Abstract
Polycomb complexes establish chromatin modifications for maintaining gene repression and are essential for embryonic development in mice. Here we use pluripotent embryonic stem (ES) cells to demonstrate an unexpected redundancy between Polycomb-repressive complex 1 (PRC1) and PRC2 during the formation of differentiated cells. ES cells lacking the function of either PRC1 or PRC2 can differentiate into cells of the three germ layers, whereas simultaneous loss of PRC1 and PRC2 abrogates differentiation. On the molecular level, the differentiation defect is caused by the derepression of a set of genes that is redundantly repressed by PRC1 and PRC2 in ES cells. Furthermore, we find that genomic repeats are Polycomb targets and show that, in the absence of Polycomb complexes, endogenous murine leukemia virus elements can mobilize. This indicates a contribution of the Polycomb group system to the defense against parasitic DNA, and a potential role of genomic repeats in Polycomb-mediated gene regulation.
Keywords: Polycomb, histone modification, chromatin, embryonic stem cells, tumor, retrovirus
Polycomb complexes establish epigenetic patterns for maintaining gene repression. On the cellular level, Polycomb complex-mediated epigenetic patterns are thought to be important for establishing and maintaining cellular identity. A crucial role for cell identity has been deduced from pioneering studies in flies, where Polycomb group (PcG) genes were identified as regulators of homeotic gene expression (for review, see Ringrose and Paro 2007; Schwartz and Pirrotta 2008). In the fly, Polycomb response elements that act as memory modules for maintaining specified gene expression patterns have been identified. In mammals, PcG proteins function in Hox gene regulation, development, and tumorigenesis, and contribute to genomic imprinting and dosage compensation (for reviews, see Schuettengruber et al. 2007; Pietersen and van Lohuizen 2008; Schwartz and Pirrotta 2008). Histone-modifying activities that establish chromatin modifications for the regulation of gene repression have been associated with two distinct PcG complexes. The Polycomb-repressive complex 2 (PRC2) contains the PcG proteins Ezh2, Eed, and Suz12 and mediates di- and trimethylation of histone H3 Lys 27 (H3K27me3) (Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002). H3K27me3 can act as a signal for recruitment of PRC1 (Cao et al. 2002; Fischle et al. 2003; Min et al. 2003). PRC1 composition is heterogeneous, depending on the cellular context, and contains several PcG proteins, including the RING finger protein Ring1B, which catalyzes ubiquitination of histone H2A (de Napoles et al. 2004; Fang et al. 2004), thereby inhibiting transcription (Stock et al. 2007). PRC1 has also been shown to mediate chromatin compaction (Francis et al. 2004). Currently, it is thought that PcG-mediated gene repression requires the sequential activity of PRC2 and PRC1, whereby H3K27me3 serves as a stable and self-perpetuating epigenetic mark (Hansen et al. 2008) for the recruitment of PRC1 (Fischle et al. 2003; Min et al. 2003; Wang et al. 2004). This view might be too simplistic, as considerable biochemical heterogeneity has been demonstrated for complexes containing PcG proteins. Ezh1 has been shown to form PRC2-like complexes with Eed and Suz12 that predominantly repress select genes by chromatin compaction, and, to a lesser extent, due to histone modifications (Margueron et al. 2008). In addition, Ezh2 complexes have been shown to carry histone H1 methylation activity (Kuzmichev et al. 2004). Also, the PRC1 protein Ring1B is present in different complexes, which also include the H3K36me2 demethylase Fbxl10/dKDM (Sanchez et al. 2007; Lagarou et al. 2008). Ring1B has further been observed in the E2F6.com-1 complex, which also contains non-PcG proteins (Ogawa et al. 2002). These findings suggest considerable interactions between PcG complexes and chromatin regulators outside the strict definition of the Polycomb group.
In mice, mutations in Ezh2 or Eed that abolish the function of PRC2 result in embryonic lethality soon after implantation (O'Carroll et al. 2001; Wang et al. 2002). Disruption of Ring1B causes a similar phenotype with arrest at gastrulation, demonstrating that both PcG complexes are essential for development (Voncken et al. 2003). The role of PcG complexes in differentiation has been investigated in pluripotent embryonic stem (ES) cells. ES cells can be maintained in culture, and retain the ability to differentiate into all embryonic cell types (for review, see Silva and Smith 2008). It is currently thought that the pluripotent state of ES cells is established by a network of transcription factors, including Oct4, Nanog, Sox2, and Klf4 (Takahashi and Yamanaka 2006; Silva et al. 2009). The contribution of epigenetic regulation to maintain ES cell identity, however, is less clear. Polycomb complexes are viewed to be critical for epigenetic regulation in ES cells, and the genome-wide distribution of PRC1 and PRC2 has been determined (Boyer et al. 2006b; Mikkelsen et al. 2007; Ku et al. 2008). In ES cells, ∼2500 Polycomb target genes have been identified, including a significant number of transcription factors with lineage-specific expression profiles (Boyer et al. 2006a; Mikkelsen et al. 2007; Ku et al. 2008).
The dynamic repression of target genes by PcG complexes has been implicated in mediating lineage decisions (Bracken et al. 2006; Mikkelsen et al. 2007). During neuronal differentiation, Polycomb targets change between ES cells, lineage-restricted progenitors, and mature neurons (Mohn et al. 2008). This indicates that the promoter regulation by PcG complexes is highly plastic in a tissue- and differentiation state-dependent manner. In the epidermis, the PcG protein Ezh2 has been shown to be required for temporal and spatial control of gene expression during differentiation in a lineage-specific manner (Ezhkova et al. 2009). Furthermore, the PcG system is central in modulating expression from the Ink4a/Arf tumor suppressor locus, linking PcG-mediated repression to cell cycle control (Voncken et al. 2003; Chen et al. 2009; Dhawan et al. 2009). The PRC1 protein Bmi1 has also been shown to regulate mitochondrial function, DNA damage response, and antioxidant defense in mice (Chatoo et al. 2009; Liu et al. 2009). This indicates that PcG proteins are important for regulating several cellular processes that contribute to development.
We and others observed previously that lineage-specific genes are derepressed in ES cells lacking either the Ring1B or the Eed (Voncken et al. 2003; Boyer et al. 2006b; Leeb and Wutz 2007; Endoh et al. 2008; van der Stoop et al. 2008). These transcriptional changes destabilize ES cells, but are compatible with ES cell self-renewal. Furthermore, ES cells lacking either PRC1 or PRC2 function are capable of differentiating in culture, and Eed-deficient ES cells have been shown to form teratomas consisting of cells from the three germ layers and to contribute to chimeric embryos (Chamberlain et al. 2008). This is surprising, given the importance of the Polycomb system for epigenetic regulation during embryogenesis. Currently, it is not clear whether Polycomb complexes are the main determinants of repression of target loci or whether they have a modulatory role for gene expression. Importantly, the extent to which they are required for the formation of differentiated cell types remains to be determined. In order to gain insight into the function of PcG-mediated transcriptional regulation, we sought to eliminate both chromatin modifications associated with the Polycomb system by simultaneous disruption of PRC1 and PRC2 function in mouse ES cells.
Results
Ring1B and Eed double-deficient (dKO) ES cells self-renew in culture
ES cells lacking PRC1 activity have been generated previously by disruption of the Ring1B gene (Leeb and Wutz 2007). Similarly, PRC2 function has been abolished in ES cells carrying a disruption of Eed (Schoeftner et al. 2006). To generate ES cells lacking both PRC1 and PRC2 activity, we followed a conditional gene targeting strategy to delete Ring1B in Eed-deficient ES cells. We disrupted the first allele of Ring1B by deleting the first three coding exons, which contain the catalytic RING domain, and inserting a splice acceptor-linked polyadenylation site to truncate the transcript. Subsequently, the selection cassette was removed using Cre recombinase-mediated excision. The second allele was targeted using a vector for flanking the first three coding exons with loxP sites. Deletion of Ring1B was then induced by transient expression of Cre recombinase. Ring1B−/− Eed−/− dKO ES cells were generated, but initially we were not able to establish lines, as dKO ES cells were extremely sensitive to culture conditions and showed a strong inclination to differentiate. However, after adaptations of the culture conditions, and using a Tamoxifen-inducible estrogen receptor fusion CreERT2 transgene to delete the second allele of the Ring1B gene, we were able to establish stable dKO ES cell lines with high efficiency. dKO ES cells self-renewed in culture and maintained a typical ES cell morphology for at least 20 passages (Fig. 1A; Supplemental Fig. 1A). We noted a pronounced dependence of dKO ES cells on fibroblast feeder cells, which was not overcome by a high dose of leukemia inhibitory factor (LIF). Apparently, lack of Ring1B and Eed caused an unstable ES cell phenotype, but did not block ES cell self-renewal.
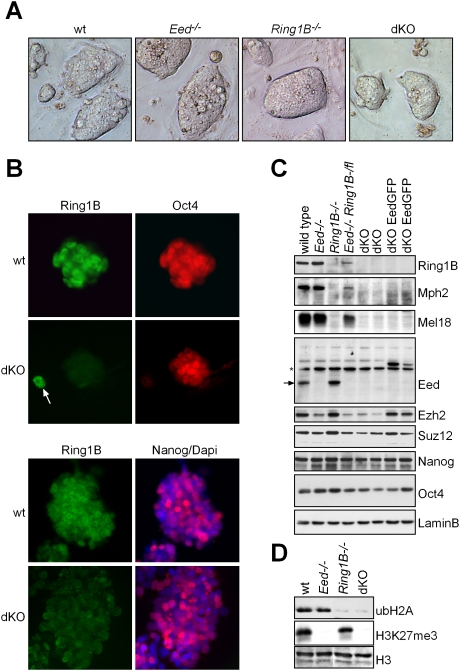
Figure 1.
Establishment of dKO ES cells lacking PRC1 and PRC2 activity. (A) Morphology of wild-type (wt), Ring1B−/−, Eed−/−, and dKO ES cell colonies. (B) Immunofluorescence staining showing Oct4 and Nanog expression in dKO and wild-type ES cell colonies. (C) Western analysis of PcG proteins and transcription factors in ES cells of indicated genotypes shows that Mph2 and Mel18 are virtually absent in Ring1B−/− and dKO ES cells. Ezh2 and Suz12 are reduced in Eed−/− and dKO ES cells. An arrow indicates Eed (asterisk; nonspecific band). The pluripotency markers Nanog and Oct4 are largely unaffected. Lamin B was used as loading control. (D) Western analysis of ubH2A and H3K27me3 showing absence of PRC1 and PRC2 activity in dKO ES cells.
dKO ES cells showed a clear nuclear immunofluorescence signal for Oct4 and Nanog, and expressed transcription factors known to be important for pluripotency of ES cells (Fig. 1B,C; Supplemental Fig. 1B). The PRC1 proteins Ring1B, Mel18, and Mph2 were abundant in control wild-type ES cells but were undetectable by Western analysis in dKO ES cells (Fig. 1C). Similarly, Eed was absent and the PRC2 proteins Suz12 and Ezh2 were reduced in dKO ES cells (Fig. 1C). Finally, dKO ES cells showed loss of genomic H3K27me3 and loss of ubiquitination of histone H2A (ubH2A) (Fig. 1D). In ES cell lines lacking Eed, we also detected a loss of dimethylation and a reduction of monomethylation of H3K27 (Supplemental Fig. 2A). Immunofluorescence analysis showed that Lys 27 monomethylation formed foci over pericentric heterochromatin, consistent with previous observations (Supplemental Fig. 2B; Schoeftner et al. 2006). Expression of an EedGFP transgene rescued histone H3 lysine methylation patterns in dKO ES cells (Supplemental Fig. 2A,B). We conclude that ES cells lacking both Ring1B and Eed can be established, and Polycomb-mediated chromatin modifications are not essential for ES cell self-renewal in culture.
Simultaneous loss of Ring1B and Eed function impairs ES cell differentiation
To investigate the function of the PcG system in differentiation, we analyzed the ability of wild-type, Eed−/−, Ring1B−/−, and dKO ES cells to form teratomas (Fig. 2A). For this, we injected 5 × 105 ES cells into the flanks of immune-deficient recipient mice using Matrigel as a carrier. Injection of wild-type, Eed-deficient, and Ring1B-deficient ES cells resulted in teratomas in all injection sites, showing that tumor formation was efficient. Tumors were excised after 3 wk and were analyzed. Histological analysis showed that wild-type, Ring1B-deficient, and Eed-deficient teratomas were composed of differentiated structures. Immunohistochemistry further showed that a range of cell types and tissues from all three germ layers was present in all teratomas (Fig. 2B). However, teratomas from Eed- and Ring1B-deficient ES cells were significantly smaller (Supplemental Fig. 3A), and we noted a larger proportion of cells with an endodermal or ectodermal origin, respectively, when compared with wild-type teratomas. The ability of wild-type, Eed-deficient, and Ring1B-deficient ES cells to form tumors consisting of cells from the three germ layers demonstrates that neither PRC1 nor PRC2 activity is essential for differentiation (Fig. 2B). In stark contrast, injection of dKO ES cells did not result in the formation of teratomas. When dKO grafts were excised after 6 mo, only Matrigel with a negligible amount of cells remained. This indicated that dKO cells were not maintained in the injection site. To ascertain that this defect is due to loss of both PcG complexes, we rescued PRC2 activity by expression of an EedGFP transgene in dKO ES cells. The resulting dKOEedGFP ES cells efficiently formed teratomas in all injection sites (Supplemental Fig. 3BC). Although these teratomas developed slower than wild-type, Eed-deficient, or Ring1B-deficient teratomas, they contained cells expressing endodermal and ectodermal markers consistent with the development of distinct lineages (Supplemental Fig. 3D). We therefore conclude that loss of Eed and Ring1B causes a synthetic phenotype, and that at least one PcG complex is required for tumor formation.
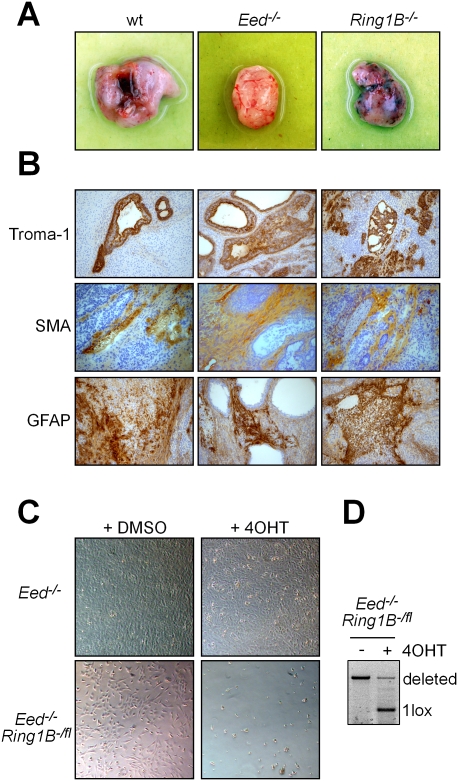
Figure 2.
Eed and Ring1B are redundantly required for differentiation. (A) Teratomas formed by wild-type (wt), Eed−/−, and Ring1B−/− ES cells 3 wk after injection. dKO ES cells did not give rise to teratomas. (B) Immunohistochemical analysis of wild-type, Ring1B−/−, and Eed−/− teratomas using markers for endoderm (Troma1), mesoderm (smooth muscle actin [SMA]), and ectoderm (GFAP). (C) Double deficiency for PRC1 and PRC2 is not compatible with NS cell viability. Deletion of Ring1B in Eed-deficient NS cells by induction of CreERT2 with 4OHT caused cell death, whereas 4OHT had no effect on control Eed-deficient NS cells. (D) PCR analysis showing that induction of CreERT2 results in deletion of the conditional Ring1Bfl allele in Eed−/− Ring1B-/fl NS cells.
To further test the differentiation potential of dKO ES cells, we used a protocol for directed differentiation into the neural lineage (Pollard et al. 2006). Nestin-positive neural stem (NS) cells could readily be established from Eed-deficient ES cells that were either wild type for Ring1B or contained a conditional Ring1B allele over a null allele (Eed−/− Ring1B-/fl), which resulted in strongly hypomorphic Ring1B expression (Fig. 2C; Supplemental Fig. 4AB). However, we did not obtain NS cells from dKO ES cells. Therefore, we aimed to delete Ring1B in Eed−/− Ring1B-/fl NS cells expressing an inducible CreERT2 recombinase. Three days to 4 d after deletion of Ring1B by addition of Tamoxifen (4-hydroxy-tamoxifen [4OHT]), we observed massive cell death (Fig. 2C). Control Eed-deficient NS cells were not affected by the addition of 4OHT. This showed that, in contrast to ES cells, the combined loss of PRC1 and PRC2 activity is not compatible with NS cell survival.
To study the differentiation defect more closely, we induced cell differentiation with retinoic acid in monolayer culture and followed the expression of pluripotency and differentiation genes (Fig. 3A). We observed the down-regulation of Oct4 on day 2 of differentiation in dKO, Eed-deficient, and wild-type ES cells. Differentiation markers Gata4, Gata6, Afp, and Brachyury were up-regulated in dKO cells on days 2 and 4, indicating that the cells had entered differentiation. The cells also showed a differentiated cell morphology consistent with entry into differentiation (Fig. 3B). However, the cell number of dKO cells was drastically reduced compared with wild-type or Eed-deficient ES cells (Fig. 3C–F). In the course of differentiation, dKO cell numbers declined sharply, and the cells were progressively lost (Fig. 3C–F). Expression of an EedGFP transgene rescued the differentiation defect of dKO ES cells to a large extent (Fig. 3A–F). Taken together, our data show that the simultaneous loss of Ring1B and Eed is not compatible with the formation of differentiated cells. Although differentiation of dKO ES cells was triggered by retinoic acid, differentiating cells were not viable and were lost.
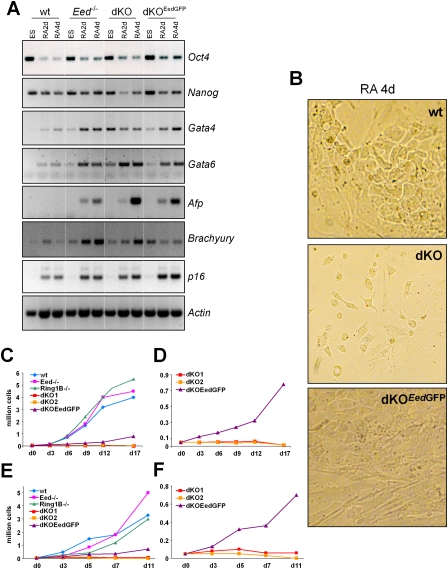
Figure 3.
Differentiation of ES cells lacking PcG activity. (A) RT–PCR expression analysis of pluripotency markers (Oct4 and Nanog) and differentiation markers (Gata4, Gata6, Afp, and Brachyury) and p16 in wild-type, Eed-deficient, dKO, and dKOEedGFP ES cells induced to differentiate with retinoic acid. Undifferentiated ES cells and cells after 2 and 4 d of differentiation are shown. Actin is used as loading control. (B) Microscopy image showing differentiated cell morphology on day 4 of retinoic acid-induced differentiation of wild-type, dKO, and dKOEedGFP ES cells. (C–F) Cell numbers in differentiating cultures of wild-type, Eed-deficient, Ring1B-deficient, dKO, and dKOEedGFP ES cells. (C,E) Two experiments are shown. (D,F) Rescaled representation of experiments in C and E showing that expression of an EedGFP transgene in dKOEedGFP ES cells rescues the differentiation defect of dKO ES cells.
To further assess the differentiation potential, we aggregated dKO ES cells into embryoid bodies. Compared with Eed-deficient and dKOEedGFP ES cells, dKO ES cells formed smaller embryoid bodies that became hollow spheres after 2 wk in suspension culture (Supplemental Fig. 5A). In these embryoid bodies, differentiation markers were up-regulated and expression of Oct4 was down-regulated (Supplemental Fig. 5B). This shows that dKO ES cells have a limited potential to form embryoid bodies and differentiate without a strong inducer like retinoic acid. Finally, we assessed the ability of dKO ES cells to contribute to embryos. We injected GFP-marked dKO ES cells into blastocysts, and observed efficient contribution to the inner cell mass similar to wild-type control ES cells (Supplemental Fig. 6). However, we could not detect a measurable contribution of dKO ES cells to chimeras at embryonic day 10.5, indicating that loss of Ring1B and Eed is not compatible with development. From our data, we conclude that, although PRC1 and PRC2 dKO ES cells can self-renew and express pluripotency markers, they are unable to differentiate properly. The aberrant differentiation potential of dKO ES cells is most likely due to a defect in maintaining differentiated cells and not a failure to enter differentiation.
PRC1 and PRC2 act in parallel to repress target genes
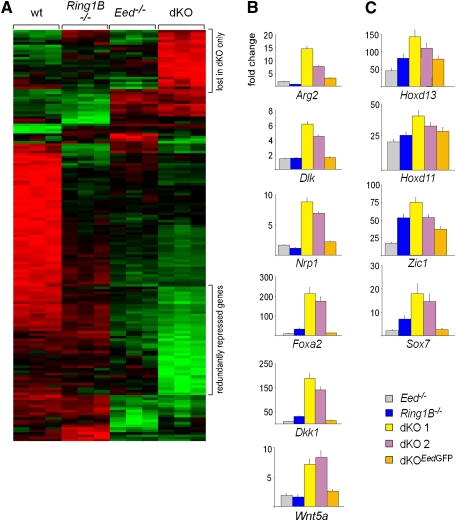
The strong phenotype caused by the combined loss of Eed and Ring1B suggested a compensatory or redundant role for Polycomb complexes in differentiation. To understand the mechanistic basis behind this synthetic phenotype, we analyzed the effect of loss of PcG complex function on gene repression. For this, we established genome-wide gene expression profiles of wild-type, Ring1B-deficient, Eed-deficient, and dKO ES cells using Affymetrix microarrays (Fig. 4A). When compared with wild-type ES cells, 814 and 1151 genes were derepressed more than twofold (P < 0.05) in Ring1B-deficient and Eed-deficient ES cells, respectively. In dKO ES cells, the number of up-regulated genes increased to 2017 (Supplemental Fig. 7A). In order to assess the direct effect of loss of PcG function, we analyzed a gene set that was reported previously to be bound by PcG complexes (Ku et al. 2008). Two-hundred-seventy-nine (10% of annotated PcG targets) and 476 (18%) PcG target genes were derepressed more than twofold (P < 0.05) in Ring1B-deficient or Eed-deficient ES cells, respectively. In dKO ES cells, the number of derepressed PcG target genes nearly doubled (757; 29%) (Supplemental Fig. 7B). A set of 329 genes was derepressed only after the combined loss of PRC1 and PRC2, but remained repressed in Ring1B-deficient and Eed-deficient ES cells (Supplemental Fig. 7B; Supplemental Table 1), suggesting that PRC1 and PRC2 are redundant for repression of these genes. We confirmed the specific derepression of the Arg2, Dlk, Nrp1, Foxa2, Dkk1, and Wnt5a genes in dKO ES cells by quantitative RT–PCR (Fig. 4B). Importantly, gene repression was restored after introducing an EedGFP transgene into dKO ES cells (Fig. 4B), indicating that derepression in dKO cells was reversible, and that genes remain marked for silencing in the absence of the PcG system in ES cells. These data show that a large set of genes is derepressed in dKO ES cells, but remains silenced in the absence of Ring1B or Eed alone. These redundantly silenced genes are repressed by both Polycomb complexes in parallel. This is in contrast to other PcG target genes such as Hoxd13, Hoxd11, Zic1, and Sox7 that were already derepressed in the absence of a single PcG complex (Fig. 4C).
Figure 4.
Analysis of transcriptional changes in ES cells lacking PcG activity. (A) A heat map showing high (green), intermediate (black), and low (red) expression of genes in wild-type, Eed−/−, Ring1B−/−, and dKO ES cells. The top regulated genes in all genotypes are shown. (B,C) Quantitative real-time PCR was used to confirm derepression of genes in PcG-deficient ES cells. Introduction of an EedGFP transgene in dKO ES cells re-establishes repression largely to the levels of Ring1B−/− ES cells. (B) A redundantly repressed set of PcG target genes is specifically derepressed in dKO ES cells. (C) PcG target genes derepressed in all PcG mutant ES cells.
In dKO but not Eed-deficient and Ring1B-deficient ES cells, a set of 94 genes—including several transcription factors such as Sall1, Rest, Pbx4, Nfat5, E2a, Aire, Hivep3, and Gli1—was more than fivefold down-regulated (Supplemental Table 2). Loss of expression of these genes is most likely a secondary effect of deregulation of Polycomb target genes in dKO ES cells.
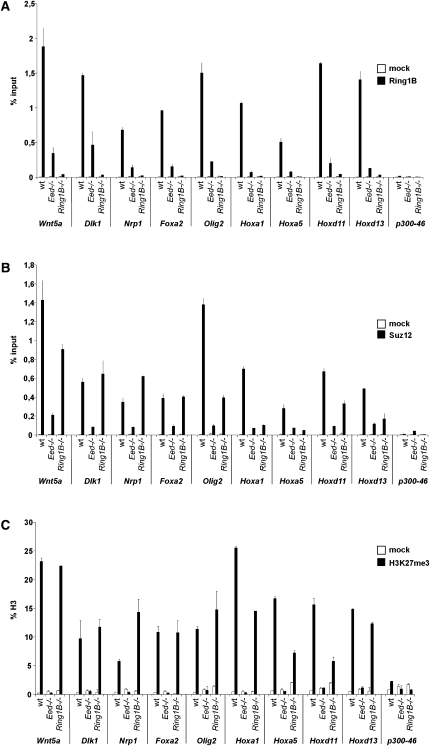
To further analyze the binding of Polycomb complexes on the promoters of redundantly silenced genes, we performed chromatin immunoprecipitation (ChIP). We detected Ring1B, Suz12, and H3K27me3 on the promoters of redundantly silenced genes as well as Hox genes in wild-type ES cells (Fig. 5). In Ring1B-deficient ES cells, the Suz12 and H3K27me3 signal was maintained at wild-type level on redundantly silenced genes, whereas Suz12 and H3K27me3 were reduced on Hox gene promoters (Fig. 5B,C). Ring1B was preferentially detected at a reduced level on the promoters of redundantly silenced genes in Eed-deficient ES cells, whereas it was lost from several Hox gene promoters (Fig. 5A). These observations show that both Polycomb complexes are recruited in parallel to redundantly silenced genes, and that recruitment of PRC1 and PRC2 is, at least in part, independent of each other. Although there is some preference for parallel recruitment of PRC1 and PRC2 to redundantly silenced genes, our data also show that the parallel recruitment is a general feature of PcG target genes, and suggest that some genes are more responsive to the loss of either of the two PcG complexes.
Figure 5.
PRC1 and PRC2 bind to the promoters of redundantly silenced genes. ChIP analysis of Ring1B (A), Suz12 (B), and H3K27me3 (C) binding to gene promoters in wild-type, Eed-deficient, and Ring1B-deficient ES cells. H3K27me3 signal is normalized to histone H3, and Ring1B and Suz12 are represented as percent input. Error bars represent standard deviation.
Stable repression of endogenous retroviruses requires the presence of PcG proteins
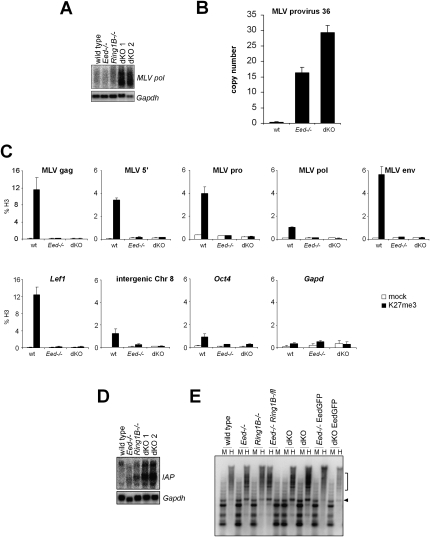
A redundant role for PRC1 and PRC2 in gene expression was unexpected, based on the existing models for PcG complex function. Hence, we wondered whether PcG complexes might contribute to a nuclear function apart from gene regulation, which could be lost in dKO ES cells. Previous studies have estimated that 15% of histone H3 is trimethylated on Lys 27 genome-wide (Peters et al. 2003; Schoeftner et al. 2006), and 10%–15% of histone H2A is ubiquitinated (de Napoles et al. 2004). Considering that genes constitute ∼2% of the genome, the amount of PcG-catalyzed histone modifications cannot be attributed solely to gene promoters. Therefore, we analyzed dKO expression profiles for transcripts originating from genomic repeats. Affymetrix arrays contain several probes that map to repeat elements. We observed a strong increase in expression of long-terminal repeat (LTR) retrotransposons in dKO ES cells when compared with wild-type, Eed-deficient, and Ring1B-deficient ES cells (Supplemental Fig. 9A). Northern analysis confirmed the derepression of murine leukemia virus (MLV) retroelements in dKO ES cells (Fig. 6A). To investigate whether derepression of MLV sequences could result in mobilization of MLV elements, we measured the copy number of a specific MLV integration on chromosome 8 (Changolkar et al. 2008). We observed a 15-fold increase in the copy number of this provirus in Eed-deficient ES cells and a 30-fold increase in dKO ES cells (Fig. 6B). In order to define whether MLV elements are targets of PcG complexes, we performed directed ChIP. Using different primer sets spanning the proviral sequence, we found that MLV elements are enriched for H3K27me3 in wild-type ES cells to a level similar to the Polycomb target gene Lef1 (Fig. 6C). The H3K27me3 enrichment on MLVs was lost in Eed-deficient ES cells, showing that the signal was specific. Enrichment of H3K27me3 over MLV elements was also maintained in Ring1B-deficient and differentiated ES cells (Supplemental Fig. 9C; data not shown). This demonstrates that Polycomb complexes contribute to the silencing of MLV retroviruses.
Figure 6.
Redundant repression of endogenous retroviruses by PRC1 and PRC2. (A) Northern analysis showing derepression of endogenous MLV retroelements in dKO ES cells. Gapdh was used as a loading control. (B) Quantitative PCR analysis showing an increase in MLV provirus copy number in Polycomb-deficient ES cells. The copy number was quantitated relative to an intergenic genomic sequence using the standard curve method. (C) Directed ChIP showing enrichment of H3K27me3 over MLV elements in ES cells. (Top row) Different regions of the MLV provirus sequence were investigated by quantitative PCR. (Bottom row) Lef1 was used as a positive control, and Oct4, Gapdh, and an intergenic sequence on chromosome 8 were used as negative controls. H3K27me3 signals are diminished in Eed-deficient ES cells demonstrating specificity. (D) Northern analysis showing the derepression of IAP retroviral sequences in dKO ES cells. Gapdh was used as loading control. (E) Restriction analysis with methylation-sensitive HpaII (H) and methylation-resistant MspI (M) and subsequent Southern analysis show loss of DNA methylation on IAP elements in Eed-deficient and dKO ES cells. Size of demethylated bands in HpaII lanes are indicated by the arrowhead and square bracket.
We also observed that intracisternal A particle (IAP) elements were derepressed in dKO ES cells (Fig. 6D). IAP repression could be restored by introduction of an EedGFP transgene in dKO ES cells (Supplemental Fig. 9B). Previously, it has been shown that DNA methylation is essential for maintaining repression of endogenous IAP retroelements during embryogenesis (Walsh et al. 1998). We investigated whether depletion of the PcG system would also affect DNA methylation of IAP elements. Using methylation-sensitive restriction enzyme analysis, we observed a reduction of DNA methylation of IAP elements in Eed-deficient and dKO ES cells (Fig. 6E). DNA methylation was largely maintained in Ring1B-deficient ES cells, which showed only slightly reduced methylation levels. This indicates that deficiency in PRC2 function leads to a loss of DNA methylation on IAP elements without affecting transcription. In conclusion, a subset of genomic repeats derived from LTR retrotransposons, including IAP and MLV elements, are repressed by PRC1 and PRC2 in ES cells. This indicates that, in ES cells, the Polycomb system contributes to the defense against parasitic DNA.
Discussion
Genomic repeats are targets of Polycomb complexes
We identify genomic repeat elements as a novel class of Polycomb targets. MLV and IAP retroelements are derepressed in dKO but not in PRC1- or PRC2-deficient ES cells, suggesting that both Polycomb complexes act in parallel in silencing of endogenous retroelements. The parallel recruitment of Polycomb complexes to genomic repeats is consistent with previous observations that genome-wide H3K27me3 is maintained in Ring1B-deficient ES cells (de Napoles et al. 2004; Schoeftner et al. 2006), and genomic ubH2A is maintained in Eed-deficient ES cells (Leeb and Wutz 2007). These data also suggest that by far the greatest proportion of Polycomb-mediated chromatin modifications is located in the nongenic regions of the genome. This might have important implications for understanding gene regulation by Polycomb complexes in mammals, as it indicates a potential role of repeat sequences in the PcG-mediated gene silencing mechanism. It is conceivable that genomic repeat sequences in mammals provide a binding platform for PcG proteins. Loss of both PRC1 and PRC2 leads to derepression of genomic repeats and, thus, could interfere with the function of such a repressive platform. We believe that this might underlie the severity of the phenotype of dKO ES cells, which show nearly twice as many genes derepressed when compared with ES cells lacking either PRC1 or PRC2 function. It will be exciting to investigate whether genomic repeats associate with Polycomb-regulated genes similar to the observed clustering of Polycomb-regulated genes and Polycomb response elements in Drosophila (Grimaud et al. 2006; Lanzuolo et al. 2007). In mammals, PRC1 and PRC2 are also recruited to the core of the inactive X chromosome, which contains genomic repeats (Chaumeil et al. 2006; Clemson et al. 2006). In X chromosome inactivation, PRC1 and PRC2 are recruited in parallel by the noncoding Xist RNA (Schoeftner et al. 2006; Leeb and Wutz 2007). Taken together, these findings suggest that Polycomb complexes might have a role in compartmentalizing the nucleus by establishing chromatin domains for gene repression.
Redundancy of PRC1 and PRC2 in gene repression
We show that the combined disruption of Ring1B and Eed results in an aberration of cell differentiation. Given the molecular differences of the activities of PRC1 and PRC2, the severity of this synthetic phenotype is unexpected. PRC1-catalyzed ubH2A has been reported to inhibit transcription elongation (Stock et al. 2007), suggesting a direct function in gene repression. Yet, the removal of ubH2A during mitosis is required for cell cycle progression, which precludes this modification as a heritable epigenetic mark (Joo et al. 2007). In contrast, H3K27me3 is transmitted through the cell cycle, but a direct function of H3K27me3 in gene repression has not been shown, and its only reported function is to recruit other Polycomb proteins such as Cbx7 or PRC2 (Fischle et al. 2003; Bernstein et al. 2006; Hansen et al. 2008; Margueron et al. 2009). The very different cell-biological properties of ubH2A and H3K27me3 indicate that the regulation of Polycomb-mediated silencing is a complex process. It is likely that considerable cross-talk exists between different Polycomb complexes, such as the reported binding of the PcG complex protein Rybp to ubH2A and Ring1B (Arrigoni et al. 2006), and other chromatin-associated complexes. PRC1 has additional functions in replication, and it has been shown that PRC1 proteins remain bound to chromatin when replicated in an in vitro system (Francis et al. 2009).
We find that Ring1B recruitment to PcG target genes depends quantitatively on PRC2, as most of the Ring1B signal on gene promoters is lost in Eed-deficient ES cells. Nevertheless, in the absence of PRC2, Ring1B remains detectable clearly over background level and is functionally relevant. In Ring1B-deficient ES cells, Suz12 and H3K27me3 signals are reduced on most nonredundantly silenced genes, but remain at wild-type levels on redundant gene promoters. These data could indicate that, to some extent, redundant gene silencing by PcG complexes reflects the recruitment of PRC1 and PRC2. However, we believe that additional factors might also contribute to and determine gene repression of PcG target genes.
Our data show that the deletion of Ring1B and Eed in ES cells disrupts the catalytic function of PRC1 or PRC2, respectively. Although we did not detect H3K27me3, we note that Ezh2 protein is still present at a reduced amount and could be functional to an extent below our detection. Since the same level of Ezh2 is also present in dKO cells, the differentiation defect does not arise due to further loss of Ezh2. Ring1B−/− ES cells are deficient for PRC1 function, as Ring1A, a functional homolog of Ring1B, is not expressed in ES cells and does not restore genomic ubH2A levels (Leeb and Wutz 2007). In addition, Ring1B deletion leads to a loss of several PRC1 proteins, including Rybp, Cbx4, Mel18, and Bmi1. At present, it is not resolved to what extent the catalytic activity toward ubH2A and the structural role of PRC1 proteins contribute to the phenotype. From our observations, we suggest that the function of PRC1 and PRC2 is largely eliminated by disruption of Ring1B and Eed, respectively.
Activity of PRC1 and PRC2 defines the differentiation potential of ES cells
Ring1B-deficient or Eed-deficient ES cells can differentiate into cell types of all three germ layers, suggesting that the dynamic modulation of gene expression required for differentiation can be performed by a single PcG complex. However, the complete loss of PcG function abolishes the tumor formation potential of dKO ES cells. The ability of an EedGFP transgene to restore tumor formation indicates that dKO ES cells can regain pluripotency. We find that, upon differentiation of dKO ES cells, pluripotency markers are down-regulated and differentiation markers are up-regulated. Thus, the differentiation defect of PcG-deficient ES cells does not result from a block to enter differentiation, but is due to a failure to maintain differentiated cells. This suggests that the Polycomb system is critical for fine-tuning gene expression, and that epigenetic patterns required to progress through differentiation cannot be set up in the absence of PcG regulation.
Polycomb complex function in ES cells
ES cells lacking Ring1B and Eed can self-renew and maintain pluripotency marker expression. We also show that they can contribute to the inner cell mass when injected into blastocysts. However, these ES cells are unstable and tend to spontaneously differentiate in culture. Furthermore, they fail to execute differentiation programs appropriately. Thus, Polycomb complexes stabilize ES cell identity. We measured only a slight decrease in the proliferation rate of dKO ES cells compared with controls, indicating that ES cell self-renewal is largely independent of epigenetic gene regulation. This is also consistent with the finding that ES cells lacking DNA cytosine methyltransferases are viable (Tsumura et al. 2006). Recently, it has been suggested that ES cells represent the ground state of pluripotency, which would not require epigenetic regulation (Silva and Smith 2008). Our data support this notion by showing that the network of transcription factors in ES cells is stable enough to maintain self renewal in the absence of Polycomb regulation. The ability to maintain ES cells in the absence of PRC1 and PRC2 catalytic activity now provides an opportunity for studying the function of the PcG system in gene repression, chromatin organization, and genome stability. In the future, this will facilitate characterizing the role of the Polycomb system for the nuclear architecture of mammals.
Materials and methods
Cell culture and generation of ES cell lines
ES cells were cultured as described (Leeb and Wutz 2007). Cells were counted with a Casy cell counter (Schaerfe System GmbH). Targeting of Ring1B was performed in Eed−/− ES cells following a conditional strategy as described previously (Leeb and Wutz 2007). For deletion of Ring1B, an inducible pCAG-CreERT2:EF1-BSD transgene was randomly inserted into Eed−/− Ring1B-/fl ES cells. The Blasticidin concentration used for selection was 5 μg/mL. For establishing dKO cells, Ring1B deletion was induced with 1 μM 4OHT for 48 h. dKO ES cells were established with a frequency of 85%–95% and subcloned before further analysis. dKO ES cells were maintained on high-density mitotically inactivated low-passage mouse embryonic fibroblast feeder cells in ES-DMEM supplemented with 1000 U/mL LIF and 15% of a special selected batch of fetal bovine serum (FBS) (PAA) tested for optimal growth of mouse ES cells. To reconstitute Eed expression, dKO ES cells were transfected with a pCAG-EGFP-Eed-IRES-HygpA transgene (Schoeftner et al. 2006). Selection was performed using 140 μg/mL Hygromycin. NS cells were derived from ES cells using a monolayer differentiation protocol (Pollard et al. 2006). Differentiation of ES cells was induced by 100 nM all-trans retinoic acid in medium containing 10% FBS and no LIF. Embryoid body cultures were established by aggregation of ES cells in a low-adherent tissue culture dish in the absence of LIF. ES cells for blastocyst injection experiments were GFP-labeled by transfection using Lipofectamine2000 (Invitrogen) with a linearized pCAC-EGFP-pA transgene. Stable ES cell lines were generated by FACS sorting 3 and 12 d after transfection.
Teratoma formation assay
We injected 5 × 105 ES cells into the flanks of nude mice in Matrigel Basement Membrane Matrix (BD Bioscience). We performed three injections each for wild-type, Eed−/−, Ring1B−/−, two independent dKO, and dKOEedGFP ES cell lines. After 3 wk, teratomas were excised, embedded in O.C.T. compound (Tissue Tek) for cryosections, and subsequently analyzed using immunohistochemistry. dKOEedGFP teratomas grew slowly and appeared after ∼10 wk of growth. Further characterization included H3K27me3 immunofluorescence analysis and PCR genotyping. dKO ES cells did not give rise to teratomas within 6 mo after injection.
Immunofluorescence, immunohistochemistry, and image acquisition
For immunofluorescence, ES cells were grown on Roboz slides (CellPoint Scientific). Immunostaining was performed as described previously (Leeb and Wutz 2007). Briefly, cells were fixed for 10 min in 4% PFA in PBS, permeabilized for 5 min in 0.1% Na citrate/0.5% Triton-X100, and blocked for 30 min in PBS containing 5% BSA and 0.1% Tween-20. DAPI (4′,6′-diamidino-2-phenylindole) was used to stain the DNA. Vectashield (Vector Laboratories) was used as imaging medium. Immunohistochemistry was performed on frozen sections using the Ventana Discovery System. Immunofluorescence images were obtained at 20× magnification at room temperature with a fluorescence microscope (Zeiss Axioplan 2) using a Coolsnap fx CCD camera (Photometrics) and the MetaMorph image analysis software (Universal Imaging). Images of ES cell colonies and retinoic acid-differentiated cells were taken with a Zeiss Axiovert 200 microscope. A Zeiss Axioplan 2 microscope was used to analyze immunohistochemistry staining at a 20× magnification.
RNA and protein analysis
Northern analysis was performed using 5–15 μg of RNA purified with Trizol (Invitrogen) as described previously (Leeb and Wutz 2007). Primers used to generate Northern probes are listed in Supplemental Table 3. Images were obtained using a Storm 860 Scanner (Molecular Dynamics) and Image Quant TL software. Proteins were extracted in RIPA buffer. Histones were acid-extracted in 0.2N HCl. α-Lamin B1, α-H3, and Ponceau S stainings were used to control for loading. The following antibodies were used: α-Ring1B (Atsuta et al. 2001), α-Mph2 (Isono et al. 2005), α-Mel18 (Santa Cruz Biotechnologies), α-Suz12 (Upstate Biotechnologies), α-Ezh2, α-H3K27me3 (Kohlmaier et al. 2004), α-H2AK119ub1 (Upstate Biotechnologies), α-H3 (Abcam), α-Nestin (Developmental Studies Hybridoma Bank), α-Lamin B1 (Abcam), smooth muscle actin (Thermo Scientific), glial fibrillary acidic protein (GFAP; Dako), α-1-Fetoprotein (Dako), and Troma-1 (Developmental Studies Hybridoma Bank). Secondary antibodies used were Alexa Fluor 488 goat anti-rabbit IgG, Alexa Fluor 488 goat anti-mouse IgG (Molecular Probes), HRP-conjugated Affinipure goat α-rabbit IgG, and HRP-conjugated, Affinipure goat α-mouse IgG (Jackson ImmunoResearch Laboratories).
Quantitative gene expression analysis
cDNA was generated from 2–4 μg of DNase-treated total RNA using the SuperScript II Reverse Transcription kit (Invitrogen) and random hexamere primers. Quantitative real-time PCR analysis was performed in triplicate using the Bio-Rad iQ Taq SYBR Green master mix on a Bio-Rad iCycler machine. The standard curve method was used for quantification of gene expression. Expression levels were normalized to L32 ribosomal protein. Error bars represent the standard deviation. Sequences for all primers used in this study are listed in Supplemental Table 3.
Affymetrix transcription analysis
ES cell cultures were carefully analyzed for ES cell morphology before microarray analysis. Gene expression profiles were established from Trizol-extracted total RNA of biological triplicates by Atlas Genomics using Affymetrix GeneChip 430 2.0 arrays, and were deposited in Gene Expression Omnibus (GEO) with the accession code GSE19076 (http://www.ncbi.nlm.nih.gov/geo/querry/acc.cgi?acc=GSE19076). Normalization was performed using the MAS 5 algorithm. Pairwise comparisons between all genotypes were performed. Genes regulated at least twofold with a P-value of <0.05 were analyzed further. PcG target gene sets, CpG promoter classes, and pluripotency transcription factor-bound genes were annotated from published data sets (Boyer et al. 2006b; Kim et al. 2008; Ku et al. 2008). PcG target genes were defined as genes with a bivalent promoter status (Ku et al. 2008). One-hundred-sixty-eight genes, which were most regulated between the different genotypes, were selected, and a hierarchical clustering of the log-transformed, median-centered, and normalized expression was performed and represented as a heat map.
ChIP
ChIP was performed as described previously with minor modifications (Sado et al. 2005). For each experiment, 2 × 106 ES cells were used. Rabbit IgG was used as mock control. Immunoprecipitation was performed using Dynabeads Protein G (Invitrogen). Primer sequences for MLV provirus sequences were reported previously (Changolkar et al. 2008). Quantitative PCR was performed in duplicates. Error bars represent standard deviation. ChIP results were confirmed in two independent experiments. The ChIP data presented in Figure 5 was generated as described previously (Pasini et al. 2008).
The following antibodies were used: rabbit H3K27me3 (Upstate Biotechnologies), rabbit monoclonal H3K27me3 (Cell Signaling), rabbit H3 (Abcam), rabbit IgG (Sigma), rabbit Suz12 (Cell Signaling), Ring1B (K Helin and D Pasini, in prep.).
MLV copy number analysis
The copy number was analyzed using primers specific for one MLV integration site on chromosome 8. The fold change between wild-type and Eed-deficient or dKO ES cells was calculated using the standard curve method. Normalization between samples was performed using an intergenic region on chromosome 8. The MLV copy number was then set to 1 in wild-type ES cells.
DNA methylation analysis
DNA samples of wild-type, Eed−/−, Ring1B−/−, Eed−/− Ring1B-/fl, dKO, Eed−/− EedGFP , and dKOEedGFP ES cells were digested with MspI and the CpG methylation-sensitive isoschizomere HapII. Primers used to generate the IAP-specific Southern probe can be found in Supplemental Table 3.
Accession of data sets
Gene expression profiles were deposited at GEO with the accession code GSE19076 (http://www.ncbi.nlm.nih.gov/geo/querry/acc.cgi?acc=GSE19076).
Acknowledgments
We thank Ruben Agrelo and Hiroyuki Kishimoto for help with ES cell injections, Christian Theussl and Jacek Wojciechowski for blastocyst injections, Andreas Bichl and Denise Imre for maintenance of the mouse colony, Vukoslav Komnenovic for histology service, and Mediyha Saltik for technical assistance. This research was supported by the IMP through Boehringer Ingelheim, and by grants from the Austrian genome initiative GEN-AU, the Vienna Science and Technology Fund (WWTF), and the Austrian Science Fund (FWF). A.W. is supported by a Wellcome Trust Senior Research Fellowship (grant reference 087530/Z/08/A).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.544410.
Supplemental material is available at http://www.genesdev.org.
References
- Arrigoni R, Alam SL, Wamstad JA, Bardwell VJ, Sundquist WI, Schreiber-Agus N. The Polycomb-associated protein Rybp is a ubiquitin binding protein. FEBS Lett. 2006;580:6233–6241. doi: 10.1016/j.febslet.2006.10.027. [DOI] [PubMed] [Google Scholar]
- Atsuta T, Fujimura S, Moriya H, Vidal M, Akasaka T, Koseki H. Production of monoclonal antibodies against mammalian Ring1B proteins. Hybridoma. 2001;20:43–46. doi: 10.1089/027245701300060427. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr Opin Genet Dev. 2006a;16:455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006b;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Changolkar LN, Singh G, Pehrson JR. MacroH2A1-dependent silencing of endogenous murine leukemia viruses. Mol Cell Biol. 2008;28:2059–2065. doi: 10.1128/MCB.01362-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatoo W, Abdouh M, David J, Champagne MP, Ferreira J, Rodier F, Bernier G. The polycomb group gene Bmi1 regulates antioxidant defenses in neurons by repressing p53 pro-oxidant activity. J Neurosci. 2009;29:529–542. doi: 10.1523/JNEUROSCI.5303-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes & Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Gu X, Su IH, Bottino R, Contreras JL, Tarakhovsky A, Kim SK. Polycomb protein Ezh2 regulates pancreatic β-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes & Dev. 2009;23:975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Dhawan S, Tschen SI, Bhushan A. Bmi-1 regulates the Ink4a/Arf locus to control pancreatic β-cell proliferation. Genes & Dev. 2009;23:906–911. doi: 10.1101/gad.1742609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Endoh T, Fujimura Y, Ohara O, Toyoda T, Otte AP, Okano M, Brockdorff N, Vidal M, et al. Polycomb group proteins Ring1A/B are functionally linked to the core transcriptional regulatory circuitry to maintain ES cell identity. Development. 2008;135:1513–1524. doi: 10.1242/dev.014340. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Chen T, Chadwick B, Li E, Zhang Y. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation. J Biol Chem. 2004;279:52812–52815. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes & Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Isono K, Fujimura Y, Shinga J, Yamaki M, O-Wang J, Takihara Y, Murahashi Y, Takada Y, Mizutani-Koseki Y, Koseki H. Mammalian polyhomeotic homologues Phc2 and Phc1 act in synergy to mediate polycomb repression of Hox genes. Mol Cell Biol. 2005;25:6694–6706. doi: 10.1128/MCB.25.15.6694-6706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14:183–193. doi: 10.1016/s1097-2765(04)00185-6. [DOI] [PubMed] [Google Scholar]
- Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, Verrijzer CP. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes & Dev. 2008;22:2799–2810. doi: 10.1101/gad.484208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- Leeb M, Wutz A. Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J Cell Biol. 2007;178:219–229. doi: 10.1083/jcb.200612127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Cao L, Chen J, Song S, Lee IH, Quijano C, Liu H, Keyvanfar K, Chen H, Cao L-Y, et al. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459:387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury Iii WJ, Voigt P, Martin SR, Taylor WR, De Marco V, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes & Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes & Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: Postponing commitment. Curr Opin Cell Biol. 2008;20:201–207. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Pollard SM, Benchoua A, Lowell S. Neural stem cells, neurons, and glia. Methods Enzymol. 2006;418:151–169. doi: 10.1016/S0076-6879(06)18010-6. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- Sado T, Hoki Y, Sasaki H. Tsix silences Xist through modification of chromatin structure. Dev Cell. 2005;9:159–165. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–536. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A. Nanog is the gateway to the pluripotent ground state. Cell. 2009;138:722–737. doi: 10.1016/j.cell.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- van der Stoop P, Boutsma EA, Hulsman D, Noback S, Heimerikx M, Kerkhoven RM, Voncken JW, Wessels LF, van Lohuizen M. Ubiquitin E3 ligase Ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouse embryonic stem cells. PLoS One. 2008;3:e2235. doi: 10.1371/journal.pone.0002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, Deschamps J, van Lohuizen M. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci. 2003;100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Wang J, Mager J, Schnedier E, Magnuson T. The mouse PcG gene eed is required for Hox gene repression and extraembryonic development. Mamm Genome. 2002;13:493–503. doi: 10.1007/s00335-002-2182-7. [DOI] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]