 space group. Two monomers of nitrobindin from adjacent asymmetric units interact to form a 920 Å2 interface as calculated by the PISA server [39]. The extensive crystallographic interface suggests that nitrobindin forms a dimer in the crystal. By gel filtration the molecular mass was determined to be approximately 26 kDa, which is significantly more than the calculated mass of 19 kDa. The mass as measured by dynamic light scattering was in between 26 kDa and 32 kDa. Based on the gelfiltration, dynamic light scattering, and the crystal structure it is probable that nitrobindin also forms a dimer in solution.
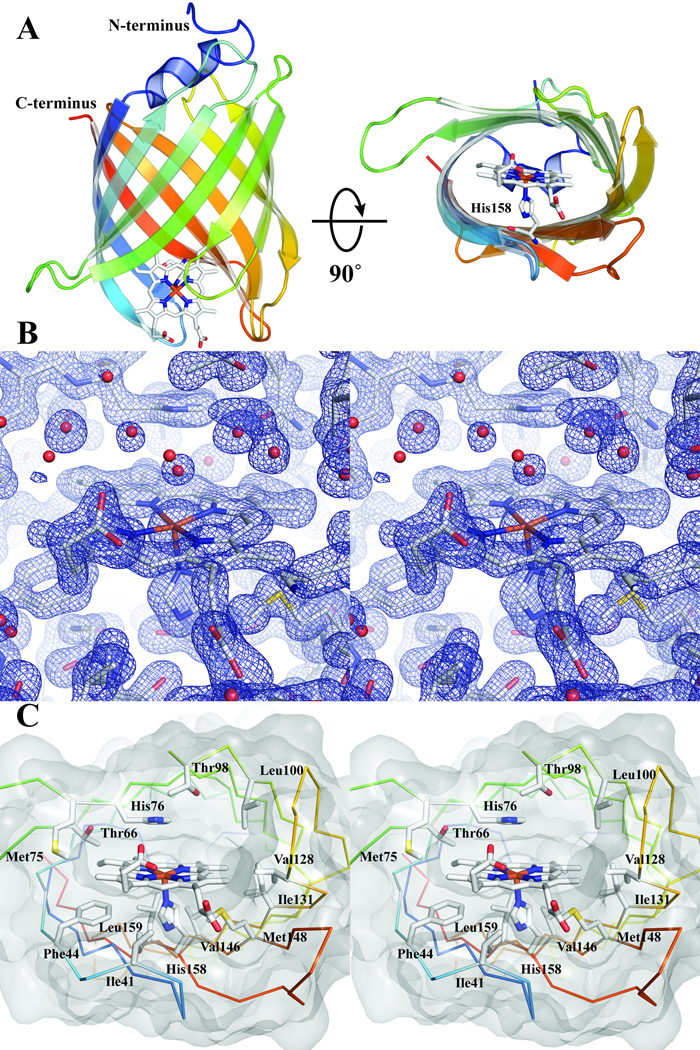
Nitrobindin is a single domain protein composed primarily of β-sheets. Ten antiparallel β-strands are connected by short loop regions (2–6 residues each) that form a compact β-barrel (Figure 1A). One end of the β-barrel is capped by a 310 α-helix, while the other end of the β-barrel is solvent accessible. The solvent accessible portion of the β-barrel forms a hydrophobic cavity lined by Phe44, Met75, Thr98, Leu100, Val128, Ile131, Met148, and Leu159. Entry to the hydrophobic cavity is defined by five loops: Tyr37–Ile40, Lys68–Ala73, Ser97–Leu100, Asn124–Lys127, and Thr151–Leu155.
Figure 1.
A. Cartoon representation of the overall structure of nitrobindin is shown going from blue at the N-terminus to red at the C-terminus. The bound heme is shown as sticks with carbon in white, nitrogen in blue, oxygen in red, and iron in orange. A view along the barrel axis (right) shows the cavity formed by the β-barrel. His158 coordinates the heme iron and is shown in the same color scheme as above. B. Stereo view of the 2Fo–Fc map contoured at 1.3 σ of the heme pocket. nitrobindin and the bound heme are shown as sticks with the same color scheme as above with waters represented as red spheres. C. Stereo view of the hydrophobic cavity. The Cα ribbon diagram is shown going from blue at the N-terminus to red at the C-terminus. Amino acids that are within 4 Å of the heme are shown as sticks and labeled.
The crystals that were used to solve the apo structure exhibited a slight reddish tint. Surprisingly, however, the apo structure did not contain any significant electron density indicative of a metal or large organic cofactor. An inductively coupled metal analysis revealed 0.13 mol of iron per mol of protein. A data set was collected at 1.5 Å utilizing a rotating Cu anode X-ray generator (data not shown) and did reveal a significant anomalous peak located 2.8 Å from the Nε of His158. We attribute this anomalous peak to the presence of iron due to the strength of the peak and the results of the mass spectrometry metal analysis. Based on the size of the hydrophobic cavity and the position of a potential proximal histidine (His158), we speculated that nitrobindin was capable of binding heme, which would dock in the hydrophobic cavity and coordinate to His158. To test this hypothesis we added hemin to purified nitrobindin as described in Materials and Methods.
Crystals of the holoprotein generated by hemin addition are dark red, and the heme moiety is clearly visible in the electron density map (Figure 1B). As predicted, the heme is coordinated by the Nε of His158, is bound in the hydrophobic cavity, and has significant solvent exposure (Figure 1A). Unlike the globins, there is no obvious distal histidine within hydrogen bonding distance of potential sixth ligands in holo- nitrobindin; however, there is a histidine at position 76 in the cavity with an imidazole oriented parallel to the heme plane (Fig 1C). In addition to these histidines, the heme is held in place by multiple van der Waals interactions with the surrounding protein. Most of the residues that form the entrance of the hydrophobic cavity are within 4 Å of the heme and include Phe44, Met75, Thr98, Leu100, Val128, Ile131, Met148, and Leu159. Other contact residues are located deeper in the hydrophobic pocket and include Ile41, Thr66, His76, and Val146 (Figure 1C). One of the heme propionate groups is within hydrogen bonding distance of Thr40 (2.5 Å) and could interact electrostatically with the terminal amine of Lys127 (3.8 Å). Similar salt bridges stabilize heme in mammalian Mbs and Hbs. The other propionate is entirely solvent-exposed and does not appear to interact directly with any amino acid side chain. The heme pocket of nitrobindin is relatively open and exposed to solvent, and not surprisingly, a water molecule is coordinated on the distal side of the heme iron.
Structural Comparison to the Lipocalins
Of the three structurally conserved regions outlined by Flower, D.R. et al., two are observed in the nitrobindin structure [13]. Specifically, the 310 helix located adjacent to the first strand in the β-barrel, and a lysine (Lys165), which packs against a conserved tryptophan (Trp30) located at the start of the first strand. Unlike the lipocalins there are no disulfide bonds observed in the nitrobindin structure. These structurally conserved regions could suggest an evolutionary link between nitrobindin and lipocalins.
Sequence Homology Search
A BLAST search using the sequence of nitrobindin revealed several plant proteins with a sequence identity of greater than 50%. The three proteins with the highest alignment scores correspond to hypothetical proteins from Vitis vinifera (grape, Uniprot code A5BBZ0), Musa balbisiana (banana, Uniprot code Q1EP49), and Oryza sativa (rice, Uniprot code Q0JKE7). The three hypothetical proteins had identities and similarities to nitrobindin ranging from 76%- 67% and 89%- 86%, respectively. The BLAST search also revealed additional proteins with less than 50% identity to nitrobindin from species ranging from bacteria to humans. None of the related proteins have been functionally annotated with the exception of a human THAP4 containing protein.
The N-terminus of the 577-amino acid human homolog contains a THAP domain. Thanatos-associated protein domains (Thanatos is the Greek God of death) are composed of a modified zinc finger, which is suspected to bind DNA in a manner similar to that in P element transposases and may be involved in apoptosis of Hela cells [40–42]. The C-terminal domain of this human homolog is predicted by sequence homology to be similar in structure to nitrobindin. The second human homolog is a truncated version of the THAP domain protein, which only contains the165-residues of the nitrobindin-like domain. Sequence alignments of the two human homologues and nitrobindin reveal that His158 is conserved in both of the human proteins.
Structure Homology Search
A number of structural homologs were identified by the Vector Alignment Search Tool (VAST) [43]. The top hits from VAST were Rv2717c (PDB ID 2FR2) and Rv0813c (PDB ID 2FWV) from Mycobacterium tuberculosis, which were previously solved in the absence of heme by structural genomic groups and were hypothesized to be FABPs based on structural homology [44]. Structural homologous residues to those in the nitrobindin heme-binding pocket are found in 2FR2, including the proximal histidine. VAST also identifies a range of functionally characterized homologs with sequence identities ranging from 4%–18%. Most of the additional identified structural homologous proteins belong to the FABP family. However, the only structural homologs presently known to bind heme specifically are the insect NPs that are involved in NO transport [45, 46].
Oxidation State of Nitrobindin
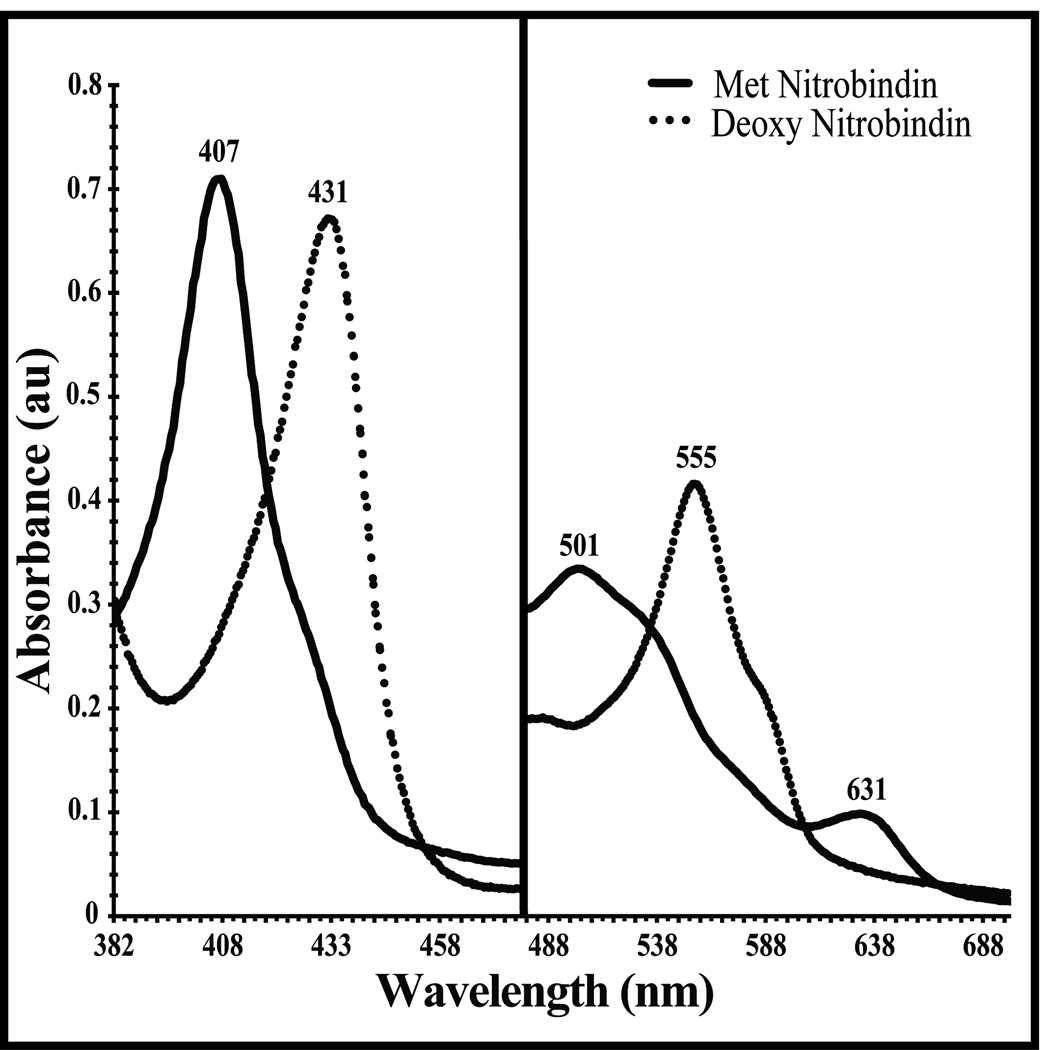
The UV-Vis spectrum of purified nitrobindin is shown in Figure 2. The spectrum has the characteristic aquohemin Soret band at 407 nm with high spin bands at 501 nm and 631 nm and weak α and β Q-bands in the 530–570 nm region. The spectrum of oxidized nitrobindin is similar to that of the neutral pH, aquomet forms of mammalian Mbs and Hbs. Anaerobic reduction with sodium dithionite produces a "classic" pentacoordinate deoxyheme spectrum, with bands at 431 nm and 555 nm. Except for the small shoulder at 585 nm, this reduced deoxygenated spectrum is similar that of deoxyMbs and deoxyHbs.
Figure 2.
UV-Vis spectrum of nitrobindin in 5 mM MES-HCl buffer at pH 6.0 containing NaCl (50mM), and TCEP (0.3mM). The solid line is for protein exposed to air and the dotted line is after nitrobindin is reduced with sodium dithionite.
Exposure of the reduced form of nitrobindin to air resulted in immediate oxidation to the ferric form, and a stable Fe2+O2 complex could not be formed, even transiently, in simple hand mixing experiments as is the case for the insect NPs [47–49]. Thus the rate of autooxidation must be greater than ≥ 0.1 s−1 and is at least 104 to 105 times faster than the autooxidation rates of MbO2 and HbO2 under physiological conditions. The preparation of Fe2+CO complexes required the continued presence of excess sodium dithionite, even under the micro-anaerobic conditions obtained by purging buffers with N2 ([O2]≤ 1 µM). Even minimal exposure to air brings about rapid oxidation of the nitrobindin CO complex, which implies a large CO dissociation rate. Again, these properties are very similar to those of the NPs [47–49] and five-coordinate model heme compounds with solvent exposed distal surfaces [50]. In contrast, Mb mutants in which the distal histidine is replaced by Gly, Ala, Val, and Leu form CO complexes that are stable for minutes to hours when exposed to oxygen, and their rates of autooxidation can be easily measured [51].
High Resistance to Hemin Dissociation
Hargrove et al. showed that both solvent exposure and the number and types of protein–heme contacts markedly influence the rate of hemin loss from Mbs and Hbs, which for the wild-type proteins is on the order of 10−4 to 10−6 s−1 (1 to 0.01 hr−1, Table II) [52, 53]. In contrast, the bimolecular rate of monomeric heme binding to apoglobins is relatively invariant, 10 to 100 × 106 M−1 s−1, and driven by non-specific hydrophobic partitioning from the solvent phase to the apolar interior of the protein [54]. As a result, the overall affinities of these globins for hemin are on the order of 1011 to 1014 M−1 with picomolar equilibrium dissociation constants. To estimate the affinity of nitrobindin for hemin, we measured the rate of hemin dissociation from the ferric form of the protein using the "green" H64Y/V68F apoMb reagent as described by Hargrove et al. [11]. Table II shows the hemin loss rate of nitrobindin compared to a number of Mb variants, adult human hemoglobin (HbA), and soybean leghemoglobin (Lba). The observed rate for hemin loss from nitrobindin was determined to be 0.126 hr−1 or 3.6 ×10−5 s−1, which is intermediate between the hemin loss rates from wild-type metSWMb and native adult human metHb and Lba (Table II) [53]. Assuming a hemin association rate constant similar to the globins, this slow rate of hemin loss suggests a hemin equilibrium dissociation constant on the order of 10−12 M. Thus hemin binding to the protein appears to be highly specific, similar in affinity to that for plant Lbs and mammalian Hbs, and physiologically relevant.
Table II.
Hemin loss of nitrobindin compared to sperm whale myoglobin and leghemoglobin
Nitrobindin–CO Ligand Binding and FTIR Spectra
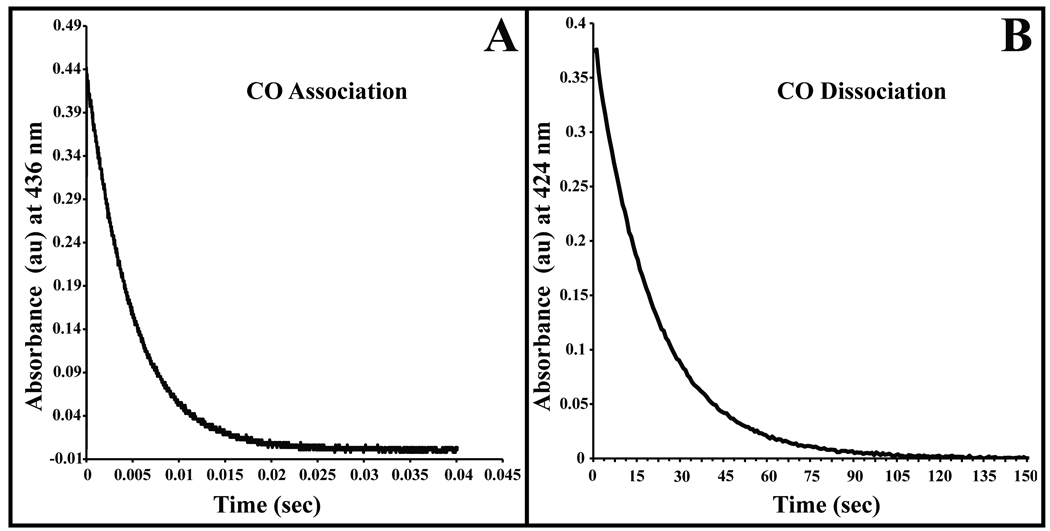
The association rate constant for CO binding to reduced nitrobindin was measured by analyzing time courses for bimolecular rebinding after laser photolysis (Fig. 3A). The absorbance traces were monophasic and fit well to a first order exponential expression. The slope of a plot of kobs vs. [CO] gave k’CO = 0.23 µM−1 s−1 (Figure 3A, Table III). This value is approximately two fold smaller than that for SWMb (k’CO = 0.53 µM−1 s−1) [55]. The rate constant for dissociation of CO from reduced nitrobindin was measured in a stopped flow, rapid mixing spectrometer by analyzing time courses for the displacement of bound CO by excess NO. The observed time course for this reaction was also monophasic and kCO = 0.05 s−1 (Figure 3B, Table III). The association equilibrium constant for CO binding was calculated as k'CO/kCO and equals to 4.5 µM−1, which is 5 to 500 times smaller than that for Mb and soybean Lba, respectively.
Figure 3.
Time courses for CO association and dissociation. A. Absorbance trace for bimolecular CO rebinding measured at 436 nm after flash photolysis of the Fe-CO complex. The CO concentration for this trace was 1 atmosphere (1000 µM), and the protein concentration was approximately 100 µM. For all CO association and dissociation experiments nitrobindin was in 100 mM potassium phosphate buffer at pH 7 containing EDTA (1 mM) at 20 °C. The overall association rate (k’CO) was computed as the slope of a linear fit to a plot of kobs versus the CO concentration. B. Measurement of the CO dissociation constant by stopped-flow spectrophotometry. CO–nitrobindin was mixed with 100 mM potassium phosphate buffer at pH 7 containing EDTA (1 mM), and 1 atmosphere (2000 µM) of NO at 20 °C. The displacement of the CO–nitrobindin complex was measured by monitoring the absorbance decrease at 424 nm.
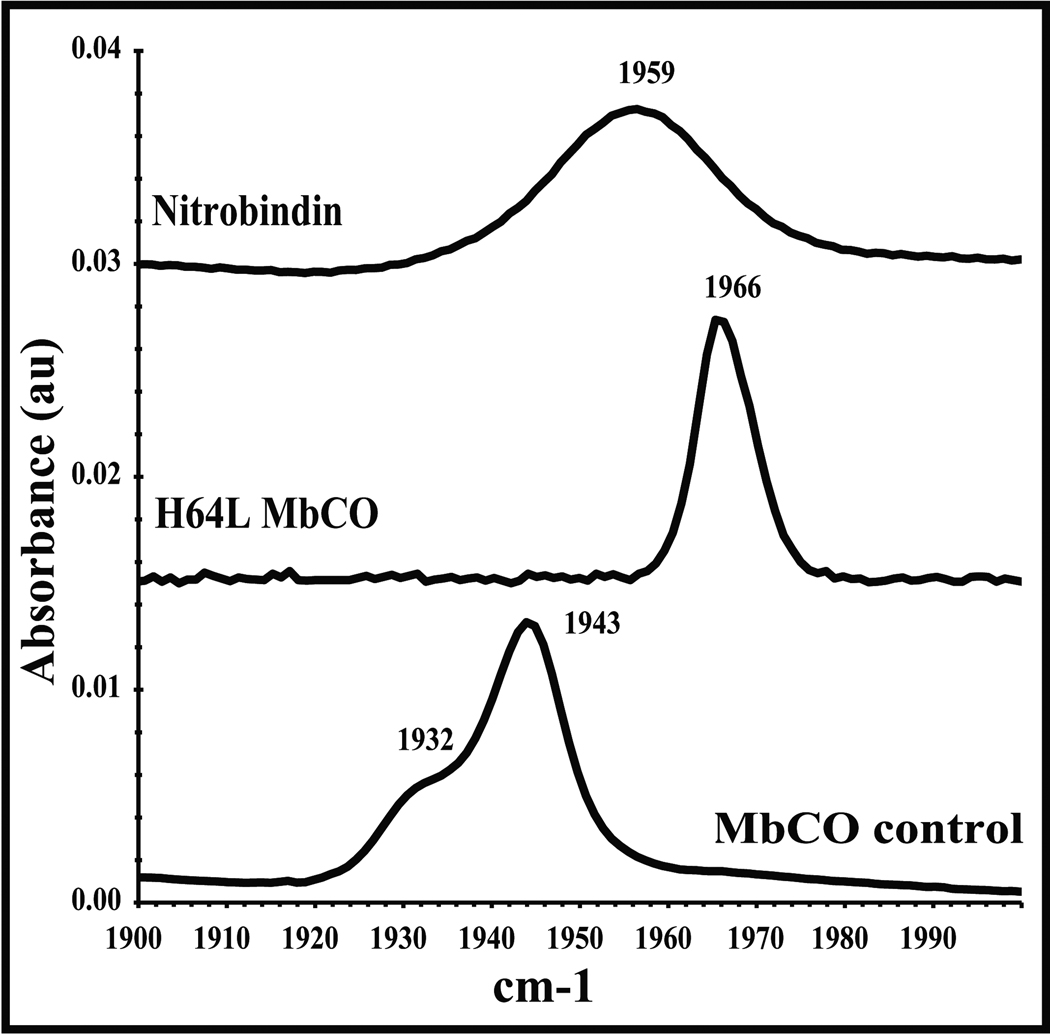
The infrared (IR) stretching frequency of heme-bound CO, ν C-O, is a sensitive probe of the electrostatic environment surrounding the ligand. Low frequency bands (1910–1950 cm−1) indicate a positive field and hydrogen bonding; high frequency bands (1960–1970 cm−1) indicate a neutral field; and even higher frequency peaks (1980–2000 cm−1) indicate a negative field [56–61]. In SWMbCO, a strong hydrogen bond donated by the distal histidine, H64, to the ligand results in a peak at ~1940 cm−1. The binding pocket of H64L Mb is completely apolar, and the νC-O occurs at 1966 cm−1. The FTIR spectrum of nitrobindin shows one broad, symmetric peak with a maximum centered at 1959 cm−1 (Figure 4), suggesting a neutral field with little hydrogen bond donation as compared to MbCO. In the heme-bound structure of nitrobindin, several water molecules are observed in close proximity to the iron atom and penetrate into the interior of the distal portion of the pocket. These waters create many microenvironments resulting in peak broadening but little net positive or negative fields. A similar FTIR spectrum was observed for H64G MbCO in which the myoglobin distal pocket is open to solvent and a similar band position (1960 cm−1) and broadening is observed [59].
Figure 4.
Iron-carbonyl IR spectrum in the range of 1900–2000 cm−1 for nitrobindin, compared to wild-type sperm whale myoglobin, and a distal histidine variant myoglobin (H64L). The IR spectra were measured at room temperature in 100 mM potassium phosphate buffer at pH 7 containing EDTA (1 mM).
NO Binding to the Ferric and Ferrous Forms
The association and dissociation rate constants for NO binding to ferric nitrobindin were measured by analysis of both flash photolysis and stopped flow, rapid mixing time courses (Table III). Unlike the NPs, which exhibit biphasic time courses for NO binding, only monophasic absorbance changes were observed after photolysis of the Fe3+NO complex of nitrobindin (Figure 5) [47]. Similar time courses were observed in rapid mixing experiments at lower NO concentration. A plot of the observed pseudo first order rate constant versus [NO] yields a slope equal to the association rate constant, k'NO = 1.2 µM−1 s−1 for the ferric form of nitrobindin, and an y-intercept equal to the dissociation rate constant, kNO = 73 s−1. The association equilibrium constant was calculated to be 0.016 µM−1, which is ~3 fold higher than that for NO binding to metMb and 3-fold smaller than that for met Lba (Table III).
The observed association rate constant for NO binding to ferric nitrobindin is similar to those observed for NO binding to ferric NP1 and NP4 (1.5 ± 0.1 µM−1 s−1 and 2.3 µM−1s−1 respectively) [47]. However, the dissociation rate constant for nitrobindin is 2 to 30 times higher than those observed for the NPs, and as result, the affinity of ferric nitrobindin for NO is 10 to 100-fold smaller (Kd of ~50 µM) than that for the NPs (Table III).
When the ferric form of nitrobindin is reacted anaerobically with buffer equilibrated with 1 atmosphere of NO (2000 µM), the Fe3+NO complex forms immediately but then slowly autoreduces over several hours to the Fe2+NO complex, with a UV-visible spectrum similar to that of Mb(Fe2+)NO. Under these conditions, the rate of autoreduction of nitrobindin is 1.0 ×10−3 s−1, which is 10 times faster than that for mammalian metMbNO complexes but similar to rates of autoreduction of metLbNO and metHbNO [36]. In the absence of oxygen, this Fe2+NO complex is stable and can be examined by laser photolysis to measure bimolecular rate constants for NO binding to the ferrous form of nitrobindin and by rapid mixing with CO and excess (~25 mM) sodium dithionite to measure the rate constant for NO dissociation [62]. These methods gave values of k'NO = 81 µM−1s−1 and kNO = ~0.08 s−1 for NO binding to ferrous nitrobindin. The association equilibrium constant (k'NO/kNO) for the ferrous protein is 1,000 µM−1. This value is 200 to 10,000 fold smaller than those for Mb and Lb (Table III). The NO dissociation constant for ferrous nitrophorins has been estimated to be 5–90 femtomolar using electrochemical measurements [47].
Nitrobindin Knockout Plants
Six seed lines (SALK_144412, SALK_088124, SALK_140278, CS100050, SALK_151606, and SALK_094969C) containing a T-DNA insert were germinated, grown, and visually inspected for any observable phenotype. There were no observable phenotypes associated with any of the plants from the six seed lines when compared to wild-type A. thalania. The SALK_144412 and SALK_151606 plants were screened for the presence of the T-DNA insert to eliminate the possibility that a nitrobindin knockout was lethal. Both the SALK_144412 and the SALK_151606 seeds produced viable plants that were homozygous for the T-DNA insert (data not shown), suggesting that the loss of nitrobindin is not lethal.
Discussion
Comparison of Nitrobindin with the Globins
Nitrobindin and the NPs are the only structurally confirmed heme-proteins in which the binding pocket is a complete β-barrel [45, 46, 63]. In both proteins, the heme iron is coordinated to a proximal histidine, which is positioned at the rim of one end of the β-barrel. In contrast, most animal globins contain a three-on-three (A, B, E, F, G, H) helical "globin fold", which binds heme between the E and F helices [64, 65]. Most Mbs and Hbs also contain two additional short helices (C, D) in the region connecting the B and E helices. The heme is coordinated to a proximal His at the F8 helical position, and a distal His or Gln residue is normally present at E7 helical position. In mammalian Mb and Hb, the distal histidine is close enough to the heme iron atom to stabilize bound O2 by hydrogen bonding but far enough away to prevent direct coordination and hemochrome formation.
His76 is present in the distal pocket of nitrobindin but is too far away to interact directly with bound ligands (Figure 1). This lack of interaction with bound ligands is confirmed by the FTIR spectrum of the CO form of nitrobindin, which presents a broad νC-O peak at 1959 cm−1 indicative of a solvated but apolar distal pocket. However, His76 probably facilitates water penetration into the open distal cavity, which in turn accounts for the markedly accelerated rate of autooxidation of reduced nitrobindin. The presence of distal pocket water and the absence of stabilizing hydrogen bonding interactions between bound O2 and a distal Gln or His side chain accounts for the ≥10,000-fold larger rate constant for autooxidation of oxygenated nitrobindin compared to those for mammalian MbO2 and HbO2 and plant LbO2. The net result is that nitrobindin does not even form a transient Fe2+O2 complex on ordinary time scales (lifetimes ≥ 10 seconds). In presence of oxygen, the protein is always oxidized.
Nitrobindin can be reduced anaerobically with dithionite and does bind CO. The association rate constant for CO binding is small, 0.23 µM−1s−1, indicating either low iron reactivity or a sterically hindered ligand binding site. The structure indicates a wide-open distal pocket with no steric restrictions, suggesting that the low reactivity is due to proximal constraints that restrict in-plane movement of the Fe2+ atom. The proximal His is held in place by the extensive hydrogen bonding network that forms the β-barrel. This idea of proximal strain is supported by the large rate constants for CO and NO dissociation from the ferrous form. These values, kCO = 0.05 s−1; kNO = 0.08 s−1, are 3 to 300 time greater than the corresponding rate constants for most globins. In contrast, the rate constant for NO binding to the ferrous form of nitrobindin is very large and comparable to sterically unhindered Mb mutants and Lba, which also have open distal pockets. In the case of ferrous heme proteins, the rate of NO binding is determined by the speed of ligand entry into the protein because the NO radical reacts extremely rapidly with high spin iron, and the rate of bond formation from within the protein occurs on the picosecond time scales [5]. Thus, the large rate constant (k'NO,Fe2+≈ 80 × 106 M−1s−1) for NO binding to ferrous nitrobindin confirms an open distal pocket.
In the ferric form of nitrobindin, a water is coordinated to the distal or sixth axial position of the heme iron but can be rapidly displaced by NO. The association rate constant for NO binding to ferric nitrobindin is 1.2 µM−1s−1 and very similar to those observed for the ferric forms of NP1 and NP4 (Table III). However, the NO dissociation rate constant for the Fe3+NO complex of nitrobindin is ~70 s−1. This value is significantly greater than those for the NPs but similar to those for Mb mutants lacking a distal histidine to stabilize the bound ligand by hydrogen bonding. The net result is a Kd (kNO/k'NO) for NO binding to ferric nitrobindin is ~50 µM and ~10 fold higher than those observed for the NPs.
We were unable to detect any binding of azide or imidazole to ferric nitrobindin, despite its open distal pocket. The lack of azide binding is not a steric effect and is probably due to poor iron reactivity and the lack of stabilizing interactions by nearby polar amino acids (i.e. His(E7) and Gln(E7) found in most globins). Brancaccio et al. [66] showed that replacing the distal histidine with an apolar amino acid decreases the affinity of Mb for azide 20 to 100 fold. The lack of imidazole binding is remarkable, and no spectral shifts were observed at levels as high as 0.1 M. However, a low spin Fe3+CN complex can be made at high cyanide concentrations.
Comparison of Nitrobindin with the Nitrophorins
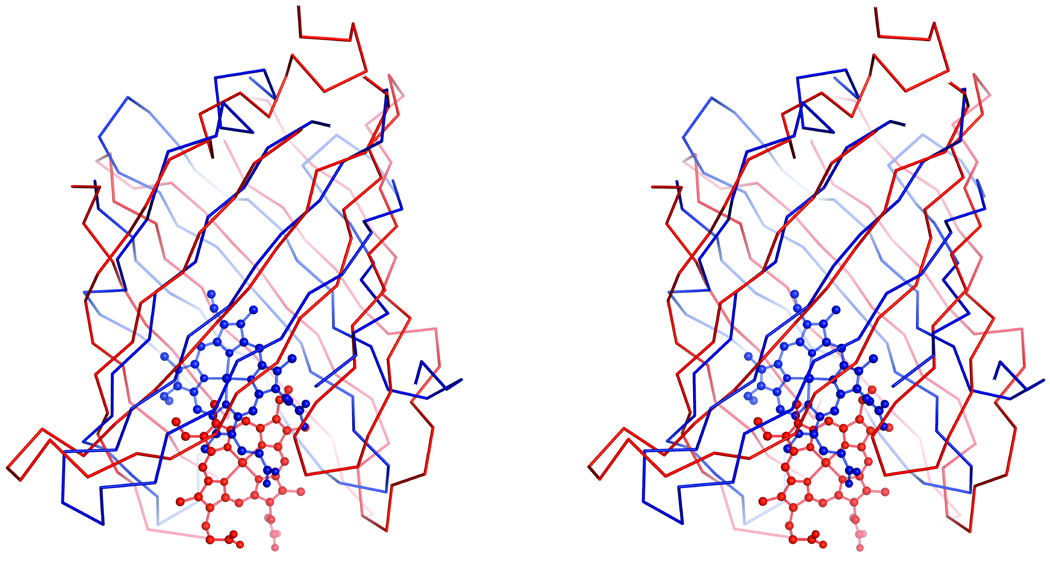
The major structural difference between nitrobindin and the NPs is the number of strands that compose the β-barrel and the position of the heme in the hydrophobic cavity. Nitrobindin is composed of ten β-strands while the NPs contain eight β-strands (Figure 6). The heme of the NPs is buried deeper within the hydrophobic cavity, while the bound heme of nitrobindin is positioned at the rim of the cavity. In both proteins, compact barrels are formed with an extensive hydrophobic cavity; the heme iron is coordinated by a proximal histidine; and the end opposite of the heme-binding site is enclosed by a 310 helix. Two key structural difference are: (1) entry to the heme pocket is more open in nitrobindin than in the nitrophorins, which is semi-enclosed, and (2) an Asp is present in the distal pocket to stabilize the protonated primary amine of bound histamine in the latter [46]. Glu78 is present in the distal pocket of nitrobindin, but does not appear to stabilize bound imidazole.
Figure 6.
Stereo view of a secondary structural alignment of nitrobindin in red and residues 13–139 of NP4 in blue (PDB ID 1YWD). The bound heme of nitrobindin is shown in red and the bound heme of NP4 is shown in blue.
The physiological function of the insect NPs has been established. Hemin is utilized to bind and transport NO to a host. NO is released when the NPs enter the host to facilitate feeding by disrupting hemostasis [15, 16]. Histamine is taken up by the protein reducing inflammation and, at the same time, displaces NO, which in turn facilitates vasodilation, allowing the insect to ingest more blood. The ability to reversibly bind NO and histamine are the result of a stable ferriheme complex with anionic character and a low rate of heme autoreduction [67].
Nitrobindin is structurally similar to proteins from the FABP and the lipocalin families, which contain the NPs. Both the lipocalins and FABPs are known to be involved in ligand transport [12, 13, 68, 69], whereas the insect NPs are specialized for NO transport and release. Nitrobindin and the NPs have similar ultrafast autooxidation rates, CO binding environments as measured by FTIR spectra (broad νC-O bands centered at 1959 –1962 cm−1[67]), and rates of NO binding and release from the ferric forms, all of which are quite different from those observed for mammalian Mbs and Hbs and plant Lbs (Table III). The time courses for NO rebinding to ferric nitrobindin after photolysis are monophasic whereas at least two phases are observed for NO rebinding to the NPs. The biphasic kinetics for NO binding to the NPs have been attributed to a structural rearrangement of two flexible loops near the distal pocket after NO is bound [17, 70, 71]. These flexible loops serve to enclose the ligand in the interior cavity and enhance affinity. No evidence for additional conformational stabilization of bound NO is observed for nitrobindin, which has a 10-fold lower affinity for this ligand than the NPs (Table III)
Possible Physiological Role of Nitrobindin
Based on the structural similarity of nitrobindin to FABPs and lipocalins, it is reasonable to assume that nitrobindin functions as a transport protein, but what it transports is less clear. Its high affinity for hemin and slow rate of release suggest that it functions as a heme protein in vivo. Nitrobindin cannot form stable O2, CO, or NO complexes in the presence of air and, thus, clearly does not have a role in O2 storage or transport. Its similarity in structural, oxidation, and ligand binding properties to the insect NPs suggest some common or related physiological role, possibly in NO metabolism or transport.
At present, little is known about NO metabolism in plants, but NO is made in relatively large amounts in response to wounding and pathogenic infection in a variety of plants including A. thaliana [72–74]. It is possible that the ferric form of nitrobindin plays a role in transporting NO to the site of infection and releasing it quickly (i.e. kNO ≈ 70 s−1). Alternatively, the reduced form might act as storage protein to bind NO in an anaerobic environment and then release it in an aerobic environment. NO dissociation from ferrous nitrobindin is much slower (t1/2 ≈ 7 s) than from the ferric form (t1/2 ≈ 0.010 s) but is still roughly 100 to 10,000 times faster than that from most reduced globins, which might be sufficient for fighting off infections in plants. Another possible function could be rapid reduction of O2 to superoxide radicals, which can dismute to hydrogen peroxide. This generation of reactive oxygen species might also be part of a host defense mechanism against infection. These ideas need to be tested by genetic analysis of nitrobindin mutants in A. thalania.
Acknowledgments
CESG has been funded by the following NIH/NIGMS grant numbers GM074901 (John L. Markley; 07/01/05-06/30/10) and GM064598 (John L. Markley; 01/01/02-08/31/05). This work was supported by a supplement from NIH GM074901, for functional studies. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. The kinetic studies at Rice University were supported by NIH/NIGMS and NIH/NIHLB grants GM 35649 (JSO) and HL 47020 (JSO), and Grant C-612 (JSO) from the Robert A.Welch Foundation. We acknowledge the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor for the support of this research program (Grant 085P1000817). The authors would like to thank Dr. Thomas J. Rutkoski, and Dr. Craig A. Bingman for many helpful discussions. We are grateful to Dr. Richard M. Amasion and Dr. Yosuke Tamada for growing and providing assistance with interpreting results from the nitrobindin knockout plants. The authors would also like to thank all the members of CESG.
References
- 1.Gray HB, Winkler JR. Electron transfer in proteins. Annu Rev Biochem. 1996;65:537–561. doi: 10.1146/annurev.bi.65.070196.002541. [DOI] [PubMed] [Google Scholar]
- 2.Ortiz de Montellano PR. Cytochrome P450: Structure, Mechanism, and Biochemistry. New York: Plenium press; 1995. [Google Scholar]
- 3.Rodgers KR. Heme-based sensors in biological systems. Curr Opin Chem Biol. 1999;3(2):158–167. doi: 10.1016/S1367-5931(99)80028-3. [DOI] [PubMed] [Google Scholar]
- 4.Chan MK. Recent advances in heme-protein sensors. Curr Opin Chem Biol. 2001;5(2):216–222. doi: 10.1016/s1367-5931(00)00193-9. [DOI] [PubMed] [Google Scholar]
- 5.Olson JS, Phillips GN., Jr Kinetic pathways and barriers for ligand binding to myoglobin. J Biol Chem. 1996;271(30):17593–17596. doi: 10.1074/jbc.271.30.17593. [DOI] [PubMed] [Google Scholar]
- 6.Yoshikawa S. Cytochrome c oxidase. Adv Protein Chem. 2002;60:341–395. doi: 10.1016/s0065-3233(02)60057-9. [DOI] [PubMed] [Google Scholar]
- 7.Maines MD. The heme oxygenase system: a regulator of second messenger gases. Annu Rev Pharmacol Toxicol. 1997;37:517–554. doi: 10.1146/annurev.pharmtox.37.1.517. [DOI] [PubMed] [Google Scholar]
- 8.Maines MD. The heme oxygenase system: update 2005. Antioxid Redox Signal. 2005;7(11–12):1761–1766. doi: 10.1089/ars.2005.7.1761. [DOI] [PubMed] [Google Scholar]
- 9.Olson JS, Phillips GN. Myoglobin discriminates between O-2, NO, and CO by electrostatic interactions with the bound ligand. Journal of Biological Inorganic Chemistry. 1997;2(4):544–552. [Google Scholar]
- 10.Springer BA, et al. Mechanisms of Ligand Recognition in Myoglobin. Chemical Reviews. 1994;94(3):699–714. [Google Scholar]
- 11.Hargrove MS, et al. His64(E7)-->Tyr apomyoglobin as a reagent for measuring rates of hemin dissociation. J Biol Chem. 1994;269(6):4207–4214. doi: 10.2210/pdb1mgn/pdb. [DOI] [PubMed] [Google Scholar]
- 12.Ockner RK, et al. A binding protein for fatty acids in cytosol of intestinal mucosa, liver, myocardium, and other tissues. Science. 1972;177(43):56–58. doi: 10.1126/science.177.4043.56. [DOI] [PubMed] [Google Scholar]
- 13.Flower DR, North AC, Attwood TK. Structure and sequence relationships in the lipocalins and related proteins. Protein Sci. 1993;2(5):753–761. doi: 10.1002/pro.5560020507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirchhoff LV. American trypanosomiasis (Chagas' disease)--a tropical disease now in the United States. N Engl J Med. 1993;329(9):639–644. doi: 10.1056/NEJM199308263290909. [DOI] [PubMed] [Google Scholar]
- 15.Law JH, Ribeiro JM, Wells MA. Biochemical insights derived from insect diversity. Annu Rev Biochem. 1992;61:87–111. doi: 10.1146/annurev.bi.61.070192.000511. [DOI] [PubMed] [Google Scholar]
- 16.Montfort WR, Weichsel A, Andersen JF. Nitrophorins and related antihemostatic lipocalins from Rhodnius prolixus and other blood-sucking arthropods. Biochim Biophys Acta. 2000;1482(1–2):110–118. doi: 10.1016/s0167-4838(00)00165-5. [DOI] [PubMed] [Google Scholar]
- 17.Marti MA, et al. Bond or cage effect: how nitrophorins transport and release nitric oxide. J Am Chem Soc. 2008;130(5):1611–1618. doi: 10.1021/ja075565a. [DOI] [PubMed] [Google Scholar]
- 18.Jeon WB, et al. High-throughput purification and quality assurance of Arabidopsis thaliana proteins for eukaryotic structural genomics. J Struct Funct Genomics. 2005;6(2–3):143–147. doi: 10.1007/s10969-005-1908-7. [DOI] [PubMed] [Google Scholar]
- 19.Sreenath HK, et al. Protocols for production of selenomethionine-labeled proteins in 2-L polyethylene terephthalate bottles using auto-induction medium. Protein Expr Purif. 2005;40(2):256–267. doi: 10.1016/j.pep.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 20.Thao S, et al. Results from high-throughput DNA cloning of Arabidopsis thaliana target genes using site-specific recombination. J Struct Funct Genomics. 2004;5(4):267–276. doi: 10.1007/s10969-004-7148-4. [DOI] [PubMed] [Google Scholar]
- 21.Zolnai Z, et al. Project management system for structural and functional proteomics: Sesame. J Struct Funct Genomics. 2003;4(1):11–23. doi: 10.1023/a:1024684404761. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski ZM, Wladek Processing of x-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Grosse-Kunstleve RW, Adams PD. Substructure search procedures for macromolecular structures. Acta Crystallogr D Biol Crystallogr. 2003;59(Pt 11):1966–1973. doi: 10.1107/s0907444903018043. [DOI] [PubMed] [Google Scholar]
- 24.Vonrhein C, et al. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 25.Perrakis AMR, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nature Structural Biology. 1999;5:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 26.McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125(2–3):156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallographica Section D - Biological Crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. Journal of Applied Crystallography. 1997;30:1022–1025. [Google Scholar]
- 29.Cowtan PEaK. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallographica Section D - Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 4):439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski RA, et al. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 32.Lovell Simon C, W.D I, Bryan Arendall W, III, de Bakker Paul I W, Michael Word J, Prisant Michael G, Richardson Jane S, Richardson David C. Structure validation by C-alpha geometry: phi, psi, and C-beta deviation. Proteins: Structure, Function, and Genetics. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 33.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific LLC; 2002. [Google Scholar]
- 34.Krysan PJ, et al. Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc Natl Acad Sci U S A. 1996;93(15):8145–8150. doi: 10.1073/pnas.93.15.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohlfs RJ, et al. The effects of amino acid substitution at position E7 (residue 64) on the kinetics of ligand binding to sperm whale myoglobin. J Biol Chem. 1990;265(6):3168–3176. [PubMed] [Google Scholar]
- 36.Foley EW. Physiologically Relevant Reactions of Myoglobin and Hemoglobin with NO. Houston: Rice University; 2005. [Google Scholar]
- 37.Ford PC, Fernandez BO, Lim MD. Mechanisms of reductive nitrosylation in iron and copper models relevant to biological systems. Chem Rev. 2005;105(6):2439–2455. doi: 10.1021/cr0307289. [DOI] [PubMed] [Google Scholar]
- 38.Pesce A, et al. Thr-E11 regulates O2 affinity in Cerebratulus lacteus mini-hemoglobin. J Biol Chem. 2004;279(32):33662–33672. doi: 10.1074/jbc.M403597200. [DOI] [PubMed] [Google Scholar]
- 39.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372(3):774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 40.Clouaire T, et al. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc Natl Acad Sci U S A. 2005;102(19):6907–6912. doi: 10.1073/pnas.0406882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deiss LP, et al. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 1995;9(1):15–30. doi: 10.1101/gad.9.1.15. [DOI] [PubMed] [Google Scholar]
- 42.Roussigne M, et al. The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem Sci. 2003;28(2):66–69. doi: 10.1016/S0968-0004(02)00013-0. [DOI] [PubMed] [Google Scholar]
- 43.Madej T, Gibrat JF, Bryant SH. Threading a database of protein cores. Proteins. 1995;23(3):356–369. doi: 10.1002/prot.340230309. [DOI] [PubMed] [Google Scholar]
- 44.Shepard W, et al. The crystal structure of Rv0813c from Mycobacterium tuberculosis reveals a new family of fatty acid-binding protein-like proteins in bacteria. J Bacteriol. 2007;189(5):1899–1904. doi: 10.1128/JB.01435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andersen JF, et al. The crystal structure of nitrophorin 4 at 1.5 A resolution: transport of nitric oxide by a lipocalin-based heme protein. Structure. 1998;6(10):1315–1327. doi: 10.1016/s0969-2126(98)00131-2. [DOI] [PubMed] [Google Scholar]
- 46.Weichsel A, et al. Crystal structures of a nitric oxide transport protein from a blood-sucking insect. Nat Struct Biol. 1998;5(4):304–309. doi: 10.1038/nsb0498-304. [DOI] [PubMed] [Google Scholar]
- 47.Andersen JF, et al. Kinetics and equilibria in ligand binding by nitrophorins 1–4: evidence for stabilization of a nitric oxide-ferriheme complex through a ligand-induced conformational trap. Biochemistry. 2000;39(33):10118–10131. doi: 10.1021/bi000766b. [DOI] [PubMed] [Google Scholar]
- 48.Maes EM, et al. Ultrahigh resolution structures of nitrophorin 4: heme distortion in ferrous CO and NO complexes. Biochemistry. 2005;44(38):12690–12699. doi: 10.1021/bi0506573. [DOI] [PubMed] [Google Scholar]
- 49.Nienhaus K, et al. Structural dynamics controls nitric oxide affinity in nitrophorin 4. J Biol Chem. 2004;279(38):39401–3407. doi: 10.1074/jbc.M406178200. [DOI] [PubMed] [Google Scholar]
- 50.Traylor TG, Berzinis AP. Binding of O2 and CO to hemes and hemoproteins. Proc Natl Acad Sci U S A. 1980;77(6):3171–3175. doi: 10.1073/pnas.77.6.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brantley RE, Jr, et al. The mechanism of autooxidation of myoglobin. J Biol Chem. 1993;268(10):6995–7010. [PubMed] [Google Scholar]
- 52.Hargrove MS, et al. Quaternary structure regulates hemin dissociation from human hemoglobin. J Biol Chem. 1997;272(28):17385–17389. doi: 10.1074/jbc.272.28.17385. [DOI] [PubMed] [Google Scholar]
- 53.Hargrove MS, Wilkinson AJ, Olson JS. Structural factors governing hemin dissociation from metmyoglobin. Biochemistry. 1996;35(35):11300–11309. doi: 10.1021/bi960372d. [DOI] [PubMed] [Google Scholar]
- 54.Hargrove MS, Olson JS. The stability of holomyoglobin is determined by heme affinity. Biochemistry. 1996;35(35):11310–11318. doi: 10.1021/bi9603736. [DOI] [PubMed] [Google Scholar]
- 55.Thorsteinsson MV, et al. A cyanobacterial hemoglobin with unusual ligand binding kinetics and stability properties. Biochemistry. 1999;38(7):2117–2126. doi: 10.1021/bi9819172. [DOI] [PubMed] [Google Scholar]
- 56.Alben JO, Caughey WS. An infrared study of bound carbon monoxide in the human red blood cell, isolated hemoglobin, and heme carbonyls. Biochemistry. 1968;7(1):175–183. doi: 10.1021/bi00841a022. [DOI] [PubMed] [Google Scholar]
- 57.Balasubramanian S, Lambright DG, Boxer SG. Perturbations of the distal heme pocket in human myoglobin mutants probed by infrared spectroscopy of bound CO: correlation with ligand binding kinetics. Proc Natl Acad Sci U S A. 1993;90(10):4718–4722. doi: 10.1073/pnas.90.10.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cameron AD, et al. Distal pocket polarity in ligand binding to myoglobin: deoxy and carbonmonoxy forms of a threonine68(E11) mutant investigated by X-ray crystallography and infrared spectroscopy. Biochemistry. 1993;32(48):13061–13070. doi: 10.1021/bi00211a016. [DOI] [PubMed] [Google Scholar]
- 59.Li T, et al. Structural determinants of the stretching frequency of CO bound to myoglobin. Biochemistry. 1994;33(6):1433–1446. doi: 10.1021/bi00172a021. [DOI] [PubMed] [Google Scholar]
- 60.Phillips GN, et al. Bound CO is a molecular probe of electrostatic potential in the distal pocket of myoglobin. Journal of Physical Chemistry B. 1999;103(42):8817–8829. [Google Scholar]
- 61.Shimada H, Caughey WS. Dynamic protein structures. Effects of pH on conformer stabilities at the ligand-binding site of bovine heart myoglobin carbonyl. J Biol Chem. 1982;257(20):11893–11900. [PubMed] [Google Scholar]
- 62.Moore EG, Gibson QH. Cooperativity in the dissociation of nitric oxide from hemoglobin. J Biol Chem. 1976;251(9):2788–2794. [PubMed] [Google Scholar]
- 63.Andersen JF, Montfort WR. The crystal structure of nitrophorin 2. A trifunctional antihemostatic protein from the saliva of Rhodnius prolixus. J Biol Chem. 2000;275(39):30496–30503. doi: 10.1074/jbc.M002857200. [DOI] [PubMed] [Google Scholar]
- 64.Lecomte JT, Vuletich DA, Lesk AM. Structural divergence and distant relationships in proteins: evolution of the globins. Curr Opin Struct Biol. 2005;15(3):290–301. doi: 10.1016/j.sbi.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 65.Lesk AM, Chothia C. How different amino acid sequences determine similar protein structures: the structure and evolutionary dynamics of the globins. J Mol Biol. 1980;136(3):225–270. doi: 10.1016/0022-2836(80)90373-3. [DOI] [PubMed] [Google Scholar]
- 66.Brancaccio A, et al. Structural factors governing azide and cyanide binding to mammalian metmyoglobins. J Biol Chem. 1994;269(19):13843–13853. [PubMed] [Google Scholar]
- 67.Ding XD, Weichsel A, Andersen JF, Shokhireva TKh, Balfour C, Pierik AJ, Averill BA, Montfort WR, Walker FA. Nitric Oxide Binding to the Ferri- and Ferroheme States of Nitrophorin 1, a Reversible NO-Binding Heme Protein from the Saliva of the Blood-Sucking Insect, Rhodnius prolixus. J. Am. Chem. Soc. 1999;121(1):128–138. [Google Scholar]
- 68.Balendiran GK, et al. Crystal structure and thermodynamic analysis of human brain fatty acid-binding protein. J Biol Chem. 2000;275(35):27045–27054. doi: 10.1074/jbc.M003001200. [DOI] [PubMed] [Google Scholar]
- 69.Coe NR, Bernlohr DA. Physiological properties and functions of intracellular fatty acid-binding proteins. Biochim Biophys Acta. 1998;1391(3):287–306. doi: 10.1016/s0005-2760(97)00205-1. [DOI] [PubMed] [Google Scholar]
- 70.Kondrashov DA, Montfort WR. Nonequilibrium dynamics simulations of nitric oxide release: comparative study of nitrophorin and myoglobin. J Phys Chem B. 2007;111(31):9244–9252. doi: 10.1021/jp071136n. [DOI] [PubMed] [Google Scholar]
- 71.Maes EM, et al. Role of binding site loops in controlling nitric oxide release: structure and kinetics of mutant forms of nitrophorin 4. Biochemistry. 2004;43(21):6679–6690. doi: 10.1021/bi049748a. [DOI] [PubMed] [Google Scholar]
- 72.Dangl J. Innate immunity. Plants just say NO to pathogens. Nature. 1998;394(6693):525–527. doi: 10.1038/28958. [DOI] [PubMed] [Google Scholar]
- 73.Ma W, et al. Innate immunity signaling: cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiol. 2008;148(2):818–828. doi: 10.1104/pp.108.125104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wendehenne D, Durner J, Klessig DF. Nitric oxide: a new player in plant signalling and defence responses. Curr Opin Plant Biol. 2004;7(4):449–455. doi: 10.1016/j.pbi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 75.Hargrove MS, et al. Characterization of recombinant soybean leghemoglobin a and apolar distal histidine mutants. J Mol Biol. 1997;266(5):1032–1042. doi: 10.1006/jmbi.1996.0833. [DOI] [PubMed] [Google Scholar]
- 76.Eich RF, et al. Mechanism of NO-induced oxidation of myoglobin and hemoglobin. Biochemistry. 1996;35(22):6976–6983. doi: 10.1021/bi960442g. [DOI] [PubMed] [Google Scholar]
- 77.Herold S, Puppo A. Kinetics and mechanistic studies of the reactions of metleghemoglobin, ferrylleghemoglobin, and nitrosylleghemoglobin with reactive nitrogen species. J Biol Inorg Chem. 2005;10(8):946–957. doi: 10.1007/s00775-005-0047-8. [DOI] [PubMed] [Google Scholar]