Abstract
The Yes-associated protein (YAP) transcriptional coactivator is a key regulator of organ size and a candidate human oncogene inhibited by the Hippo tumor suppressor pathway. The TEAD family of transcription factors binds directly to and mediates YAP-induced gene expression. Here we report the three-dimensional structure of the YAP (residues 50–171)–TEAD1 (residues 194–411) complex, in which YAP wraps around the globular structure of TEAD1 and forms extensive interactions via three highly conserved interfaces. Interface 3, including YAP residues 86–100, is most critical for complex formation. Our study reveals the biochemical nature of the YAP–TEAD interaction, and provides a basis for pharmacological intervention of YAP–TEAD hyperactivation in human diseases.
Keywords: YAP, TEAD, structure, Hippo
The Yes-associated protein (YAP) is a transcriptional coactivator (Yagi et al. 1999). YAP knockout in mice causes embryonic lethality (Morin-Kensicki et al. 2006), indicating its critical role in development. In contrast, transgenic expression of YAP dramatically increases mouse liver size in a reversible fashion (Camargo et al. 2007; Dong et al. 2007), suggesting a key role of YAP in organ size regulation. Consistent with its growth-promoting function, yap genomic amplification and elevated protein levels have been observed in several human cancers (Overholtzer et al. 2006; Zender et al. 2006; Dong et al. 2007; Zhao et al. 2007; Steinhardt et al. 2008). Furthermore, expression of active YAP potently induces transformation in both NIH3T3 and MCF10A cells (Overholtzer et al. 2006; Zhao et al. 2009), and liver-specific transgenic expression of YAP causes tumor formation in vivo (Camargo et al. 2007; Dong et al. 2007). These observations support the function of YAP as a human oncogene.
As a transcriptional coactivator, YAP needs to bind transcription factors to stimulate gene expression. Reported YAP target transcription factors include TEAD, p73, Runx2, and the ErbB4 cytoplasmic domain (Yagi et al. 1999; Strano et al. 2001; Vassilev et al. 2001; Komuro et al. 2003). However, only TEAD has been demonstrated to be important for the growth-promoting function of YAP (Ota and Sasaki 2008; Zhao et al. 2008). In humans, the TEAD family has four highly homologous proteins sharing a conserved DNA-binding TEA domain. YAP and TEAD1 bind to a common set of promoters in MCF10A cells (Zhao et al. 2008). Knockdown of TEAD aborts expression of the majority of YAP-inducible genes and largely attenuates YAP-induced overgrowth, epithelial–mesenchymal transition (EMT), and oncogenic transformation (Zhao et al. 2008). Furthermore, the phenotype of TEAD1/TEAD2 double-knockout mice resembles YAP knockout mice (Sawada et al. 2008). Notably, the role of the YAP and TEAD complex in growth promotion is implicated in Sveinsson's chorioretinal atrophy caused by the TEAD1 Y406H mutation, which abolishes its interaction with and activation by YAP (Kitagawa 2007; Zhao et al. 2008). Consistently, Scalloped (Sd), the Drosophila homolog of TEAD, directly mediates Yorkie (Yki)-induced gene expression and overgrowth phenotypes (Zhao et al. 2007; Goulev et al. 2008; Wu et al. 2008; Zhang et al. 2008). These observations strongly indicate that the TEAD family of transcription factors plays a key role in mediating YAP function. The three-dimensional structure of the TEAD1 DNA-binding domain has been solved (Anbanandam et al. 2006). However, the molecular basis of how TEAD recruits YAP is unclear.
In this study, we present the crystal structure of the YAP2 (residues 50–171) and TEAD1 (residues 194–411) complex. We chose TEAD1 in our study because it is mutated in human disease and the DNA-binding domain structure of TEAD1 is available. In the YAP–TEAD1 complex, the extended structure of YAP—which includes a β strand, an α helix, and a twisted-coil region—wraps around the globular surface of TEAD, forming three highly conserved interfaces. Structural and functional analyses indicate that the coil region (residues 86–100) in YAP is most important for TEAD binding, while α helix 1 (residues 61–73) and the β strand (residues 52–58) in YAP play limited roles in complex formation with TEAD. Residues critical for complex formation are evolutionarily conserved in YAP and TEAD. The structure presented in this study provides a molecular understanding of YAP–TEAD interaction, and establishes a structural basis for pharmacological intervention of diseases associated with YAP overactivation.
Results and Discussion
Assembly and crystallization of the YAP–TEAD1 complex
YAP has a TEAD-binding domain at the N terminus followed by two WW domains and a C-terminal transactivation domain (Supplemental Fig. S1A). TEAD contains an N-terminal TEA DNA-binding domain (Anbanandam et al. 2006) and a C-terminal region responsible for YAP interaction (Supplemental Fig. S1A; Vassilev et al. 2001).
To make the YAP–TEAD complex for X-ray crystallography studies, we further narrowed the YAP interaction domain in TEAD1 using an in vitro pull-down assay (Supplemental Fig. S1B). The TEAD1 fragment with residues 194–411 displayed strong binding affinity to YAP 50–171 and was used for complex assembly. Three YAP fragments (residues 50–114, 50–159, and 50–171) were also verified to assemble stable complexes with TEAD1 (residues 194–411) (Supplemental Fig. S1C). Crystals of the YAP 50–171 and TEAD1 194–411 (referred to as YAP and TEAD hereafter) complex were used for data collection. The crystal structure of YAP–TEAD was solved by Se-SAD (single-wavelength anomalous diffraction), and the final model was refined to 2.8 Å resolution. Statistics of the structure determination and refinement are summarized in Supplemental Table S1.
Overall structure of the YAP–TEAD complex
In the 2.8 Å resolution structure (Fig. 1), YAP and TEAD form a heterodimer. In one asymmetric unit of the crystal structure, two YAP–TEAD complexes form a dimerized heterodimer (data not shown). However, gel filtration chromatography showed that the complex was a heterodimer in solution (data not shown), suggesting that further dimerization in the asymmetric unit may result from crystal packing. Therefore, the structure is presented as a heterodimer below. There is no electron density for residues 230–238 of TEAD and residues 101–171 of YAP, and therefore these were not built in the model.
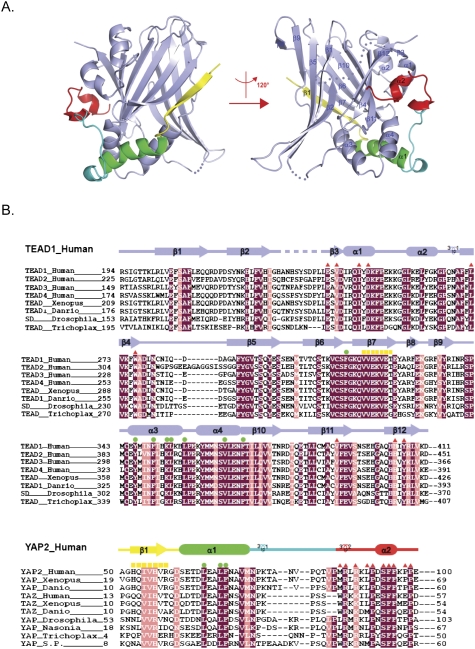
Figure 1.
Overall structure of the YAP–TEAD complex and their sequence conservation. (A) Overall structure of the human YAP–TEAD complex shown as a ribbon representation. TEAD is shown in light blue, and different YAP elements are shown in yellow, green, cyan, and red. Secondary structural elements are labeled, and two different views of the complex structure are shown. (B) Sequence alignment of TEAD and YAP across isoforms and species. TEAD, YAP, and TAZ from indicated species are included. Identical residues are highlighted with a purple background, and highly conserved residues are highlighted with a pink background. Secondary structural elements are colored as in A and are indicated above the sequences. Residues that are involved in interactions on interfaces 1, 2, and 3 (Fig. 2A) are indicated by yellow squares, green dots, and red triangles, respectively.
The structure shows that the complex has overall dimensions of ∼50 × 60 × 40 Å3 (Fig. 1A) composed of four α helices and 12 β strands in TEAD (Fig. 1A, shown in light blue), and two α helices, one β strand, and a coil in YAP (Fig. 1A, shown in yellow, green, cyan, and red, respectively). In the TEAD structure, two β sheets are packed against each other, forming a β-sandwich fold, which is surrounded by four α helices on one side. One of the β sheets is formed by strands β1, β2, β5, β8, and β9, and the other is formed by strands β3, β4, β6, β7, β10, β11, and β12. α helices α1 and α2 connect β3 and β4, and pack on the surface of β sandwich on one side. α helices α3 and α4, which lie between β9 and β10, further stabilize the β-sandwich core domain by closing up the open end between two β sheets. The extended YAP structure wraps around TEAD by interacting with α1, α3, α4, and one of the β sheets of TEAD.
TEAD is highly conserved from Trichoplax to mammals. Structurally based sequence alignment of the TEAD family is shown in Figure 1B. The TEAD interaction domain of YAP is highly conserved, indicating that the YAP–TEAD partnership is evolutionarily conserved (Fig. 1B). Furthermore, residues critical for TEAD interaction are highly conserved between YAP and TAZ (Fig. 1B), a YAP paralog that also interacts with TEAD (Mahoney et al. 2005; Chan et al. 2009; Zhang et al. 2009).
Interfaces between YAP and TEAD
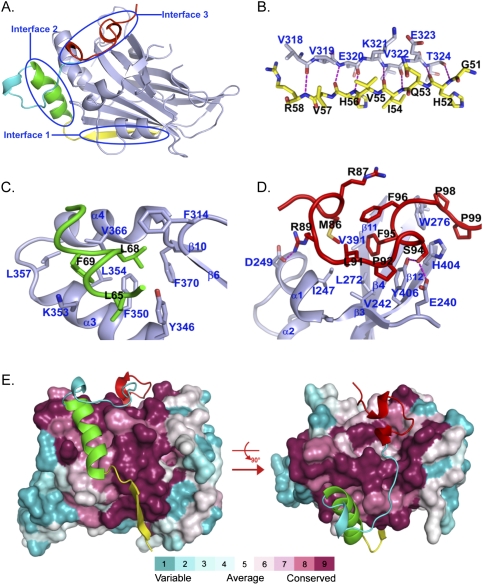
The TEAD-binding domain of YAP wraps around the globular structure of TEAD via three interfaces (Fig. 2A). Interface 1 is mediated by seven intermolecular hydrogen bonds between the peptide backbones of YAP β1 (residues 52–58) and TEAD β7 (residues 318–324), forming an anti-parallel β sheet (Fig. 2B; Supplemental Fig. S3). Compared with YAP residues in the other two TEAD-binding interfaces, β1 of YAP is more variable across species (marked by yellow squares in Fig. 1B), consistent with the limited participation of side chains in the anti-parallel β sheet.
Figure 2.
Binding interfaces between YAP and TEAD. (A) Overall interaction between YAP and TEAD. The color scheme is the same as in Figure 1A. Major binding interfaces between YAP and TEAD are highlighted as interface 1, interface 2, and interface 3. (B–D) Detailed interactions between YAP and TEAD in interfaces 1, 2, and 3. Side chains involved in interaction are shown and colored as in Figure 1A. Residue numbers for YAP and TEAD are labeled in black and blue, respectively. Hydrogen bonds are represented by magenta dotted lines. (E) Sequence conservation projected on the surface of TEAD. TEAD is shown as a surface model and is colored according to the conservation scores. YAP is shown as a ribbon and is colored as in Figure 1A.
The second interface is dominated by the YAP α1 helix (residues 61–73), which packs into a binding groove formed by TEAD α3 and α4 (residues 345–369) (Fig. 2C; Supplemental Fig. S3). Here the binding is mediated mainly by hydrophobic interactions between L65, L68, and F69 of YAP and F314, Y346, F350, K353, L354, L357, V366, and F370 of TEAD. The three residues L65, L68, and F69 of YAP form a conserved LXXLF motif, which is well known as a binding module for a hydrophobic groove (Darimont et al. 1998; Nolte et al. 1998; Westin et al. 1998). In particular, the L65-binding site is formed by Y346, F350, and K353; the L68-binding site is formed by F314, F350, and F370; and the F69-binding site is formed by F350, K353, L354, L357, and V366. The three residues of YAP also interact with each other, forming a very compact hydrophobic patch that binds to the hydrophobic groove of TEAD. Residues participating in the hydrophobic interaction from both YAP and TEAD are completely conserved, as marked by green dots in Figure 1B.
In the third interface, the twisted-coil region of YAP (residues 86–100) interacts with TEAD by fitting side chains into the deep pocket, which is formed by β4, β11, β12, α1, and α4 of TEAD (Fig. 2D; Supplemental Fig. S3). Particularly, the hydrophobic side chains M86, L91, and F95 of YAP make multiple van der Waals contacts with I247, V242, L272, V391, and Y406 of TEAD. The interaction is further strengthened by hydrogen bonds between the guanidinium group of YAP R89 and the carboxylate oxygen of TEAD D249, and two hydrogen bonds formed by the side chain of YAP S94: one with Y406, and the other with E240 of TEAD. F96 of YAP has no direct interaction with TEAD but has internal hydrophobic interaction with M86, R87, L91, and F95 of YAP, and stabilizes this compact hydrophobic coil region. P98 and P99 of YAP are separated from the coil region by W276 and H404 of TEAD and are stabilized by the outside surface of the pocket, mainly through hydrophobic interaction with W276 and H404 of TEAD.
A Dali search (Holm et al. 2008) of TEAD structure versus the Protein Data Bank yielded 669 entries with structural similarity (Z score >2.0), among which PDEδ (phosphodiesterase) was the best, with a Z score of 9.8 (Supplemental Table S3; Hanzal-Bayer et al. 2002). Most structures produced from the Dali search adopt an immunoglobulin-like fold, but none of them share similar functional properties with TEAD. To investigate the sequence conservation at the YAP–TEAD interfaces, TEAD surface residues are colored according to conservation scores, which were calculated using the ConSurf Server (Fig. 2E; Landau et al. 2005; http://consurf.tau.ac.il). Strikingly, most of the invariant residues of TEAD map to a continuous surface to which YAP binds, including β7 corresponding to interface 1; α3 and α4 corresponding to interface 2; and β4, β11, β12, and α1 corresponding to interface 3. This analysis supports the importance of the YAP–TEAD interaction from an evolutionary point of view.
Functional validation of YAP residues in TEAD-binding interfaces
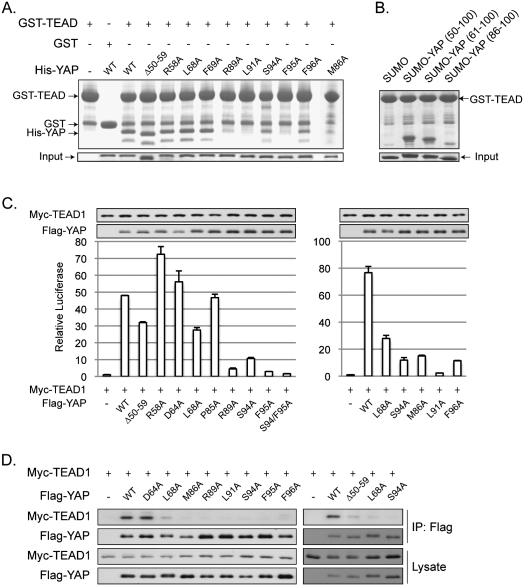
The YAP–TEAD cocrystal structure indicates three major interfaces between the two proteins (Fig. 2). To test the contribution of each interface in YAP–TEAD binding and function, we performed mutagenesis studies. Individual YAP mutants were tested in an in vitro GST pull-down assay. Deletion of β1 (50–59) in YAP had little effect on its interaction with TEAD (Fig. 3A), indicating that the anti-parallel β sheet between YAP and TEAD in interface 1 is not critical for their association. In addition, mutations of L68A and F69A, which are involved in a hydrophobic interaction with TEAD in interface 2, did not reduce YAP interaction with TEAD (Fig. 3A). However, S94A and F96A mutations significantly reduced the binding affinity between YAP and TEAD. YAP–TEAD interaction was most dramatically decreased by M86A, R89A, L91A, and F95A mutations (Fig. 3A). To further confirm the significance of interface 3 in YAP–TEAD interaction, we expressed three YAP fragments as a SUMO fusion. Our data showed that YAP containing residues 50–100 (β1, α1, and the coil region) and residues 61–100 (α1 and the coil region) strongly interacted with GST-TEAD (Fig. 3B). Interestingly, the YAP 86–100 fragment, which has only the coil region, could also interact with TEAD, although the interaction was weaker than the longer YAP fragments. These results suggest that, under in vitro conditions, interface 3 is the most important to YAP–TEAD interaction, and it alone is sufficient for complex formation. In contrast, interface 2 is less critical than interface 3, while interface 1 is the least important for interaction with TEAD.
Figure 3.
Effect of YAP mutations on YAP–TEAD-binding affinity. (A) In vitro pull-down assay. His-tagged YAP (50–171) and mutants were tested in a GST pull-down assay using GST-TEAD (194–411) immobilized on glutathione resin. Pull-down products were analyzed by Tricine-SDS-PAGE and Coomassie blue staining. (B) In vitro GST-TEAD pull-down of different YAP fragments. Three YAP fragments, as indicated, were expressed and purified as SUMO fusions. In vitro pull-down was performed as described in A and was analyzed by SDS-PAGE and Coomassie blue staining. (C) CTGF reporter assay. Indicated plasmids were cotransfected with a CTGF reporter and a CMV–β-gal construct into 293T cells. Luciferase activity was measured and normalized to β-galactosidase activity. YAP and TEAD expression levels were determined by Western blot with anti-Flag and anti-Myc antibodies, respectively. (D) Coimmunoprecipitation. Flag-YAP wild type or mutants were cotransfected with Myc-TEAD1 wild type into HEK293 cells. Flag-YAP was immunoprecipitated, and coimmunoprecipitated TEAD1 was determined by anti-Myc Western blot.
The YAP–TEAD interfaces were further evaluated in functional assays in cells using a luciferase reporter driven by the CTGF promoter, which contains three TEAD-binding sites and is potently activated by YAP (Zhao et al. 2008). Activation of this reporter requires the binding of YAP to TEAD, which then binds to the CTGF reporter. Deletion of YAP residues 50–59 caused a moderate reduction in CTGF reporter activity, while mutation of R58 to alanine did not inhibit the reporter, indicating a limited role of this interface in TEAD activation by YAP in vivo (Fig. 3C). Although the D64A mutation had little effect, mutation of L68A in interface 2 did cause a moderate reduction in the ability of YAP to stimulate the CTGF reporter. Notably, mutation of M86A, R89A, L91A, S94A, F95A, and F96A in interface 3 strongly diminished YAP activity on the CTGF reporter (Fig. 3C). Similar results were obtained without TEAD1 cotransfection, although the overall activity was lower (data not shown). We confirmed our observation of the functional importance of interface 3 using another luciferase reporter driven by five Gal4-binding elements. In this system, TEAD binds to the reporter through the fused Gal4 DNA-binding domain, but not its own TEA DNA-binding domain. The reporter activity depends on the interaction between TEAD and YAP. As expected, we obtained results similar to those for the CTGF reporter (Supplemental Fig. S2A). These data further support the important role of interface 3 in the function of YAP–TEAD interaction, and also suggest that YAP–TEAD interaction is independent of TEAD binding to DNA. We further tested if lentiviral expression of YAP fragment 86–100, corresponding to interface 3, could inhibit YAP function in vivo. No significant effect on the mRNA level of either CTGF or ITGB2, two YAP target genes (Zhao et al. 2009), was observed (data not shown). This could be due to a combination of weak interaction of the YAP 86–100 fragment with TEAD and an insufficient level of expression.
Mutations that reduced YAP activity in reporter assays were further tested for their interaction with TEAD in cells by a coimmunoprecipitation assay. Consistent with the reporter assay, M86A, R89A, L91A, S94A, F95A, and F96A mutations in interface 3 abolished YAP interaction with TEAD1 (Fig. 3D). Deletion of residues 50–59 in interface 1 or mutation of L68 in interface 2 also attenuated YAP–TEAD1 interaction, consistent with the YAP reporter assay data. As a negative control, mutation of D64, which did not decrease the reporter activity, had no effect on YAP–TEAD interaction (Fig. 3D). Based on the above data (summarized in Supplemental Table S2), it appears that interface 3 between YAP and TEAD is the most critical contact for YAP–TEAD interaction, although the other two interfaces also contribute to YAP–TEAD interaction in vivo.
Functional validation of TEAD residues in YAP-binding interfaces
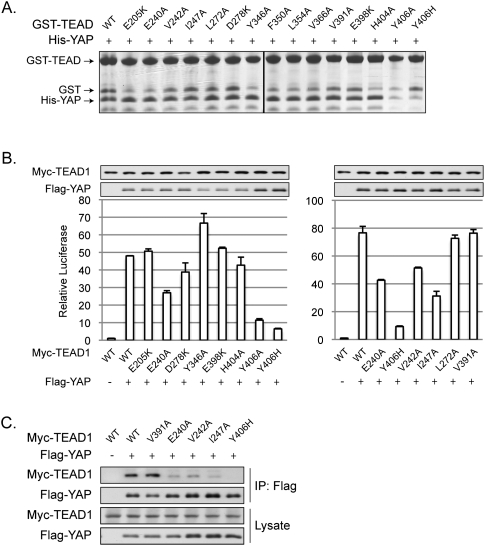
Fifteen point mutations were made in TEAD to determine their role in YAP interaction. In vitro GST pull-down experiments indicated that only the Y406A and Y406H mutations strongly reduced the interaction with YAP, while the other mutations showed modest or no effects (Fig. 4A). Because Y406 is in interface 3, the above data are consistent with the functional analyses performed with YAP mutants, which also demonstrate the critical role of interface 3 in the YAP–TEAD association.
Figure 4.
Effect of TEAD mutations on YAP–TEAD-binding affinity. (A) In vitro pull-down assay. Experiments were similar to Figure 3A except His-tagged YAP (50–171) wild-type protein and GST-TEAD1 (194–411) mutants were used. The GST fragment present in the samples of GST-TEAD1 protein preparation is indicated. (B) CTGF reporter assay. Experiments were performed as in Figure 3C with TEAD1 mutants and wide-type YAP. (C) Coimmunoprecipitation. Experiments were performed as in Figure 3D with TEAD1 mutants and wide-type YAP.
We examined TEAD mutations for their ability to be activated by YAP. The ability of Y406A and Y406H to be activated by YAP was abolished in both Gal4-TEAD1 and CTGF reporter assays (Fig. 4B; Supplemental Fig. S2B). Mutation of E240A, V242A, and I247A, three other residues in interface 3, partially compromised their activation by YAP (Fig. 4B; Supplemental Fig. S2B). Consistent with the reporter assay data, coimmunoprecipitation between YAP and TEAD1 mutants showed that TEAD1-Y406H was unable to interact with YAP, while the E240A, V242A, and I247A mutations partially decreased TEAD1 interaction with YAP (Fig. 4C). These results further support the essential function of interface 3 in YAP–TEAD1 interaction.
The functions of TEAD/Sd in development have been convincingly established by genetic studies in Drosophila (Halder et al. 1998; Simmonds et al. 1998) and knockout mouse models (Yagi et al. 2007; Nishioka et al. 2008; Sawada et al. 2008). Recent studies have also suggested that TEAD functions in progenitor cells and stem cells (Cao et al. 2008; Nishioka et al. 2009). The four TEAD proteins in humans are highly homologous and may function similarly at the molecular level, while their physiological functions may be influenced by spatial and temporal expression patterns.
Both TEAD and YAP are highly conserved in diverse species, indicating the functional conservation of the YAP–TEAD complex. In TEAD, the highly conserved residues map to the surface formed by β7, α3, α4, β4, β11, β12, and α1, corresponding to the three interfaces with YAP (Fig. 2E). In YAP, residues involved in TEAD binding are located mainly on α1 and the coil region connected with α2 (Fig. 1B). Mutational analyses show that M86, R89, L91, S94, F95, and F96 are essential for interaction with TEAD. Among these residues, S94, F95, and F96 are invariable in YAP across species. Residues corresponding to M86 and L91 are always hydrophobic, while the residue corresponding to R89 is either R or K (Fig. 1B). TAZ is a YAP homolog that also activates TEAD (Chan et al. 2009; Zhang et al. 2009). Consistently, critical residues for TEAD binding are conserved in TAZ (Fig. 1B).
The TEAD1 Y406H mutation has been implicated as a cause of Sveinsson's chorioretinal atrophy, a rare genetic disease (Fossdal et al. 2004). Interestingly, Y406 directly forms a hydrogen bond with S94 in YAP. The importance of this hydrogen bond is supported by biochemical characterizations that mutation of either Y406 in TEAD1 or S94 in YAP strongly disrupts YAP–TEAD1 interaction and abolishes TEAD1 activation by YAP. Given the fact that YAP is known to promote tissue growth, the YAP–TEAD complex structure provides molecular insights into the mechanism of the TEAD1 mutation in causing Sveinsson's chorioretinal atrophy.
An independent study by Chen et al. (2010) reports the structure of mouse TEAD4 (residues 210–427) in complex with YAP (residues 35–92). These two studies reveal a similar structural fold of the TEAD–YAP complex. Chen et al. (2010) noted that the loop with the PXXΦP motif is essential for TEAD4 interaction and transforming activity. Intriguingly, however, PXXΦP is not conserved in TAZ, and we observed that the YAP fragment (residues 86–100) without the PXXΦP motif could still bind to TEAD, although more weakly (Fig. 3B), indicating that the PXXΦP motif is not essential for interaction with TEAD.
YAP has been established as a human oncogene acting downstream from the Hippo tumor suppressor pathway. Our structural studies provide valuable information for potential drug design. For example, the most critical interaction between YAP and TEAD involves a twisted-coil region of YAP in interface 3. It is appealing to design inhibitors to disrupt the YAP–TEAD interaction based on our structural studies. Such inhibitors could be potential drugs for cancer or hypertrophic disease caused by mutations of the Hippo pathway.
Materials and methods
Structure determination
Phases for the YAP–TEAD complex were initially determined by SAD using the phasing module Autosol, and density modification and automatic model building were performed using the AutoBuild of program package PHENIX (Adams et al. 2002). More than 50% of residues were autotraced into the experimental electron density map. The remaining models were built manually with COOT (Emsley and Cowtan 2004). A native data set with a maximum resolution of 2.8 Å was used for refinement. All refinements were performed using the refinement module phenix.refine of the PHENIX package (Adams et al. 2002). The model quality was checked with the PROCHECK program, which shows a good stereochemistry according to the Ramachandran plot. The structure similarity search was performed with the Dali server (Holm et al. 2008).
Accession number
The atomic coordinates of the YAP–TEAD1 complex have been deposited in the Protein Data Bank with accession number 3KYS.
Acknowledgments
We thank all of the staff at the beamline BL17U at Shanghai Synchrotron Radiation Facility (China) and BL17A at Photon Factory (Japan) for assistance in data collection, and Karen Tumaneng for critical reading of the manuscript. This work was supported by grants from the National Natural Science Foundation of China (30870493), National Basic Research Program of China (2009CB918600), Shanghai Pujiang Program (08PJ14010), and Shanghai Leading Academic Discipline Project (B111) to Y.X., and grants from the NIH to K.L.G.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1865810.
Supplemental material is available at http://www.genesdev.org.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Anbanandam A, Albarado DC, Nguyen CT, Halder G, Gao X, Veeraraghavan S. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc Natl Acad Sci. 2006;103:17225–17230. doi: 10.1073/pnas.0607171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes & Dev. 2008;22:3320–3334. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Loo LS, Chong YF, Huang C, Hong W. TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. J Biol Chem. 2009;284:14347–14358. doi: 10.1074/jbc.M901568200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chan SW, Zhang XQ, Walsh M, Lim CJ, Hong W, Song H. Structural basis of YAP recognition by TEAD4 in the Hippo pathway. Genes & Dev. 2010 doi: 10.1101/gad.1865310. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR. Structure and specificity of nuclear receptor-coactivator interactions. Genes & Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fossdal R, Jonasson F, Kristjansdottir GT, Kong A, Stefansson H, Gosh S, Gulcher JR, Stefansson K. A novel TEAD1 mutation is the causative allele in Sveinsson's chorioretinal atrophy (helicoid peripapillary chorioretinal degeneration) Hum Mol Genet. 2004;13:975–981. doi: 10.1093/hmg/ddh106. [DOI] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the Hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Halder G, Polaczyk P, Kraus ME, Hudson A, Kim J, Laughon A, Carroll S. The Vestigial and Scalloped proteins act together to directly regulate wing-specific gene expression in Drosophila. Genes & Dev. 1998;12:3900–3909. doi: 10.1101/gad.12.24.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzal-Bayer M, Renault L, Roversi P, Wittinghofer A, Hillig RC. The complex of Arl2-GTP and PDEδ: From structure to function. EMBO J. 2002;21:2095–2106. doi: 10.1093/emboj/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M. A Sveinsson's chorioretinal atrophy-associated missense mutation in mouse Tead1 affects its interaction with the co-factors YAP and TAZ. Biochem Biophys Res Commun. 2007;361:1022–1026. doi: 10.1016/j.bbrc.2007.07.129. [DOI] [PubMed] [Google Scholar]
- Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney WM, Jr, Hong JH, Yaffe MB, Farrance IK. The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem J. 2005;388:217–225. doi: 10.1042/BJ20041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, Magnuson TR, O'Neal W, Milgram SL. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol. 2006;26:77–87. doi: 10.1128/MCB.26.1.77-87.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, Nakao K, Sasaki H. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. 2008;125:270–283. doi: 10.1016/j.mod.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Nishioka N, Inoue K, Adachi K, Kiyonari H, Ota M, Ralston A, Yabuta N, Hirahara S, Stephenson RO, Ogonuki N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada A, Kiyonari H, Ukita K, Nishioka N, Imuta Y, Sasaki H. Redundant roles of Tead1 and Tead2 in notochord development and the regulation of cell proliferation and survival. Mol Cell Biol. 2008;28:3177–3189. doi: 10.1128/MCB.01759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds AJ, Liu X, Soanes KH, Krause HM, Irvine KD, Bell JB. Molecular interactions between Vestigial and Scalloped promote wing formation in Drosophila. Genes & Dev. 1998;12:3815–3820. doi: 10.1101/gad.12.24.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D, Montgomery EA, Anders RA. Expression of Yes-associated protein in common solid tumors. Hum Pathol. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, Oren M, Sudol M, Cesareni G, Blandino G. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem. 2001;276:15164–15173. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes & Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin S, Kurokawa R, Nolte RT, Wisely GB, McInerney EM, Rose DW, Milburn MV, Rosenfeld MG, Glass CK. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature. 1998;395:199–202. doi: 10.1038/26040. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, Buonanno A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827–3836. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- Zender L, Spector MS, Xue W, Flemming P, Cordon-Cardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu CY, Zha ZY, Zhao B, Yao J, Zhao S, Xiong Y, Lei QY, Guan KL. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes & Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes & Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Kim J, Ye X, Lai ZC, Guan KL. Both TEAD-binding and WW domains are required for the growth stimulation and oncogenic transformation activity of yes-associated protein. Cancer Res. 2009;69:1089–1098. doi: 10.1158/0008-5472.CAN-08-2997. [DOI] [PubMed] [Google Scholar]