Abstract
Phospholipids are key components of cellular membrane and signaling. Among cellular phospholipids, phosphoinositides, phosphorylated derivatives of phosphatidylinositol are important as a participant in essential metabolic processes in animals. However, due to its low abundance in cells and tissues, it is difficult to identify the composition of phosphoinositides. Recent advances in mass spectrometric techniques, combined with established separation methods, have allowed the rapid and sensitive detection and quantification of a variety of lipid species including phosphoinositides. In this mini review, we briefly introduce progress in profiling of cellular phosphoinositides using mass spectrometry. We also summarize current progress of matrices development for the analysis of cellular phospholipids using matrix-assisted laser desorption/ionization mass spectrometry. The phosphoinositides profiling and phospholipids imaging will help us to understand how they function in a biological system and will provide a powerful tool for elucidating the mechanism of diseases such as diabetes, cancer and neurodegenerative diseases. The investigation of cellular phospholipids including phosphoinositides using electrospray ionization mass spectrometry and matrix-assisted laser desorption/ionization mass spectrometry will suggest new insights on human diseases, and on clinical application through drug development of lipid related diseases.
Keywords: lipids; lipid metabolism disorders; phospholipids; phosphatidylinositols; spectrometry, mass, electrospray ionization; spectrometry, mass, matrix-assisted laser desorption-ionization
Introduction
Lipids are an important class of cellular components and play diverse roles in human physiology. Although they are important, the study of lipids has been limited by analytical difficulties. However, recently improved analytical methods, such as mass spectrometry (MS) and liquid chromatography (LC) provide systems-level investigation in lipid research (Wenk, 2005). This systems-level analysis of lipids (lipidomics) is a growing field with different applications in drug and biomarker development. There are various efforts to explore abnormal lipid metabolism in human diseases, such as insulin-resistant diabetes, Alzheimer's disease, schizophrenia, cancer, atherosclerosis, and infectious diseases through lipidomics approach using mass spectrometry (Oresic et al., 2008). Lipidomics will contribute to drug development for pharmaceutical therapy of lipid related diseases through mostly presenting cellular lipid profile and lipid distribution in a tissue.
Electrospray ionization (ESI)-MS was initially developed by Fenn and colleagues for analysis of biomolecules (Fenn et al., 1989). It depends on the formation of gaseous ions from polar, thermally labile and mostly non-volatile molecules. The "soft" electrospray ionization causes little or no fragmentation and virtually all phospholipid species are detected as molecular ion species. The identification and structural information about a certain peak is acquired by fragmentation (tandem mass spectrometry, MS/MS) (Han and Gross, 1994; Kim et al., 1994). Advances in mass spectrometry (MS) have created powerful new avenues for the measurement of global changes in the cellular lipid inventory.
Electrospray ionization (ESI) mass spectrometry has been successfully used to analyze lipids in various cell types. However, for less abundant and more complicated chemical compositions, HPLC is needed for upfront separation before ionization by ESI and detection by MS (Watson, 2006). In the field of lipidomics HPLC or TLC have been coupled with electrospray ionization (ESI)/atmospheric pressure chemical ionization (APCI)-MS and matrix-assisted laser desorption/ionization (MALDI)-MS, respectively (Wenk, 2005). These methods provide the identification of lipid composition and quantification of cellular lipids, such as fatty acids, phospholipids, cholesterols, and sphingolipids.
The identification of lipid composition and quantification of cellular lipids provide molecular signature for a lipid related pathway. One of the most obvious applications of lipidomics will include profiling lipid extracts in order to identify metabolic pathways and enzymes that are affected (Wenk, 2005). A major advantage of such lipidomics-based discovery is that, together with our relatively good understanding of many biosynthetic and metabolic pathways, it will lead to the identification of pathways and enzymes governing human diseases (Cascante et al., 2002). A recent review (Wenk, 2005) well summarized diverse aspects and fundamental principles of lipidomics.
Among lipidomics technologies, phosphoinositide profiling using electrospray ionization mass spectrometry (ESI-MS) and phospholipid analysis using matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) are quite challenging due to their technical difficulties although they could suggest more information on lipid related diseases. In these fields, several attempts have been reported in order to beat our technical limitation. We are bringing out the technical progress of cellular phosphoinositide profiling with ESI-MS and phospholipid analysis using MALDI-MS in this review. A brief suggestion on clinical applications of phosphoinositide profiling and phospholipid analysis using mass spectrometry is additionally given.
Profiling of cellular phosphoinositides using ESI mass spectrometry
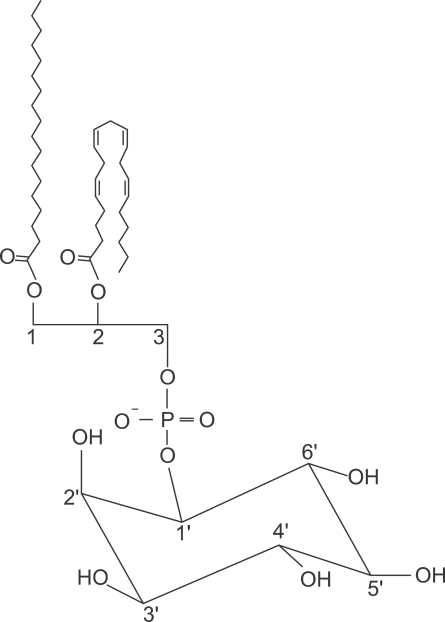
Phosphoinositides are phospholipids with an inositol head groups. They have a central position in the fields of cell signaling and regulation, including calcium homeostasis, membrane trafficking and cytoskeletal dynamics (Di Paolo and De Camilli, 2006). They are able to achieve signaling effects directly by binding to cytosolic proteins or cytosolic domains of membrane proteins via their polar head groups (Figure 1). In this way, they can regulate the function of proteins integral to membranes, or they can attract cytoskeletal and signaling components to the membrane (Di Paolo and De Camilli, 2006).
Figure 1.
A phosphatidylinositol structure. This figure is originated from http://en.wikipedia.org/wiki/File:Phosphatidylinositol.jpg. The hydroxyl moieties at 3', 4', and 5' in the head group can be phosphorylated to produce seven phosphoinositides, phosphatidylinositol 3-phosphate (PI3P), phosphatidylinositol 4-phosphate (PI4P), phosphatidylinositol 5-phosphate (PI5P), phosphatidylinositol (3,4)-bisphosphate (PI(3,4)P2), phosphatidylinositol (3,5)-bisphosphate (PI(3,5)P2), phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2), and phosphatidylinositol (3,4,5)-trisphosphate (PI(3,4,5)P3, PIP3).
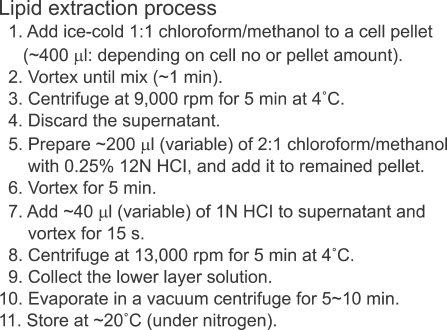
Phosphoinositides are less abundant lipids in eukaryotic cell membranes. Therefore, analysis of phosphoinositides requires special cautions during sample treatment. According to several reports (Hsu and Turk, 2000; Wenk et al., 2003; Milne et al 2005; Shui et al., 2007) to minimize the interference of other phospholipids during analysis, a selective two-step extraction was used (Scheme 1). First, the cell material was extracted with neutral solvents (mixture of chloroform (CHCl3) and methanol (MeOH)) and the resulting pellet was extracted with acidified solvents (containing HCl or citric acid) for quantitative recovery of phosphoinositides.
Scheme 1.
A two-step extraction for the analysis of phosphoinositides from a cell material. This process is slightly modified from the method in Wenk et al., 2003.
After extraction of phosphoinositides, HPLC and TLC separation have been employed in order to classify molecular species such as phosphatidylinositol (PI), phosphatidylinositol 3-phosphate (PI3P), phosphatidylinositol 4-phosphate (PI4P), phosphatidylinositol 5-phosphate (PI5P), phosphatidylinositol (3,4)-bisphosphate (PI(3,4)P2), phosphatidylinositol (3,5)-bisphosphate (PI(3,5)P2), phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2), and phosphatidylinositol (3,4,5)-trisphosphate (PI(3,4,5)P3, PIP3). Traditionally, the molecular species have been detected, identified and quantified by autoradiograph (Wenk et al., 2003). However, the identification based on radio-labeling could not provide information of fatty acyl composition of phosphoinositides that can be solved by the application of modernized mass spectrometry methods. Current mass spectrometry is able to analyze lipid extracts of phosphoinositides without prior separation through HPLC or TLC due to its high resolving power.
Several methods such as fast atom bombardment mass spectrometry (FAB-MS), matrix-assisted laser desorption and ionization/time of flight mass spectrometry (MALDI-TOF-MS) and ESI-MS have been used for the analysis of phosphoinositides. MALDI-TOF-MS has been recently applied for phosphatidylinositols in murine brain extracts (Berry et al., 2004) and ESI-MS has been used for PIPs and PIP2s in lipid extracts from cultures cells and rat brain (Wenk et al., 2003). ESI-MS has also been utilized for PIPs, PIP2s and PIP3s in murine macrophage extracts (Milne et al., 2005). These biological applications of ESI-MS can provide us of understanding of the acyl chain contents of phosphoinositides due to its technical advance and advantage. The fatty acyl composition of phosphoinositides is assigned by precursor ion scan or fragmentation analysis in ESI-MS in negative ion mode. Among these applications for biological samples, a report (Milne et al., 2005) for the analysis of phosphoinositides in murine macrophage cells presented the identification of 28 PIP and PIP2 compounds as well as 8 PIP3 compounds. Additionally, fatty acid composition of lyso-phosphoinositides in plasma from patients with ovarian cancer was identified through the application of ESI-MS (Sutphen et al., 2004). According to a report (Milne et al., 2005), the limit of detection using ESI-MS is < 9 pmol/ml for 38:4 PIP3.
In general, single-stage ESI-MS is enough to investigate the fatty acid composition of relatively abundant phosphoinositides from biological samples. The inspection of samples using tandem mass spectrometer (MS/MS) combined with collision-induced dissociation (CID), can identify and quantify less abundant phosphoinositides with their fatty acid composition and degree of unsaturation (Wenk et al., 2003). MS/MS can monitor transition from a parent ion to a product ion of small quantity of phosphoinositides and provide increasing sensitivity.
In the analysis of phosphoinositides using MS, we need to consider suppression effect of ion signal of phosphoinositides by other components in lipid extracts. In order to solve this effect, they also presented the effect of the addition of piperidine onto cellular extracts on the change of signal to noise (S/N) ratio (Wenk et al., 2003; Milne et al., 2005). This addition of a organic buffer, piperidine results in enhancement of ion signal of phosphoinositides, similarly as in the analysis of phosphopeptides and oligonucleotides.
Using these methods, there are several investigations on detailed analysis of cellular phosphoinositides. They identified approximately 75 species of phosphoinositides with detailed fatty acyl chain composition and profiled them from mouse brain tissue, yeast mutant cells (Wenk et al., 2003), RAW 264.7 cell, primary murine peritoneal macrophage (Milne et al., 2005) and Mycobaterium bovis (Shui et al., 2007). Interestingly, they detected elevated concentrations of PIP2 in human fibroblasts from patients with Lowe syndrome (Wenk et al., 2003). Lowe syndrome is a genetic disorder characterized by bilateral congenital cataracts, renal Fanconi syndrome, and mental retardation. It is coming from mutations of a phosphoinositide 5-phosphatase. Thus, it results in the disruption of phosphoinositide metabolism and increment of PIP2 concentrations in human cells.
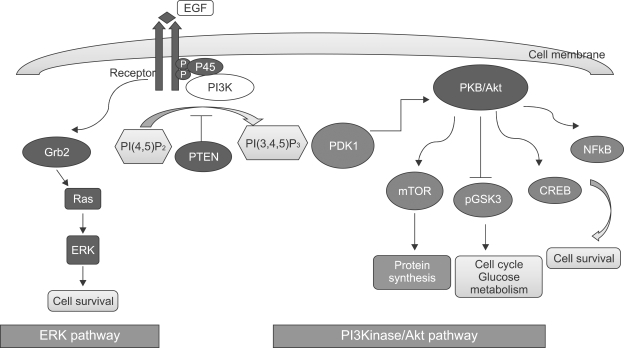
In order to suggest the significance of phosphoinositides profiling, we shortly summarize cellular signaling mechanism of phosphoinositides. The various phosphoinositides are maintained at steady state levels in the inner leaflet of the plasma membrane by a continuous and sequential series of phosphorylation and dephosphorylation reactions by specific kinases and phosphatases, respectively, which are regulated and/or relocated through cell surface receptors for extracellular ligands (Di Paolo and De Camilli, 2006). This has been termed a 'futile cycle', and can consume a significant proportion of cellular ATP production. Controlled synthesis of these different phosphoinositides can occur in different intracellular compartments for distinct and independently regulated functions with differing target enzymes (Roth, 2004). Among the enzymes relating to phosphoinositides phosphorylation and dephosphorylation, phosphoinositide 3-kinase (PI3K) related pathway is well characterized (Vivanco and Sawyers, 2002). The lipid kinase, PI3K is involved in the regulation of a number of cellular processes, like transcription, migration, angiogenesis, cell growth, proliferation, apoptosis and glucose metabolism. PI3K phosphorylates phosphatidylinositols of the cell membrane, thus generating, e.g., phosphatidylinositol-3,4,5-trisphosphate (PIP3) from phosphatidylinositol 4,5-bisphosphate (PIP2). PIP3 at the cell membrane recruits protein kinases such as protein kinase B or Akt (PKB/Akt) and phosphoinositide-dependent kinase-1 (PDK1) which bind with their PH-(pleckstrin homology)-domain to PIP3. The downstream signaling of PI3K phosphorylate a wide variety of cellular signaling molecules relevant for the regulation of cell growth, cell cycle and cell proliferation, for apoptosis, or for transport (Figure 2). PKB/Akt further activates mammalian target of rapamycin (mTOR), which is a kinase stimulating the uptake of nutrients such as glucose, amino acids, cholesterol and iron. mTOR further activates eIF4E-binding protein-1 and thus is involved in the regulation of translation (Pitt and Chen, 2008). Besides, phosphoinositides are key components of the nucleus of the cell, where they have many essential functions, such as DNA repair, transcription regulation and RNA dynamics (Hammond et al., 2004). They are considered as activity switches responsible for such processes, with the phosphorylation state of the inositol ring. Different forms of phosphoinositides appear to have specific functions at each level of gene expression, so extracellular events must coordinate the production of these compounds in a highly organized manner (Han and Gross, 2005).
Figure 2.
A cellular signaling mechanism of phosphoinositides.
As presented above, phosphoinositides are involved in many cellular processes through their kinase and phosphatase. Therefore, the profiling of phosphoinositides will lead us to understand the extraordinary roles of phosphoinositides relevant to major human diseases, including cancer and diabetes, making them important targets for clinical application and intervention. Although they have huge potentials on pharmacological application, profiling of phosphoinositides has been applied for elucidation of several host-pathogen interactions (Hernandez et al., 2004). Thus far, there are few trials using phosphoinositides profiling by mass spectrometry. Further investigation on different application would be mandatory in near future with clinical advantages of phosphoinositides profiling.
Phospholipid analysis in MALDI mass spectrometry
The analyses of cellular components, such as protein, peptide, and metabolites in several tissues using MALDI-MS have been developed. However, the lipid identification using MALDI-MS has been limited due to less existence of proper matrix for lipid identification. Therefore, we mentioned here about some efforts to find a proper matrix for phospholipid imaging.
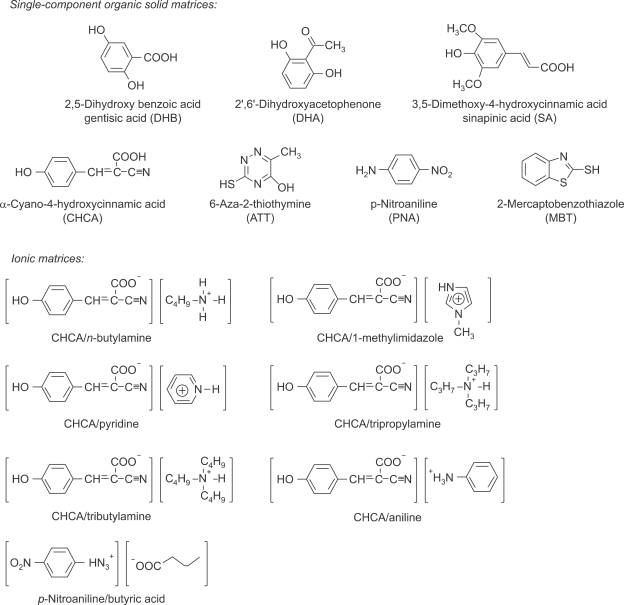
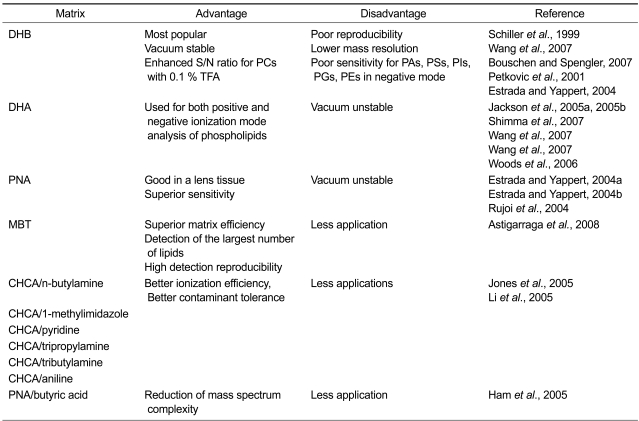
The choice of matrix, one of important issues for MALDI-MS, is not simple for in vitro and in situ lipid analysis due to the special properties of lipids, including lower molecular weights (<1,500 Da), rather wide molecular diversity, very close presented m/z ratio, relative insolubility in aqueous systems and the obvious concentration difference within a crude sample. Various matrices had been screened and suggested to be useful for phospholipid analysis (Figure 3, Table 1). Besides the low chemical noise and interferences from matrix relevant ions, the important prerequisite of a matrix for lipid analysis are namely, good absorptivity at the given laser wavelength, good solubility in analyte solvents, suitable acidity and basicity and high ionization efficiency of analyte molecules.
Figure 3.
Structures and names of organic matrices tested useful for analysis of phospholipids by MALDI-MS.
Table 1.
Advantages and disadvantages of organic matrices in the analysis of phospholipids by MALDI-MS.
DHA, 2,6-dihydroxyacetophenone; MBT, 2-mercaptobenzothiazole; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PNA, p-nitroaniline; PS, phosphatiylserine.
Among the matrices shown in Figure 3, 2,5-dihydroxybenzoic acid (DHB), so far, is regarded as the most suitable for lipid analysis in rat brain and neutrophilic granulocytes because it can produce cleaner spectra and fewer fragments of analyte ions (Schiller et al., 1999). Furthermore relevant matrix peaks do not interfere with analyte identification (Petkovic et al., 2001). Additionally, suitable vacuum stability of DHB is a key factor for its popular application in imaging mass spectrometry (IMS) of lipids (Wang et al., 2007). When used for phospholipids analysis in mouse brain, rat brain, mouse cerebellum, mouse retina, single zooplankter individuals, and lipid mixtures extracted from tissues, matrix solution can be prepared with a number of solvent system, such as ethanol/water (1:1, v/v with or without 0.1% trifluoroacetic acid (TFA)) (Schwartz et al., 2003; Jackson et al., 2005b; Woods et al., 2006; Puolitaival et al., 2008) ethanol/water (9:1, v/v, with or without 0.1% TFA) (Fujiwaki et al., 2002) acetonitrile/water (1:1, v/v, with 0.1% TFA) (Chen et al., 2008) methanol/water (7:3, v/v) (Hayasaka et al., 2008; Shimma et al., 2008) and chloroform/methanol (2:1 v/v ) (Ishida et al., 2003). Addition of small amounts of TFA in the matrix solution was found to enhance considerably signal to noise (S/N) ratio in the case of phosphatidylcholines and phosphoinositides, most likely due to enhanced solubility of lipids (Schiller et al., 1999; Petkovic et al., 2001).
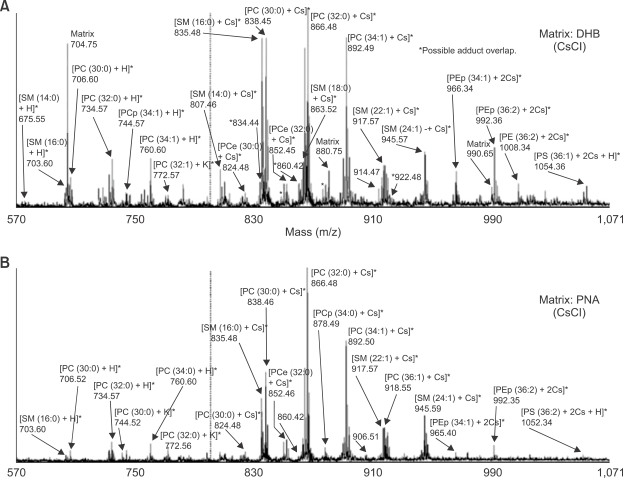
One obvious drawback of DHB as MALDI matrix is to its tendency to form large crystals, where the analyte is not evenly distributed, leading to poor spot-to-spot reproducibility and lowering mass resolution (Bouschen and Spengler, 2007). DHB has some disadvantages for phospholipid analysis as shown in Figure 4. Due to its relatively higher acidity, DHB produced weak or no signal intensity and exhibited poor sensitivity for phospholipids, especially for phosphatidic acids (PAs), phosphatiylserines, phosphatidylinositols, phosphatidylglycerols, phosphatidylethanolamines in negative-ion mode (Petkovic et al., 2001; Estrada and Yappert, 2004a).
Figure 4.
Positive-ion mass spectra of phospholipids extracted from the cortical region of porcine lenses, (A) matrix: 0.5 M DHB in methanol after addition of CsCl crystals, (B) matrix: 0.17 M p-nitroaniline in chloroform/methanol=2/1 after addition of CsCl crystals. (Adapted from Estrada and Yappert, 2004b, with slight modifications). The appearance of some matrix-related peaks (m/z 704.75, 880.75 and 990,65) from DHB in (A) increased the complexity of the spectrum. Because of the characteristic of p-nitroaniline to favor the ionization of phosphatidylcholines with respect to other phospholipids, including sphingomyelins, it is much easier to assign some phosphatidylcholine-related peaks (labeled with asterisks in (A)) using p-nitroaniline, instead of DHB, as matrix to ionize CsCl-treated samples. The decrease in relative intensity of non-phosphatidylcholine related positive-ion peaks when p-nitroaniline is used as matrix can be used as a peak assignment criterion to resolve ambiguous assignments. PC, phosphatidylcholine; SM, sphingomyelin.
Several neutral, basic or low acidic compounds were also tested their suitability for MALDI-MS analysis of phospholipids (Estrada and Yappert, 2004b; Jackson et al., 2005a; Woods et al., 2006; Wang et al., 2007) to alleviate difficulties in the detection and identification of phospholipid classes with ionization efficiencies lower than those of sphingomyelins and phosphatidylcholines and to improve the sensitivity of negative-ion mass spectra. Among the matrices shown in Figure 3, 2,6-dihydroxyacetophenone is one of most employed matrices. 2,6-Dihydroxyacetophenone in 50-70% ethanol or methanol with or without 0.1%TFA is commonly used for lipid analysis (Jackson et al., 2005a; Shimma et al., 2007). Cesium iodide (Wang et al., 2007) and lithium chloride (Jackson et al., 2005a) can be added. 2,6-Dihydroxyacetophenone can be used for both positive and negative ionization mode analysis of phospholipids (Woods et al., 2006; Jackson et al., 2007; Wang et al., 2007), and works more efficient than DHB in terms of detection sensitivity, resolution and signal-to-noise of mass spectra. 2,6-Dihydroxyacetophenone and another neutral matrix of 6-aza-2-thiothymine (ATT) also can be used to detect noncovalent complexes between chlorisondamine and phosphatidylcholine molecular species phosphatidylcholine32:0 and phosphatidylcholine34: 1 on a rat brain tissue added with 1 nmol of chlorisondamine. Noncovalent complex formation was not observed when highly acidic matrices such as sinapinic acid (SA) and cyano-4-hydroxycinnamic acid (CHCA) were used (Jackson et al., 2005a). p-Nitroaniline proved to be another good option for the use of DHB for MALDI-MS analysis of phospholipid in a lens tissue (Figure 4). Due to its higher absorptivity and its lower acidity than that of DHB, p-nitroaniline provided superior sensitivity for analysis of phospholipids either positive-ion or negative-ion modes and increased the relative amount of deprotonated species in the negative-ion mode than DHB (Estrada and Yappert, 2004a, 2004b). Therefore, it was possible to confirm peak assignments for phospholipid classes (phosphatidylglycerols and phosphatiylseriness) that normally give weak signals when DHB is used. p-Nitroaniline allowed the identification of phosphatidylethanolamines and phosphatidylethanolamines plasmalogens (PEps) even in mixtures containing sphingomyelins and phosphatidylcholines (Estrada and Yappert, 2004b). However, p-nitroaniline had the same drawback as 2,6-dihydroxyacetophenone, a high vapor pressure. Although addition of CsCl in p-nitroaniline solution can minimize matrix (Rujoi et al., 2004), p-nitroaniline is precluded in MALDI-MS of phospholipids due to poor vacuum stability.
Most recently, 2-mercaptobenzothiazole as an advantageous alternative to the use of DHB for MALDI-MS of phospholipids in a brain and liver tissue was developed (Astigarraga et al, 2008). Several special features of 2-mercaptobenzothiazole allows its superior matrix efficiency to that of the commonly used matrices of DHB, 2,6-dihydroxyacetophenone and p-nitroaniline for profiling and imaging phospholipids in liver and brain samples both in vitro and in situ, directly on tissue slices. Its low vapor pressure allows acquisition times of hours, which makes it possible to create well-defined mass images by scanning the whole tissue slices. Its low acidity allows identification of several additional species in negative-ion mode and enables the detection of the largest number of lipids from liver and brain samples. The formation of small and uniform crystals of 2-mercaptobenzothiazole in the IMS sample preparation is a paramount factor for the achievement of good spatial resolution and uniform spectra. Its crystallization in very small and homogeneous crystals also allows high detection reproducibility.
Some ionic liquid or solid matrices have exhibited some advantages over traditional single solid matrixes such as DHB for in vitro or in situ phospholipid analysis by MALDI-MS in a brain tissue and biological fluids. Due to their notable features of strong absorptivity, low vacuum volatility (negligible vapor pressure), promote ionization and homogeneous solution, CHCA-based ionic matrices, such as CHCA/n-butylamine (nBA-CHCA) (Jones et al., 2005; Li, et al., 2005), CHCA/1-methylimidazole, CHCA/pyridine, CHCA/tripropylamine, CHCA/tributylamine and CHCA/aniline (Li et al., 2005), have been reported to give better quality data in terms of ionization efficiency, signal intensities, sensitivity, resolution, signal reproducibility, number of compounds detected, and contaminant tolerance. The utilization of p-nitroaniline-based solid ionic matrix, p-nitroaniline/butyric acid, can reduce mass spectrum complexity by reliably appearing of only protonated molecules [M + H]+ of lipids containing phosphatidylcholine head groups, such as lysophosphatidylcholine, phosphatidylcholine, and platelet-activating factor (PAF) and of monosodium adducts [M + Na]+ as the major molecular ions for anionic phospholipids, such as phosphatidylglycerol, PA, and phosphatiylserine in the mass spectra (Ham et al., 2005). Another feature of p-nitroaniline/butyric acid is its ability to simultaneously detect phosphorylated lipids of phosphatidylcholines, sphingomyelins, PAFs and phosphatidylethanolamines in the positive-ion mode.
As summarized above, there are several efforts to analyze phospholipid profiles in several tissues using MALDI-MS. Even though current MALDI-MS has certain technical drawbacks, it will become more reliable methodology for phospholipid imaging sooner or later because a lot of trials to conquer the limitations are currently in progress. Therefore, phospholipid imaging by MALDI-MS could be located in a central position of pharmacological investigation of lipid related diseases, such as diabetes, cancer, and obesity.
Conclusion
A full understanding of biological models of cell signaling requires detailed knowledge of lipid composition. We are having the ability to assess the metabolic dynamics of hundreds of phospholipid species including phosphoinositides through development of mass spectrometry. The changes of phosphoinositides composition by various agonists in a cellular signaling are subtle and under the tight control, so phosphoinositide profiling requires very sensitive measurement and dependable quantification. This requirement might be reached by current development of mass spectrometry. Therefore, a clinical application of current methodology of phosphoinositides profiling is demanded for evaluating relationships among multiple phosphoinositide classes and species in conjunction with other cellular functions.
As one of efforts in the field of lipidomics, phospholipids imaging by MALDI mass spectrometry will provide distribution of phospholipids in several tissues of animals. The pattern data of lipid organization including phospholipids are informative because it will suggest lipid composition affecting pathologic states in animal tissues. Furthermore, therapeutics aimed at interfering with lipid metabolism will become more focused. Therefore, more clinical applications using MALDI imaging mass spectrometry of lipid profiling will be expected.
Acknowledgements
This work was supported by the faculty research fund of Konkuk University in 2008.
Abbreviations
- APCI
atmospheric pressure chemical ionization
- ATT
6-aza-2-thiothymine
- CHCA
cyano-4-hydroxycinnamic acid
- CHCl3
chloroform
- CID
collision-induced dissociation
- DHB
2,5-dihydroxybenzoic acid
- eIF4E
eukaryotic translation initiation factor 4E
- ESI-MS
electrospray ionization mass spectrometry
- FAB-MS
fast atom bombardment mass spectrometry
- IMS
imaging mass spectrometry
- LC
liquid chromatography
- MALDI-MS
matrix-assisted laser desorption/ionization mass spectrometry
- MALDI-TOF-MS
matrix-assisted laser desorption and ionization/time of flight mass spectrometry
- MeOH
methanol
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- mTOR
mammalian target of rapamycin
- PA
phosphatidic acids
- PAF
platelet-activating factor
- PDK1
phosphoinositide-dependent kinase-1
- PEps
phosphatidylethanolamine plasmalogens
- PH
pleckstrin homology
- PI3K
phosphoinositide 3-kinase
- PIP
phosphatidylinositol phosphate
- PIP2
phosphatidylinositol bisphosphate
- PIP3
phosphatidylinositol-3,4,5-trisphosphate
- PKB/Akt
protein kinase B or Akt
- S/N
signal to noise
- SA
sinapinic acid
- TFA
trifluoroacetic acid
- TLC
thin layer chromatography
References
- 1.Astigarraga E, Gómez GB, Lombardero L, Gresnedo O, Castaño F, Giralt MT, Ochoa B, Puertas RR, Fernandez JA. Profiling and Imaging of Lipids on Brain and Liver Tissue by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Using 2-Mercaptobenzothiazole as a Matrix. Anal Chem. 2008;80:9105–9114. doi: 10.1021/ac801662n. [DOI] [PubMed] [Google Scholar]
- 2.Berry GT, Buccafusca R, Greer JJ, Eccleston E. Phosphoinositide deficiency due to inositol depletion is not a mechanism of lithium action in brain. Mol Genet Metab. 2004;82:87–92. doi: 10.1016/j.ymgme.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Bouschen W, Spengler B. Artifacts of MALDI sample preparation investigated by high-resolution scanning microprobe matrix-assisted laser desorption/ionization (SMALDI) imaging mass spectrometry. Int J Mass Spectrom. 2007;266:129–137. [Google Scholar]
- 4.Cascante M, Boros LG, Comin-Anduix B, de Atauri P, Centelles JJ, Lee PW. Metabolic control analysis in drug discovery and disease. Nat Biotechnol. 2002;20:243–249. doi: 10.1038/nbt0302-243. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Allegood J, Liu Y, Wang E, Cachon-Gonzalez B, Cox TM, Merrill AH, Jr, Sullards MC. Imaging MALDI mass spectrometry using an oscillating capillary nebulizer matrix coating system and its application to analysis of lipids in brain from a mouse model of Tay-Sachs/Sandhoff disease. Anal Chem. 2008;80:2780–2788. doi: 10.1021/ac702350g. [DOI] [PubMed] [Google Scholar]
- 6.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 7.Estrada R, Yappert MC. Alternative approaches for the detection of various phospholipid classes by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Mass Spectrom. 2004a;39:412–422. doi: 10.1002/jms.603. [DOI] [PubMed] [Google Scholar]
- 8.Estrada R, Yappert MC. Regional phospholipid analysis of porcine lens membranes by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Mass Spectrom. 2004b;39:1531–1540. doi: 10.1002/jms.759. [DOI] [PubMed] [Google Scholar]
- 9.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 10.Fujiwaki T, Yamaguchi S, Tasaka M, Sakura N, Taketomi T. Application of delayed extraction-matrix-assisted laser desorption ionization time-of-flight mass spectrometry for analysis of sphingolipids in pericardial fluid, peritoneal fluid and serum from Gaucher disease patients. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;776:115–123. doi: 10.1016/s1570-0232(02)00177-0. [DOI] [PubMed] [Google Scholar]
- 11.Ham BM, Jacob JT, Cole RB. MALDI-TOF MS of phosphorylated lipids in biological fluids using immobilized metal affinity chromatography and a solid ionic crystal matrix. Anal Chem. 2005;77:4439–4447. doi: 10.1021/ac058000a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammond G, Thomas CL, Schiavo G. Nuclear phosphoinositides and their functions. Curr Top Microbiol Immunol. 2004;282:177–206. doi: 10.1007/978-3-642-18805-3_7. [DOI] [PubMed] [Google Scholar]
- 13.Han X, Gross RW. Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids. Proc Natl Acad Sci USA. 1994;91:10635–10639. doi: 10.1073/pnas.91.22.10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han X, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 15.Hayasaka T, Goto-Inoue N, Sugiura Y, Zaima N, Nakanishi H, Ohishi K, Nakanishi S, Naito T, Taguchi R, Setou M. Matrix-assisted laser desorption/ionization quadrupole ion trap time-of-flight (MALDI-QIT-TOF)-based imaging mass spectrometry reveals a layered distribution of phospholipid molecular species in the mouse retina. Rapid Commun Mass Spectrom. 2008;22:3415–3426. doi: 10.1002/rcm.3751. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez LD, Hueffer K, Wenk MR, Galan JE. Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science. 2004;304:1805–1807. doi: 10.1126/science.1098188. [DOI] [PubMed] [Google Scholar]
- 17.Hsu FF, Turk J. Characterization of phosphatidylinositol,phosphatidylinositol-4-phosphate, and phosphatidylinositol-4,5-bisphosphate by electrospray ionization tandem mass spectrometry: a mechanistic study. J Am Soc Mass Spectrom. 2000;11:986–999. doi: 10.1016/S1044-0305(00)00172-0. [DOI] [PubMed] [Google Scholar]
- 18.Ishida Y, Nakanishi O, Hirao S, Tsuge S, Urabe J, Sekino T, Nakanishi M, Kimoto T, Ohtani H. Direct analysis of lipids in single zooplankter individuals by matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem. 2003;75:4514–4518. doi: 10.1021/ac030072j. [DOI] [PubMed] [Google Scholar]
- 19.Jackson SN, Wang HY, Woods AS. In situ structural characterization of phosphatidylcholines in brain tissue using MALDI-MS/MS. J Am Soc Mass Spectrom. 2005a;16:2052–2056. doi: 10.1016/j.jasms.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 20.Jackson SN, Wang HY, Woods AS, Ugarov M, Egan T, Schultz JA. Direct tissue analysis of phospholipids in rat brain using MALDI-TOFMS and MALDI-ion mobility-TOFMS. J Am Soc Mass Spectrom. 2005b;16:133–138. doi: 10.1016/j.jasms.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Jackson SN, Wang HY, Woods AS. In situ structural characterization of glycerophospholipids and sulfatides in brain tissue using MALDI-MS/MS. J Am Soc Mass Spectrom. 2007;18:17–26. doi: 10.1016/j.jasms.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 22.Jones JJ, Batoy SM, Wilkins CL, Liyanage R, Lay JO., Jr Ionic liquid matrix-induced metastable decay of peptides and oligonucleotides and stabilization of phospholipids in MALDI FTMS analyses. J Am Soc Mass Spectrom. 2005;16:2000–2008. doi: 10.1016/j.jasms.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Kim HY, Wang TC, Ma YC. Liquid chromatography/mass spectrometry of phospholipids using electrospray ionization. Anal Chem. 1994;66:3977–3982. doi: 10.1021/ac00094a020. [DOI] [PubMed] [Google Scholar]
- 24.Li YL, Gross ML, Hsu FF. Ionic-liquid matrices for improved analysis of phospholipids by MALDI-TOF mass spectrometry. J Am Soc Mass Spectrom. 2005;16:679–682. doi: 10.1016/j.jasms.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 25.Milne SB, Ivanova PT, DeCamp D, Hsueh RC, Brown HA. A targeted mass spectrometric analysis of phosphatidylinositol phosphate species. J Lipid Res. 2005;46:1796–1802. doi: 10.1194/jlr.D500010-JLR200. [DOI] [PubMed] [Google Scholar]
- 26.Oresic M, Hanninen VA, Vidal-Puig A. Lipidomics: a new window to biomedical frontiers. Trends Biotechnol. 2008;26:647–652. doi: 10.1016/j.tibtech.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Petkovic M, Schiller J, Muller M, Benard S, Reichl S, Arnold K, Arnhold J. Detection of individual phospholipids in lipid mixtures by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry: phosphatidylcholine prevents the detection of further species. Anal Biochem. 2001;289:202–216. doi: 10.1006/abio.2000.4926. [DOI] [PubMed] [Google Scholar]
- 28.Pitt SC, Chen H. The phosphatidylinositol 3-kinase/akt signaling pathway in medullary thyroid cancer. Surgery. 2008;144:721–724. doi: 10.1016/j.surg.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puolitaival SM, Burnum KE, Cornett DS, Caprioli RM. Solvent-free matrix dry-coating for MALDI imaging of phospholipids. J Am Soc Mass Spectrom. 2008;19:882–886. doi: 10.1016/j.jasms.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roth MG. Phosphoinositides in constitutive membrane traffic. Physiol Rev. 2004;84:699–730. doi: 10.1152/physrev.00033.2003. [DOI] [PubMed] [Google Scholar]
- 31.Rujoi M, Estrada R, Yappert MC. In situ MALDI-TOF MS regional analysis of neutral phospholipids in lens tissue. Anal Chem. 2004;76:1657–1663. doi: 10.1021/ac0349680. [DOI] [PubMed] [Google Scholar]
- 32.Schiller J, Arnhold J, Benard S, Muller M, Reichl S, Arnold K. Lipid analysis by matrix-assisted laser desorption and ionization mass spectrometry: A methodological approach. Anal Biochem. 1999;267:46–56. doi: 10.1006/abio.1998.3001. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz SA, Reyzer ML, Caprioli RM. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J Mass Spectrom. 2003;38:699–708. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- 34.Shimma S, Sugiura Y, Hayasaka T, Hoshikawa Y, Noda T, Setou M. MALDI-based imaging mass spectrometry revealed abnormal distribution of phospholipids in colon cancer liver metastasis. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;855:98–103. doi: 10.1016/j.jchromb.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 35.Shimma S, Sugiura Y, Hayasaka T, Zaima N, Matsumoto M, Setou M. Mass imaging and identification of biomolecules with MALDI-QIT-TOF-based system. Anal Chem. 2008;80:878–885. doi: 10.1021/ac071301v. [DOI] [PubMed] [Google Scholar]
- 36.Shui G, Bendt AK, Pethe K, Dick T, Wenk MR. Sensitive profiling of chemically diverse bioactive lipids. J Lipid Res. 2007;48:1976–1984. doi: 10.1194/jlr.M700060-JLR200. [DOI] [PubMed] [Google Scholar]
- 37.Sutphen R, Xu Y, Wilbanks GD, Fiorica J, Grendys EC, Jr, LaPolla JP, Arango H, Hoffman MS, Martino M, Wakeley K, Griffin D, Blanco RW, Cantor AB, Xiao YJ, Krischer JP. Lysophospholipids are potential biomarkers of ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:1185–1191. [PubMed] [Google Scholar]
- 38.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 39.Wang HY, Jackson SN, Woods AS. Direct MALDI-MS analysis of cardiolipin from rat organs sections. J Am Soc Mass Spectrom. 2007;18:567–577. doi: 10.1016/j.jasms.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watson AD. Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: a global approach to lipid analysis in biological systems. J Lipid Res. 2006;47:2101–2111. doi: 10.1194/jlr.R600022-JLR200. [DOI] [PubMed] [Google Scholar]
- 41.Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 42.Wenk MR, Lucast L, Di Paolo G, Romanelli AJ, Suchy SF, Nussbaum RL, Cline GW, Shulman GI, McMurray W, De Camilli P. Phosphoinositide profiling in complex lipid mixtures using electrospray ionization mass spectrometry. Nat Biotechnol. 2003;21:813–817. doi: 10.1038/nbt837. [DOI] [PubMed] [Google Scholar]
- 43.Woods AS, Ugarov M, Jackson SN, Egan T, Wang HY, Murray KK, Schultz JA. IR-MALDI-LDI combined with ion mobility orthogonal time-of-flight mass spectrometry. J Proteome Res. 2006;5:1484–1487. doi: 10.1021/pr060055l. [DOI] [PMC free article] [PubMed] [Google Scholar]