Abstract
The controlling factors that prompt mature oligodendrocytes to myelinate axons are largely undetermined. In this study, we used a forward genetics approach to identify a mutant mouse strain characterized by the absence of CNS myelin despite the presence of abundant numbers of late-stage, process-extending oligodendrocytes. Through linkage mapping and complementation testing, we identified the mutation as a single nucleotide insertion in the gene encoding zinc finger protein 191 (Zfp191), which is a widely expressed, nuclear-localized protein that belongs to a family whose members contain both DNA-binding zinc finger domains and protein–protein-interacting SCAN domains. Zfp191 mutants express an array of myelin-related genes at significantly reduced levels, and our in vitro and in vivo data indicate that mutant ZFP191 acts in a cell-autonomous fashion to disrupt oligodendrocyte function. Therefore, this study demonstrates that ZFP191 is required for the myelinating function of differentiated oligodendrocytes.
Keywords: Mouse mutant, forward genetics, hypomyelination, conditional allele, zinc finger protein, scan domain
The myelin sheath is a feature unique to vertebrates. The ensheathment of axons by myelin allows for the rapid conduction of action potentials along the nerve fiber (Baumann and Pham-Dinh 2001). In addition to its role in facilitating saltatory conduction, the myelin sheath also insulates axons and contributes to their survival (Griffiths et al. 1998). During CNS development, a majority of oligodendrocyte progenitor cells (OPCs) originate in the ventricular zone and migrate to their target axons (Richardson et al. 2006). Progenitor cell fate is determined by several transcription factors, including Olig1, Olig2, Ying Yang 1 (Yy1), and Nkx6.2 (Lu et al. 2001, 2002; Zhou and Anderson 2002; He et al. 2007; Wegner 2008). The migration pattern of OPCs is influenced by extracellular signals, such as netrin-1 and the chemokine CXCL1 (Tsai et al. 2002, 2003). Upon reaching their final destinations, these cells undergo terminal differentiation, first becoming premyelinating then myelinating oligodendrocytes (Wegner 2008).
Oligodendrocyte differentiation is characterized by changes in both cell morphology and protein expression. Since oligodendrocytes can be isolated and differentiated in culture, in vitro studies have been particularly useful in the characterization of the different stages of the oligodendrocyte maturation process (Armstrong 1998). Morphologically, OPC differentiation begins with the extension of thin processes (Armstrong 1998). This differentiation process is regulated by several transcription factors, such as Sox10, Nkx2.2, Olig1, Id2, Id4, and zinc finger protein 488 (Zfp488) (Kondo and Raff 2000; Qi et al. 2001; Wang et al. 2001, 2006; Xin et al. 2005; Li et al. 2007). As OPC differentiation progresses, the cells become immature/premyelinating oligodendrocytes, and the myelinating cell-specific marker O4 appears (Armstrong 1998). At this stage, several of the myelin genes—such as myelin-associated glycoprotein (Mag) and 2′,3′-cyclic nucleotide 3′-phosphodiesterase (Cnp)—are expressed at a low level. As maturation approaches completion, the processes thicken and form sheet-like structures (Armstrong 1998). This later stage of in vitro maturation—when the mature oligodendrocyte markers myelin basic protein (MBP), myelin oligodendrocyte glycoprotein (MOG), and myelin-associated oligodendrocyte basic protein (MOBP) are expressed (Armstrong 1998; Baumann and Pham-Dinh 2001)—is thought to correlate with the in vivo myelination of axons.
Although several regulatory factors such as Yy1 and Olig1 have been identified in the regulation of OPC differentiation into mature oligodendrocytes, little is known about the factors that control the conversion of premyelinating to myelinating oligodendrocytes (Xin et al. 2005; He et al. 2007). Recently, advances in microarray and oligodendrocyte isolation technologies have led to the identification of myelin gene regulatory factor (Mrf), also known as gene model 98, as a critical gene in this post-mitotic differentiation state (Dugas et al. 2006; Nielsen et al. 2006; Cahoy et al. 2008; Emery et al. 2009). Mrf appears to be necessary for the expression of the myelin genes after terminal differentiation of the oligodendrocyte. A better understanding of the regulatory mechanisms that control the final stages of oligodendrocyte maturation is critical, since it appears that the disruption of oligodendrocyte differentiation at the premyelinating stage is a major pathological feature of the autoimmune, demyelinating neurological disorder multiple sclerosis (MS) (Chang et al. 2002).
In the present study, we used a phenotype-driven strategy to identify a molecule critical to late-stage oligodendrocyte maturation. In mammals, the loss of CNS myelin frequently leads to tremors and seizures. We therefore used these phenotypes as the basis of our screen for mutants with myelin defects. Here, we describe a mutant mouse strain that is characterized by tremors and tonic seizures that are the result of severe hypomyelination in the CNS. These mice are homozygous for a spontaneous mutation we named hypomyelinated CNS (allele symbol: hmcns). These mutant animals contain abundant numbers of process-extending oligodendrocytes; however, these cells express an array of myelin protein genes at significantly reduced levels. Through linkage mapping and complementation testing, the causative mutation was identified in Zfp191, a gene encoding a zinc finger protein.
Results
The hmcns mutant exhibits severe CNS hypomyelination
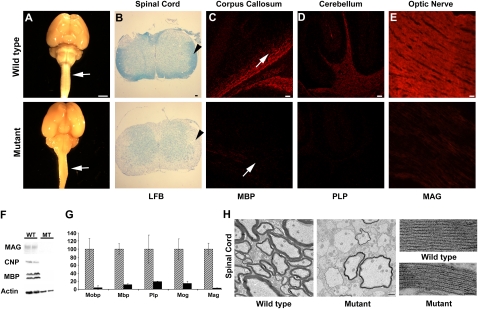
The hmcns mutant phenotype is the result of a recessive mutation that arose spontaneously on the C3H/HeJ background. Beginning at around postnatal day 14 (P14), mutants can be identified by a tremor and the occurrence of tonic seizures (Supplemental Movie 1). Typically, most mutants die by P25. Brains and spinal cords isolated from P14 mutant mice appeared to be smaller than those from wild-type mice and to lack white matter (Fig. 1A). To further investigate a potential myelin deficit in these mutants, brain, spinal cord, and optic nerve sections were stained with the lipophilic dye Luxol fast blue (LFB) and by immunohistochemistry (IHC) using antibodies against the myelin proteins MBP, proteolipid protein (PLP), and MAG. A myelin deficiency in the CNS was confirmed by reduced LFB staining (Fig. 1B), as well as by reduced levels of immunostaining for MBP, PLP, and MAG (Fig. 1C–E). Consistent with the IHC data, Western blot analysis of isolated total brain proteins demonstrated that the myelin proteins MBP, CNP, and MAG were dramatically reduced in the mutant brain (Fig. 1F). Additionally, real-time quantitative RT–PCR (qPCR) revealed a significant reduction in the mRNA expression levels of several myelin-related genes (Fig. 1G). The myelination defect remains until the death of these mice, and myelin gene expression does not increase as these mice age from 2 to 3 wk (data not shown).
Figure 1.
The hmcns mutation causes severe myelin defects in all CNS tissues. (A) Images of P14 wild-type and mutant whole-brain and spinal cord samples demonstrate the lack of white matter (arrows) in the mutants. Bar, 5 mm. (B) Spinal cord sections stained with LFB dye reveal the lack of myelin in the mutant (arrowhead). Bar, 50 μm. (C, bottom panel) Immunohistochemical staining of P14 corpus callosum with anti-MBP shows a severe myelin deficit throughout the CNS in the mutant. Arrows point at the corpus callosum. Bar, 50 μm. (D, bottom panel) Immunohistochemical staining of P14 cerebellum with anti-PLP shows a severe myelin deficit in the mutant. Bar, 50 μm. (E, bottom panel) Immunohistochemical staining of P14 optic nerve with anti-MAG shows a severe myelin deficit in the mutant. Bar, 50 μm. (F) Myelin loss was also confirmed by Western blot analysis of MBP, CNP, and MAG. Actin was used to indicate the presence of proteins in the mutant sample. (G) qPCR of Mobp, Mbp, Plp, Mog, and Mag indicates that myelin gene expression levels were severely deficient in the mutant brains. Three P14 wild-type and mutant brains were used for this study. Error bars represent standard deviation (STD). Analysis by Student's t-test determined that the reduction was statistically significant (P < 0.05). Mutant expression values are normalized to 1 for each gene, and the wild-type expression levels are shown as fold differences. Wild-type mice are represented by diagonal boxes, and mutant mice are represented by black boxes. (H) TEM reveals that most axons are unmyelinated in the mutant spinal cord. Bar, 1 μm. When viewed at a high magnification, the periodicity and compaction of the rare myelin sheaths present in the mutant appear to be comparable with the wild type. Bar, 25 nm.
Examination of spinal cord and optic nerve by transmission electron microscopy (TEM) revealed that the vast majority of the P14 mutant's axons were unmyelinated (Fig. 1H) as compared with wild type (Fig. 1H). This observation was confirmed when the percentage of myelinated axons in wild-type and mutant P14 optic nerves was quantitated (69% ± 24.5% and 3% ± 1.8%, respectively; n = 3). The rare myelin sheaths present in the mutant appeared to be normally compacted, although thinner in the mutant than in the wild-type mouse (Fig. 1I). The reduction in myelin thickness was confirmed by calculating the g-ratios of the myelinated fibers, which were 0.75 ± 0.007 in wild-type and 0.84 ± 0.016 in mutant spinal cords (n = 3).
Despite the myelin abnormalities in the CNS, axons in the peripheral nervous system (PNS) appeared to be normally myelinated, as observed by TEM (Supplemental Fig. 1). Morphometric analysis of PNS fibers produced similar g-ratio values for both mutant and wild-type mice (data not shown). Using qPCR, sciatic nerve MBP mRNA levels were found to be similar in mutant and wild-type mice (data not shown), suggesting that the myelin effects of the hmcns mutation are restricted to the CNS.
Mutant oligodendrocytes arrest at a late stage of differentiation
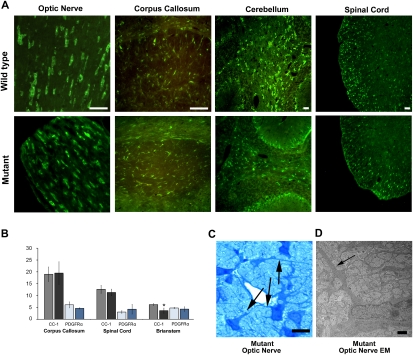
The severe CNS hypomyelination and the dramatically reduced levels of myelin gene transcription displayed by hmcns mutants raised the possibility that abnormalities in OPC differentiation might exist in these animals. Nevertheless, when different regions of the mutant P14 CNS were probed with CC-1 antibodies, a marker for post-mitotic and myelinating oligodendrocytes, many mutant oligodendrocytes were positive for the marker, indicating that they were late-stage cells (Fig. 2A). When CC-1-positive cells were counted in the corpus callosum, spinal cord, brain stem, and optic nerve (data not shown), mutant oligodendrocyte numbers were similar, although slightly diminished in the brain stem, compared with those of wild type (Fig. 2B). The abundant presence of late-stage oligodendrocytes in the mutant was confirmed by in situ hybridization against PLP mRNA (Supplemental Fig. 2). Furthermore, IHC staining to detect the platelet-derived growth factor receptor-α (PDGFRα), a marker for OPCs, did not show an alteration in the number of immature cells in the various P14 mutant CNS regions examined as compared with P14 wild type (Fig. 2B), suggesting that there are no overt alterations to the oligodendrocyte lineage in the hmcns mutants.
Figure 2.
The mutant OPCs are capable of differentiating into late-stage oligodendrocytes. (A) Late-stage oligodendrocytes (CC-1-positive cells) were identified in the mutant CNS at P14. Wild-type and mutant CNS tissue sections stained with CC-1 antibodies showed abundant numbers of positive cells in both samples. Bars, 50 μm. (B) When the number of positive CC-1 (black-shaded columns) and PDGFRα (blue-shaded columns) cells in the corpus callosum, spinal cord, and brain stem were counted, the results were similar for wild-type (gray-shaded column, CC-1; light-blue-shaded column, PDGFRα) and mutant (black-shaded column, CC-1; dark-blue-shaded column, PDGFRα) mice, with only brain stem CC-1 counts showing a slight significant difference (P < 0.05). Counts are per 10,000 μm2. N = 3 for wild type and mutant. Error bars represent STD. (C) Mutant optic nerve stained with toluidine blue revealed oligodendrocytes extending processes (arrows), suggesting an attempt to myelinate. Bar, 10 μm. (D) Higher magnification of the optic nerves using TEM revealed that the mutant axons were not myelinated. At this magnification, unmyelinated axons can be seen in the mutant optic nerve. The arrows indicate oligodendrocyte processes. Bar, 1 μm.
Mutant oligodendrocytes were further examined morphologically using light microscopy and TEM. The mutant cells were observed to be extending normal-appearing processes that were in contact with axons, although the majority of these processes did not form myelin sheaths (Fig. 2C,D). Collectively, the analyses of the hmcns mutant suggest that the CNS hypomyelination is due to a defect in the final stages of oligodendrocyte maturation that severely limits the capability of the oligodendrocytes to myelinate axons.
hmcns is a novel allele of the Zfp191 gene
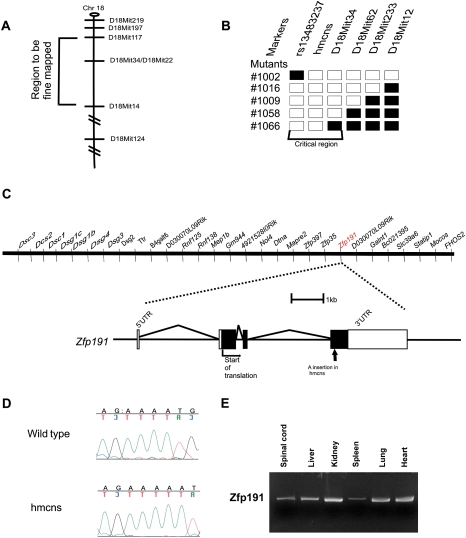
In order to understand the molecular basis of the hmcns phenotype, a linkage mapping approach was used to identify the causative mutation. Heterozygous carriers of the spontaneous mutation, which arose on the C3H/HeJ background, were outcrossed to CAST/EiJ mice to generate F1 heterozygous carriers. An intercross breeding strategy was used to produce F2 mutants, since homozygous mutants die by P25 and thus are unable to mate. Genotyping 12 F2 mutant homozygotes for polymorphic microsatellite markers revealed that the mutation is on proximal chromosome 18 (two out of 24 recombinant chromosomes with D18Mit14). Genotyping of additional mice with more densely spaced marker loci showed zero out of 42 chromosomes with recombination events between the mutant locus and either marker D18Mit117 or D18Mit22. In a second intercross with C57BL/6J mice, the genotyping of 60 affected F2 mice for polymorphic microsatellite markers spaced throughout the genome confirmed the mutation's location on the proximal end of chromosome 18 (Fig. 3A). Fine mapping with additional markers further localized the mutation to the 5.5-Mb region between the microsatellite marker D18Mit34 and the SNP marker rs13483237 (Fig. 3B). This region included 29 known and predicted genes at the time of mapping (Fig. 3C; University of California at Santa Cruz Genome Browser Assembly, February 2006). Real-time qPCR analysis of the genes within the 5.5-Mb region was performed to determine if any displayed a differential level of expression between the mutant and wild type (data not shown). When no expression differences were observed, all of the genes within this region known to be expressed in the brain were sequenced, and an adenine (A) insertion was discovered in a stretch of four As, coding nucleotides 586–589, of Zfp191, a putative transcription factor with four zinc finger domains that is expressed in multiple tissues (Fig. 3D,E; Supplemental Fig. 3). This insertion causes a frameshift in the ORF, resulting in a premature stop codon that truncates the gene product and eliminates a 155-amino-acid region that contains all four zinc finger domains (Supplemental Fig. 3, right panel).
Figure 3.
The hmcns mutation is caused by an A nucleotide insertion in the Zfp191 gene. (A) Schematic of chromosome 18 (Chr. 18) showing hmcns localized between the markers D18Mit117 and D18Mit14. (B) With the addition of markers between D18Mit117 and D18Mit14, the critical region of the mutation was mapped to the region between the SNP rs13483237 and the SSLP marker D18Mit34. Black boxes and white boxes represent alleles derived from C57BL/6J (normal) and C3H/HeJ (mutation background) mice, respectively. (C, top) The area defined by fine mapping contains 29 genes. (Bottom) Zfp191 is a gene consisting of four exons. The arrow indicates the location of the adenine insertion in the gene. (D) Chromatogram showing the A insertion in the mutant sequence. (E) RT–PCR analysis of Zfp191 expression, which revealed a transcript in several tissues in addition to the CNS.
To rule out the possibility that the A insertion represents natural variation among different strains, we sequenced 20 different mouse strains, including the parental C3H/HeJ strain. The A insertion was never detected (data not shown). Additionally, when the coding exons of the other 28 genes within the critical region were sequenced, no additional mutations were discovered (data not shown). To confirm that the insertion mutation in Zfp191 is responsible for the hypomyelinating hmcns phenotype, we performed a genetic complementation test. As described in the Materials and Methods, we used a gene targeting approach to generate a mouse strain carrying a Zfp191null allele (Supplemental Fig. 4) that we then crossed to hmcns heterozygous animals. Six out of 19 pups derived from this cross developed a tremoring phenotype. These six mice were compound heterozygotes carrying both the Zfp191hmcns insertion allele and the null allele. None of the 13 animals that displayed a normal phenotype carried both mutant alleles. IHC staining using anti-MBP antibodies confirmed that the compound heterozygous mutants were hypomyelinated in the CNS (Supplemental Fig. 5).
ZFP191 is required for myelination
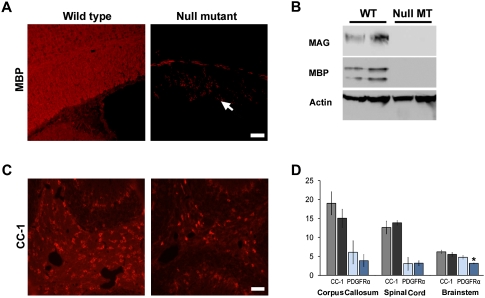
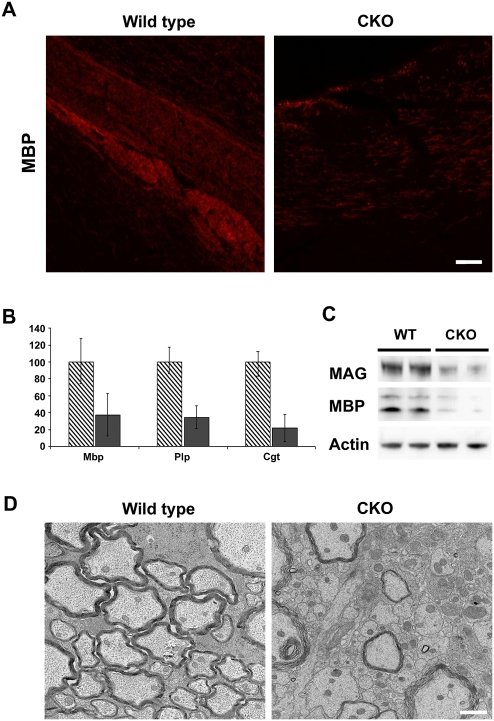
Since the mutant Zfp191 allele is expressed in homozygous hmcns mutants (data not shown) and the SCAN domain is predicted to be intact, a possibility exists that some functions of the gene remain. To better clarify the function of Zfp191 in CNS myelination, homozygous Zfp191null mutants were generated on the C57BL/6J background. Similar to the hmcns mutants, these null mutants also displayed tremors and tonic seizures. The null mutants also died prematurely at ∼3.5 wk. Detection of MBP by IHC revealed a severe hypomyelination phenotype in the CNS of the null mutants (Fig. 4A). Staining with other antibodies against CNS myelin sheath components also showed a similar dearth of myelin (data not shown). Furthermore, no MBP or MAG was detected by Western blot (Fig. 4B). Despite the absence of myelin, OPCs and late-stage oligodendrocyte numbers were comparable with wild type throughout the mutant CNS (Fig. 4C,D). Because of the similar phenotypes displayed by the hmcns and Zfp191null mutants, it is likely that the hmcns allele retains little to no function. Since the knockout of Zfp191 resulted in a hypomyelination phenotype, these data suggest that myelination is a functional role of Zfp191, and that ZFP191 is required for the conversion of oligodendrocytes from the premyelinating to the myelinating state.
Figure 4.
The null mutants recapitulate the hmcns phenotype. (A) IHC in the corpus callosum of wild-type and Zfp191null mutant mice reveals significantly decreased MBP staining in the Zfp191null mutant, indicating the lack of myelin (arrow in the mutant panel indicates corpus callosum). Bar, 50 μM. (B) Western blot analysis shows a severe decrease in myelin protein levels. (C) Despite the lack of myelin, CC-1-positive oligodendrocytes are present in the Zfp191null mutant. Bar, 50 μM. (D) When the number of positive CC-1 (black-shaded columns) and PDGFRα (blue-shaded columns) cells in the corpus callosum, spinal cord, and brain stem were counted, the results were similar for wild-type (gray-shaded column, CC-1; light-blue-shaded column, PDGFRα) and mutant (black-shaded column, CC-1; dark-blue-shaded column, PDGFRα) mice, with only brain stem PDGFRα counts showing a slight significant decrease in the mutants (P < 0.05). Counts are per 10,000 μm2. N = 3 for wild type and mutant. Error bars represent STD.
The hmcns mutation has a cell-autonomous effect in oligodendrocytes
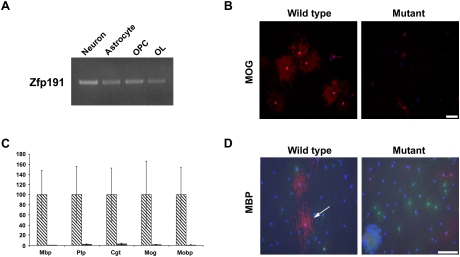
The loss of Zfp191 results in a CNS-specific defect despite expression of the gene in all tissues examined (Khalfallah et al. 2008). Furthermore, cells that have been shown to play a role in myelination—neurons, astrocytes, OPCs, and oligodendrocytes—all express Zfp191 mRNA (Fig. 5A). Since the expression pattern of Zfp191 does not reveal the cell-specific function of Zfp191, in vitro and in vivo experiments were performed to determine the cellular cause of the hypomyelination defect.
Figure 5.
The Zfp191 gene product has an oligodendrocyte-specific effect in vitro. (A) Zfp191 mRNA is expressed in isolated rat neurons, astrocytes, OPCs, and late-stage oligodendrocytes, as detected by RT–PCR. (B) OPCs isolated from P7 wild-type and Zfp191null mutant mice, which were allowed to differentiate for 5 d in culture and then were stained for MOG, showed that mutant oligodendrocytes produced less MOG than control cells. MOG staining is red, and DAPI is blue. Bar, 100 μm. (C) Examination of myelin gene transcripts by qPCR in OPCs differentiated for 5 d shows that cells isolated from Zfp191null mutants produced very little myelin gene mRNA compared with similarly differentiated wild-type cells (n = 3; P < 0.05). Error bars represent STD. Diagonally striped boxes represent wild type, and gray boxes represent mutants. (D) Oligodendrocytes isolated from P7 wild-type mice were induced to myelinate axons of RGC neurons isolated from rat in cocultures, then were stained for mature oligodendrocyte (MBP, red), OPC (NG2, green), and astrocyte (GFAP, blue) markers. Axons positively stained for MBP are ensheathed by myelin. hmcns mutant oligodendrocytes were not able to myelinate axons as readily as wild-type cells. The arrow indicates MBP-positive axons that are considered ensheathed. Bar, 100 μm.
OPCs from P7 mice were isolated using an immunopanning approach (Watkins et al. 2008). OPC numbers were determined at the time of isolation and following proliferation: No differences in cell numbers between wild-type and Zfp191null cultures were observed (data not shown). When the OPCs were allowed to differentiate for 5 d and then were examined for the late-stage oligodendrocyte markers MOG and MBP, the mutant cultures displayed a severe decrease compared with wild-type cultures (Fig. 5B; data not shown, respectively). The number of MOG-positive cells in the wild-type differentiated cultures (24.17 ± 5.18) were significantly greater than in the mutant cultures (4.97 ± 1.52; n = 4; P < 0.05). The mutant oligodendrocytes also produced significantly lower levels of myelin gene transcripts than the wild-type oligodendrocytes (Fig. 5C). Furthermore, when the mutant OPCs were cocultured with wild-type retinal ganglion cells (RGCs), they displayed a reduced ability to myelinate RGC axons, as indicated by the decrease in MBP staining (Fig. 5D). The percentage of OPCs associated with axons that were surrounded by MBP-positive sheaths was reduced fivefold in the mutant versus wild-type cocultures (0.84% ± 0.13% of the mutant cells vs. 4.84% ± 0.92% of the wild type). These studies suggest that, in culture, ZFP191 plays an intrinsic role in the oligodendrocyte's ability to mature and myelinate axons.
To determine if Zfp191 has an intrinsic oligodendrocyte effect in vivo, oligodendrocyte-specific Zfp191 knockout mice were generated. The oligodendrocyte-specific conditional mutants were generated by crossing C57BL/6J mice with a floxed Zfp191 allele (Supplemental Fig. 4) to C57BL/6J mice that express the Cre recombinase under the control of the Cnp transcriptional regulatory region, which drives oligodendrocyte-specific expression of Cre (Lappe-Siefke et al. 2003). The conditional mutants displayed mild tremors and tonic seizures beginning at P14. Histological analysis of 3-wk-old conditional mutants showed reduced staining of MBP (Fig. 6A), MAG (data not shown), and PLP (data not shown). Additionally, Western blots and real-time qPCR of myelin genes indicated a severe reduction in myelin protein levels and a fourfold decrease in the expression levels of several myelin transcripts in 3-wk-old mutant animals (Fig. 6B,C). Furthermore, TEM analysis of spinal cord from the conditional mutants confirmed that a majority of axons were not myelinated (Fig. 6D). These in vivo data concur with the in vitro data and further demonstrate that proper ZFP191 function within oligodendrocytes is essential for robust CNS myelination, indicating that Zfp191 has a cell-autonomous effect in these cells.
Figure 6.
Conditional knockout of Zfp191 in oligodendrocytes results in hypomyelination. (A) IHC of wild-type and Zfp191 conditional mutant corpus callosum using anti-MBP showed reduced myelin in the oligodendrocyte-specific Zfp191 knockout. Bar, 50 μM. Samples were from P21 mice. (B) qPCR of Mbp, Plp, and Cgt mRNA found that message expression was reduced for all three genes in Zfp191 conditional mutant brains. Diagonally striped boxes represent wild-type brains, and gray boxes represent mutants. Error bars represent STD. All expression data were significantly different (P < 0.05). Samples were from P21 mice. (C) Western blotting found that the levels of MAG and MBP protein are severely reduced in Zfp191 conditional mutant brains. Samples were from P21 mice. (D) Observation by TEM revealed that many axons in the mutant spinal cord are unmyelinated (P21). Bar, 1 nm.
The hmcns mutation down-regulates the expression of genes expressed specifically in mature myelinating oligodendrocytes
The presence of four zinc finger domains suggests that ZFP191 is likely to be a transcription factor, which would be reflected in a nuclear localization. To determine the subcellular localization of ZFP191, a plasmid containing the Zfp191 cDNA with a V5 epitope tag under the control of the CMV transcriptional regulatory region was constructed and transfected into HeLa cells. After 2 d of incubation, the localization of the tagged protein was visualized by IHC staining for the V5 epitope and observed to be localized to the nucleus (Supplemental Fig. 6).
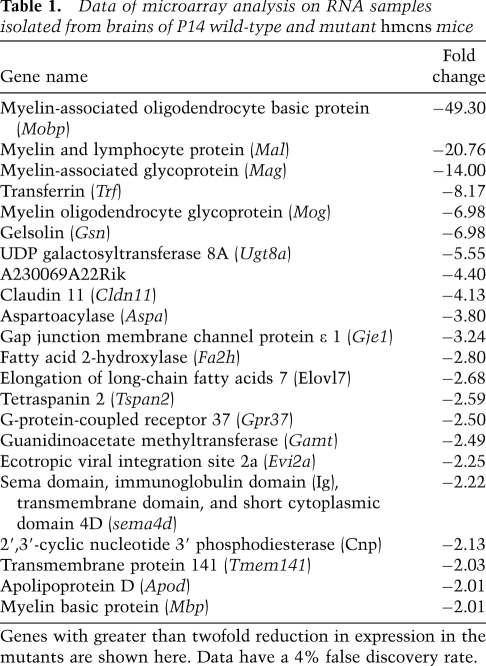
The confirmation of ZFP191's nuclear localization increases the likelihood that ZFP191 functions as a transcription factor. To identify genes whose expression levels are affected downstream from the hmcns mutation, total RNA samples isolated from P14 wild-type and mutant brains were analyzed by microarray using Affymetric Mouse Exon FIRST arrays. No genes were found to be up-regulated greater than twofold in the mutant RNA samples (data not shown). Table 1 shows the genes that displayed greater than a twofold decrease in their expression levels in the mutant as compared with the wild-type animals. This list predominantly contains genes known to play a role in myelination, including structural proteins and lipid-synthesizing enzymes. Strikingly, a comparison of this list with a database of genes expressed in the oligodendrocyte lineage (Cahoy et al. 2008) revealed that all of the genes in Table 1 are up-regulated during oligodendrocyte maturation. These findings suggest that ZFP191 is a transcription factor likely to be involved, either directly or indirectly, in coordinating the myelination program in oligodendrocytes.
Table 1.
Data of microarray analysis on RNA samples isolated from brains of P14 wild-type and mutant hmcns mice
Genes with greater than twofold reduction in expression in the mutants are shown here. Data have a 4% false discovery rate.
Discussion
In this study, we characterized a new mouse model of CNS hypomyelination, hmcns. These mutant animals exhibit a novel phenotype characterized by abundant numbers of process-extending, late-stage oligodendrocytes that express an array of genes involved in the myelination process at severely reduced levels, suggesting that the mutation lies in a gene responsible for coordinating the myelination program of oligodendrocytes. Consistent with this possibility, the causative mutation was identified in a nuclear-localized, zinc finger protein family member, ZFP191. Upon the discovery of this mutation in Zfp191, null mutants were generated that displayed a similar phenotype as the hmcns mutants. Furthermore, our in vitro and in vivo data demonstrated that oligodendrocytes require functional ZFP191 expression to properly myelinate axons. Together, these studies indicate that ZFP191 is required by oligodendrocytes for normal CNS myelination.
ZFP191 belongs to the large subset of the zinc finger protein family whose members also contain SCAN domains (Sander et al. 2003). The SCAN domain is a leucine-rich domain that is capable of facilitating protein–protein interactions (Stone et al. 2002; Edelstein and Collins 2005; Peterson et al. 2006). This subset of zinc finger proteins exists only in the mammalian lineage, indicating that they evolved relatively recently (Edelstein and Collins 2005). Despite this group's sizable presence in the mammalian genome, relatively little is known about the functions of zinc finger proteins that contain SCAN domains, although there is evidence to suggest that they can play a role in a variety of distinct biological processes (Yang et al. 1999; Gaboli et al. 2001; Edelstein and Collins 2005). ZFP191 contains one SCAN domain and four C2H2 zinc finger domains. Studies on ZNF191, the human ortholog of ZFP191 with which it shares 95% identity, show that the SCAN domain of ZNF191 can bind selectively to several different proteins and form a dimer with itself (Schumacher et al. 2000). The presence of the C2H2 domains indicates that ZFP191 can also bind to DNA, and studies have shown that the zinc finger domains of ZFP191 have the capacity to bind to TCAT motifs (Albanese et al. 2001; Wang et al. 2008). Consistent with the sequence similarity between the mouse and human, the developmental expression pattern of ZNF191 in humans is similar to developing mice (Khalfallah et al. 2009).
Outside the nervous system, Zfp191 is expressed in several tissues such as kidney, liver, heart, and spleen (Prost et al. 1999). During development, the expression of Zfp191 can be observed in the CNS as early as embryonic day 7.5 (E7.5) (Khalfallah et al. 2008). The presence of Zfp191 expression in multiple tissues and at multiple stages of development suggests that the gene participates in functions in addition to myelination. Nevertheless, abnormalities other than the CNS myelination defect were not observed in our Zfp191 mutants. Interestingly, a previously published study described the generation of a Zfp191null mouse mutant that displayed an embryonic-lethal phenotype at E7.5 (Li et al. 2006). This differs dramatically from the Zfp191null mutants we describe here, which display a similar phenotype to that of the hmcns animals. Although the reason for this difference is unclear, one possible explanation emanates from the observation that the two null alleles are not identical. In the Zfp191null mutants described here, the PGK-neo cassette used for positive selection of embryonic stem (ES) cells has been removed, whereas it remains in the Zfp191-null allele described by Li et al. (2006). Previous studies have shown that the presence of this selection cassette can modify the activity of genes in the vicinity of the targeted allele (Pham et al. 1996; Xin et al. 2005). The discrepancy between the phenotypes displayed by the two Zfp191-null mutant animals will need to be explored further.
The Zfp191 mutant animals have a unique phenotype in which oligodendrocyte differentiation arrests at a late, process-extending phase and the capacity of oligodendrocytes to myelinate axons is significantly reduced. This phenotype, along with our observation of normal OPC numbers in the mutant, indicates that Zfp191 is not required for the early differentiation of OPCs, but is required for the later stages of myelination. Moreover, our microarray studies revealed that the genes down-regulated in the hmcns mutant are ones that are up-regulated during normal oligodendrocyte maturation in vitro. Thus, it appears likely that Zfp191 regulates the final stages of oligodendrocyte development, when the bulk of the myelin structural proteins are produced.
It remains unclear how ZFP191 participates in the regulation of myelin-associated genes. When transfected into heterologous cells, ZFP191 does not appear capable of driving expression from the MBP transcriptional control region (data not shown), suggesting that ZFP191 has an indirect effect on myelin gene expression. Interestingly, recent studies in chick embryos suggest that ZNF191 exerts negative control on the Notch signaling pathway (Khalfallah et al. 2009), which has been shown to play a critical role in oligodendrocyte maturation (Wang et al. 1998). Nevertheless, in preliminary studies we did not detect alterations in the expression of Notch pathway components in the CNS of hmcns mutants (data not shown). To further elucidate the molecular mechanism of ZFP191's function, the identification of the DNA sequences and proteins to which ZFP191 binds in oligodendrocyte lineage cells should help further determine this protein's function.
It is interesting to note that, although the conditional mutants that lack functional Zfp191 expression specifically in oligodendrocytes were hypomyelinated, their clinical phenotype was not as severe as that seen in hmcns and Zfp191-null mutants. The conditional mutants survived past 3.5 wk and appeared to recover from the tremors and seizures as they aged. Although the reason for this clinical difference remains unclear, it is possible that Zfp191 expression in astrocytes and/or neurons also contributes to CNS myelination. Another possibility is that ZFP191 function is critical in early OPC development, such that the Cnp-induced expression of Cre occurs too late for the conditional mutant to fully recapitulate the hmcns and null mutants. Nevertheless, our data clearly show that ZFP191 has a cell-autonomous effect in the oligodendrocyte.
In summary, we identified a new mouse mutant, hmcns, that displays severe CNS hypomyelination as a result of a mutation in the gene encoding the nuclear protein ZFP191. Strikingly, there are abundant numbers of OPCs and late-stage oligodendrocytes present throughout the CNS of the mutants. Moreover, the mutant oligodendrocytes appear to extend processes that associate with axons. The inability of these cells to myelinate is coupled with their reduced capacity to express a wide range of myelination-associated genes. Interestingly, oligodendrocytes apparently blocked at late stages of maturation, similar to that observed in the Zfp191 mutants, can be seen in the vicinity of demyelinated axons in MS patients that display remyelination failure (Chang et al. 2002; Kuhlmann et al. 2008). Thus, the continued analyses of the underlying mechanism of myelination failure in the Zfp191 mutants may have important implications for human demyelinating disorders.
Materials and methods
Positional cloning of hmcns
hmcns arose spontaneously on the C3H/HeJ inbred strain background at the Jackson Laboratory. To map the mutation, hmcns C3H/HeJ mutants were outcrossed to the CASTEiJ and C57BL/6J strains to obtain F1 progeny. Heterozygous F1 mice were then intercrossed to produce the F2 mutants. Both crosses produced F2 mutants that were distinguishable from wild-type littermates at P14 by a visible tremor. Genotyping and mapping were performed as described in Traka et al. (2008). The results of these crosses are detailed in the Results.
Generation of Zfp191null allele
A genetically engineered mouse, Zfp191flox, containing lox P sites flanking exons 2 and 3, was generated on the C57BL/6J background at inGenious Targeting Laboratory, Inc. The null allele (Supplemental Fig. 4) was generated after crossing the Zfp191flox mouse to a mouse carrying a Cre transgene under the transcriptional control of the protamine promoter (O'Gorman et al. 1997). PCR was used to check for the absence of exons 2 and 3.
Sequence analysis
To examine for mutations in the 29 genes within the critical interval, we used both genomic DNA (gDNA) and cDNA for sequencing. The gDNA samples were isolated from tail tissue, as above. To obtain cDNA, brains were homogenized in TRIzol (Invitrogen) reagent using a Polytron to extract RNA. Isolated RNA was treated with DNaseI (Invitrogen) to eliminate genomic DNA. Reverse transcription was performed using the iScript cDNA systhesis kit (Bio-Rad). Following amplification, amplicons were cleaned with the QIA Quick PCR Purification kit (Qiagen) and sequenced using an Applied Biosystems 3730XL instrument (Applied Biosystems, Inc.). All sequencing was done at the University of Chicago Cancer Research Center DNA Sequencing Facility. Analysis of the sequence files was done using the Sequencher 4.7 (Gene Codes) program. Primers for gene amplification are available on request.
Western blot
P14 mouse brains were prepared and analyzed as describe previously (Traka et al. 2008). Antibodies against MAG (1:1000; Zymed), CNP (1:1000; Covance), MBP (1:10,000; Covance), and Actin (1:1000; Sigma) were used.
Real-time qPCR
Brains were dissected from P14 mutant and wild-type mice. RNA was extracted and cDNA was generated as described above; however, prior to the cDNA reaction, the quality of the RNA was examined using RNA Nano Chips (Agilent) on the Agilent 2100 Bioanalyzer (Agilent). Real-time qPCR was performed on first-strand cDNA using iQ Supermix or iQ Sybrgreen Supermix (Bio-Rad Laboratories) using a real-time PCR instrument (iCycler; Bio-Rad). Mbp, Mag, Mog, Mobp, Plp, Zfp191, and Gapdh primer sequences are available on request. Each sample was done in triplicate. All expression data were normalized to Gapdh.
Histology and IHC
For frozen and paraffin sections, mice were first deeply anesthetized with Avertin, then perfused with 4% paraformaldehyde in 1× PBS (pH 7.3). Samples were prepared and stained as described (Traka et al. 2008). MBP (1:1000; Covance), CC-1 (1:40; Calbiochem), PLP (1:200; Chemicon), and MAG (1:200; Zymed) stainings were performed.
LFB staining
LFB staining was performed on paraffin-embedded sections. Sections were cut to a thickness of 10 μm, deparaffinized, and immersed in 0.1% LFB stain overnight in a 55°C oven. The slides were then immersed in 0.05% lithium carbonate solution followed by 70% ethyl alcohol until a difference between the gray and white matter could be differentiated. The slides were then counterstained with 0.1% cresyl echt violet. The slides were then dehydrated using increasing concentrations of ethanol (50%, 70%, 90%, 100%) followed by xylene. Afterward, glass coverslips were mounted on the slides using Permount mounting solution (Fisher).
TEM
Preparation of the sample was described previously (Lin et al. 2005). Ultrathin sections (60- to 90-nm thickness) were mounted on grids and stained with uranyl acetate and lead citrate. Images were collected using the Tecnai electron microscope at the University of Chicago Electron Microscope facility.
HeLa cell culture and transfection
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) (GIBCO) supplemented with 10% fetal calf serum (FCS) and 50 U/mL gentamycin. Expanded cultures were then lifted from the flasks using 0.05% trypsin-EDTA solution (GIBCO) for 10 min at room temperature, followed by trypsin inactivation by the addition of an equal volume of growth media. The cells were then pelleted at 1000g for 10 min. The media was then removed and the cells were diluted to a concentration of 1 × 106 per 100 μL with Nucleofector Solution R (Amaxa). Two microliters of plasmid DNA were mixed into the solution and transfected with program I-13 on the Nucleofector device (Amaxa). After electroporation, cells were transferred to culture slides (Lab TekII) and grown in 10% FCS DMEM for 2 d before immunocytochemistry.
Rat primary cell cultures
Rat OPCs and astrocytes were purified as described (Armstrong 1998). To obtain oligodendrocytes, the isolated OPCs were allowed to differentiate for 5 d before RNA was collected. Fetal hippocampal neurons were dissociated and maintained in vitro as described (Banker and Cowan 1979). Briefly, astrocyte-enriched glial cultures from 2-d-old rats were plated onto 35-mm petri dishes in DMEM containing FBS (10%) and allowed to proliferate to confluence at 37°C in 5% CO2. The culture media was replaced with DMEM containing horse serum (10%), N2 supplement (Life Technologies), and ovalbumen (1 mg/mL). One day later, fetal rats from timed pregnancies were killed at 17 d gestation. Following trypsin digestion, hippocampi were mechanically dissociated and plated at 60,000 cells per milliliter onto poly-L-lysine-coated coverslips. Coverslips were placed face down into petri dishes containing glia and glia-conditioned media and incubated at 37°C in 5% CO2. Cytosine-β-darabinofuranoside was added 24 h later to inhibit glial growth. RNA was isolated from 14-d in vitro neurons.
Isolation and differentiation of mouse OPCs
Isolation of the mouse OPCs and RGCs for the differentiation and coculture experiments were performed as described by Watkins et al. (2008). Quantification was performed as described (Watkins et al. 2008). To estimate OPC differentiation in vitro, counts of MBP- or MOG-positive cells versus total cell numbers were averaged from 10 images taken from wild-type or mutant OPC cultures (one animal per genotype). To estimate myelination potential, OPCs were plated with RGC and immunostained. Counts of myelinating cells versus the total of cell numbers were averaged from 10 images taken from a single wild-type or control animal. Smooth MBP-positive tubes are considered ensheathed/myelinated.
Cloning of tagged proteins
Full-length Zfp191 was amplified from wild-type mouse cDNA using the HotStar HiFidelity PCR kit (Qiagen). The primers used for the full-length sequence were GTCCCGAGCCAGAGGAGTA and CACAACATTCAAAAGTTTTTCTGC. Following PCR purification, amplicons were cloned into the pcDNA 3.1/V5-His TOPO TA vector. Wild-type amplicons were cloned in-frame with the downstream V5/His6 epitope. The plasmids were transfected into Top10 cells for amplification and isolated from the cells using the Endofree Plasmid Maxi Kit (Qiagen).
Immunocytochemistry
Transfected cells were grown for 2 d, then fixed in 4% paraformaldehyde in 1× PBS for 10 min, permeabilized in 0.01% Triton X-100 for 5 min, and blocked in 1× PBS containing 1% normal goat serum (NGS) and 0.01% Triton 100-X for 30 min. Cells were then incubated with 1:200 anti-V5 (Invitrogen) diluted in blocking solution for 1 h at room temperature, followed by incubation with the secondary antibodies Alexa-fluor 488 anti-mouse and anti-rabbit (1:500; Molecular probe) and FITC anti-mouse (1:200; Vector) for 1 h at room temperature. Cells were then mounted using Vectashield with DAPI (Vector).
Toluidine blue staining
Tissues were embedded similarly to EM blocks, then cut to 1 μM and stained with toluidine blue.
Microarray
RNA was isolated from the brains of P14 hmcns mutant (n = 3) and wild-type mice (n = 3) as described above, except that samples were not treated with DNaseI. The quality of the RNA was checked with RNA Nano Chips (Agilent) on the Agilent 2100 Bioanalyzer (Agilent). Additionally, the RNA was purified using the RNeasy kit (Qiagen). The RNA was then prepared and hybridized to the Mouse Exon 1.0ST array (Affymetrix) by the University of Chicago Functional Genomic Facility, following protocols provided by Affymetrix. Data analysis was done using ChipInspector analysis software (Genomatix). The data shown have a 4% false discovery rate.
In situ hybridization
A region of the Plp gene was amplified from mouse brain cDNA using the forward primer CTGGCTGAGGGCTTCTACAC and reverse primer TTCAGGAGATGCTTGCAAAA. This fragment of the Plp gene was subsequently cloned into the pCRII-TOPO vector. Sense and antisense cRNA probes were then produced from the dual promoter vector using the Dig RNA Labeling Kit (Sp6/T7; Roche). Frozen sections were cut to 20-μM thickness. Before hybridization, the sections were fixed in RNase-free 4% PFA in PBS solution (pH 7.3) and acetylated with acetylation solution (1% triethanolamine, 0.17% HCl, 0.25% acid anhydride in DEPC-treated H2O). Sections were then prehybridized with hybridization solution (50% formamide, deionized [Sigma-Aldrich], 5× Denhart's, 500 μg/μL herring sperm [Sigma-Aldrich], 250 μg/μL bakers yeast RNA [Sigma-Aldrich] in SSC) for 2 h at 58°C. Samples were incubated in the hybridization solution containing either 400 ng/μL DIG-labeled sense or antisense RNA probe overnight at 58°C. The next day, the sections were stained with anti-DIG (1:5000) and developed using the BCIP/NBT (Vector) kit.
g-ratio measurements
The circumferences of fibers and axons were measured using ImageJ (Abramoff et al. 2004). The g-ratio was determined by (axon circumference)/(fiber circumference). The g-ratio was only calculated for myelinated axons. Images taken at 5600× magnification were used for the g-ratio measurement. The g-ratio was measured for 36 axons in each animals (n = 3).
Statistical analysis
All results except for quantification of the OPC differentiation experiment are expressed as mean ± standard deviation. The OPC differentiation experiment is expressed as mean ± standard error. The statistical significance for real-time qPCR data was determined by two-tailed Student's t-test. The statistical significance for the difference between the numbers of positive MOG cells in culture was determined by ANOVA. Differences were considered as statistically significant at P < 0.05.
Acknowledgments
The hmcns mutation was originally discovered by Lyman Corson at the Jackson Laboratory (JAX). We thank JAX Mouse Mutant Resource technicians for assistance in the initial maintenance and genotype linkage crosses of these mice. We also thank the University of Chicago Electron Microscope Facility for assistance with the TEM images, Darlene S. Douglas for critical comments on the manuscript, Gloria Wright for assistance with figure preparation, and Eliot Popko for videotaping the mutant mouse. We also thank Dr. Klaus Nave for generously providing the CNP/Cre mice. This work was supported by grants from the Myelin Repair Foundation to B.A.B. and B.P., as well as NIH grant R01NS067550 to B.P. and NIH/NCRR grant P40 RR001183 to M.D.
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1864510.
Supplemental material is available at http://www.genesdev.org.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Albanese V, Biguet NF, Kiefer H, Bayard E, Mallet J, Meloni R. Quantitative effects on gene silencing by allelic variation at a tetranucleotide microsatellite. Hum Mol Genet. 2001;10:1785–1792. doi: 10.1093/hmg/10.17.1785. [DOI] [PubMed] [Google Scholar]
- Armstrong RC. Isolation and characterization of immature oligodendrocyte lineage cells. Methods. 1998;16:282–292. doi: 10.1006/meth.1998.0685. [DOI] [PubMed] [Google Scholar]
- Banker GA, Cowan WM. Further observations on hippocampal neurons in dispersed cell culture. J Comp Neurol. 1979;187:469–493. doi: 10.1002/cne.901870302. [DOI] [PubMed] [Google Scholar]
- Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev. 2001;81:871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Tourtellotte WW, Rudick R, Trapp BD. Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. N Engl J Med. 2002;346:165–173. doi: 10.1056/NEJMoa010994. [DOI] [PubMed] [Google Scholar]
- Dugas JC, Tai YC, Speed TP, Ngai J, Barres BA. Functional genomic analysis of oligodendrocyte differentiation. J Neurosci. 2006;26:10967–10983. doi: 10.1523/JNEUROSCI.2572-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein LC, Collins T. The SCAN domain family of zinc finger transcription factors. Gene. 2005;359:1–17. doi: 10.1016/j.gene.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Emery B, Agalliu D, Cahoy JD, Watkins TA, Dugas JC, Mulinyawe SB, Ibrahim A, Ligon KL, Rowitch DH, Barres BA. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell. 2009;138:172–185. doi: 10.1016/j.cell.2009.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboli M, Kotsi PA, Gurrieri C, Cattoretti G, Ronchetti S, Cordon-Cardo C, Broxmeyer HE, Hromas R, Pandolfi PP. Mzf1 controls cell proliferation and tumorigenesis. Genes & Dev. 2001;15:1625–1630. doi: 10.1101/gad.902301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths I, Klugmann M, Anderson T, Thomson C, Vouyiouklis D, Nave KA. Current concepts of PLP and its role in the nervous system. Microsc Res Tech. 1998;41:344–358. doi: 10.1002/(SICI)1097-0029(19980601)41:5<344::AID-JEMT2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave KA, Casaccia-Bonnefil P. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalfallah O, Faucon-Biguet N, Nardelli J, Meloni R, Mallet J. Expression of the transcription factor Zfp191 during embryonic development in the mouse. Gene Expr Patterns. 2008;8:148–154. doi: 10.1016/j.gep.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Khalfallah O, Ravassard P, Lagache CS, Fligny C, Serre A, Bayard E, Faucon-Biguet N, Mallet J, Meloni R, Nardelli J. Zinc finger protein 191 (ZNF191/Zfp191) is necessary to maintain neural cells as cycling progenitors. Stem Cells. 2009;27:1643–1653. doi: 10.1002/stem.88. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M. The Id4 HLH protein and the timing of oligodendrocyte differentiation. EMBO J. 2000;19:1998–2007. doi: 10.1093/emboj/19.9.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann T, Miron V, Cuo Q, Wegner C, Antel J, Bruck W. Differentiation block of oligodendroglial progenitor cells as a cause for remyelination failure in chronic multiple sclerosis. Brain. 2008;131:1749–1758. doi: 10.1093/brain/awn096. [DOI] [PubMed] [Google Scholar]
- Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, Griffiths IR, Nave KA. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- Li J, Chen X, Yang H, Wang S, Guo B, Yu L, Wang Z, Fu J. The zinc finger transcription factor 191 is required for early embryonic development and cell proliferation. Exp Cell Res. 2006;312:3990–3998. doi: 10.1016/j.yexcr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Li H, Lu Y, Smith HK, Richardson WD. Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J Neurosci. 2007;27:14375–14382. doi: 10.1523/JNEUROSCI.4456-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Harding HP, Ron D, Popko B. Endoplasmic reticulum stress modulates the response of myelinating oligodendrocytes to the immune cytokine interferon-γ. J Cell Biol. 2005;169:603–612. doi: 10.1083/jcb.200502086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Cai L, Rowitch D, Cepko CL, Stiles CD. Ectopic expression of Olig1 promotes oligodendrocyte formation and reduces neuronal survival in developing mouse cortex. Nat Neurosci. 2001;4:973–974. doi: 10.1038/nn718. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Nielsen JA, Maric D, Lau P, Barker JL, Hudson LD. Identification of a novel oligodendrocyte cell adhesion protein using gene expression profiling. J Neurosci. 2006;26:9881–9891. doi: 10.1523/JNEUROSCI.2246-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson FC, Hayes PL, Waltner JK, Heisner AK, Jensen DR, Sander TL, Volkman BF. Structure of the SCAN domain from the tumor suppressor protein MZF1. J Mol Biol. 2006;363:137–147. doi: 10.1016/j.jmb.2006.07.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham CT, MacIvor DM, Hug BA, Heusel JW, Ley TJ. Long-range disruption of gene expression by a selectable marker cassette. Proc Natl Acad Sci. 1996;93:13090–13095. doi: 10.1073/pnas.93.23.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost JF, Negre D, Cornet-Javaux F, Cortay JC, Cozzone AJ, Herbage D, Mallein-Gerin F. Isolation, cloning, and expression of a new murine zinc finger encoding gene. Biochim Biophys Acta. 1999;1447:278–283. doi: 10.1016/s0167-4781(99)00157-8. [DOI] [PubMed] [Google Scholar]
- Qi Y, Cai J, Wu Y, Wu R, Lee J, Fu H, Rao M, Sussel L, Rubenstein J, Qiu M. Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development. 2001;128:2723–2733. doi: 10.1242/dev.128.14.2723. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat Rev Neurosci. 2006;7:11–18. doi: 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander TL, Stringer KF, Maki JL, Szauter P, Stone JR, Collins T. The SCAN domain defines a large family of zinc finger transcription factors. Gene. 2003;310:29–38. doi: 10.1016/s0378-1119(03)00509-2. [DOI] [PubMed] [Google Scholar]
- Schumacher C, Wang H, Honer C, Ding W, Koehn J, Lawrence Q, Coulis CM, Wang LL, Ballinger D, Bowen BR, et al. The SCAN domain mediates selective oligomerization. J Biol Chem. 2000;275:17173–17179. doi: 10.1074/jbc.M000119200. [DOI] [PubMed] [Google Scholar]
- Stone JR, Maki JL, Blacklow SC, Collins T. The SCAN domain of ZNF174 is a dimer. J Biol Chem. 2002;277:5448–5452. doi: 10.1074/jbc.M109815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traka M, Wollmann RL, Cerda SR, Dugas J, Barres BA, Popko B. Nur7 is a nonsense mutation in the mouse aspartoacylase gene that causes spongy degeneration of the CNS. J Neurosci. 2008;28:11537–11549. doi: 10.1523/JNEUROSCI.1490-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HH, Frost E, To V, Robinson S, Ffrench-Constant C, Geertman R, Ransohoff RM, Miller RH. The chemokine receptor CXCR2 controls positioning of oligodendrocyte precursors in developing spinal cord by arresting their migration. Cell. 2002;110:373–383. doi: 10.1016/s0092-8674(02)00838-3. [DOI] [PubMed] [Google Scholar]
- Tsai HH, Tessier-Lavigne M, Miller RH. Netrin 1 mediates spinal cord oligodendrocyte precursor dispersal. Development. 2003;130:2095–2105. doi: 10.1242/dev.00424. [DOI] [PubMed] [Google Scholar]
- Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres BA. Notch receptor activation inhibits oligodendrocyte differentiation. Neuron. 1998;21:63–75. doi: 10.1016/s0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
- Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA. A role for the helix–loop–helix protein Id2 in the control of oligodendrocyte development. Neuron. 2001;29:603–614. doi: 10.1016/s0896-6273(01)00237-9. [DOI] [PubMed] [Google Scholar]
- Wang SZ, Dulin J, Wu H, Hurlock E, Lee SE, Jansson K, Lu QR. An oligodendrocyte-specific zinc-finger transcription regulator cooperates with Olig2 to promote oligodendrocyte differentiation. Development. 2006;133:3389–3398. doi: 10.1242/dev.02522. [DOI] [PubMed] [Google Scholar]
- Wang H, Sun R, Liu G, Yao M, Fei J, Shen H. Characterization of the target DNA sequence for the DNA-binding domain of zinc finger protein 191. Acta Biochim Biophys Sin (Shanghai) 2008;40:704–710. [PubMed] [Google Scholar]
- Watkins TA, Emery B, Mulinyawe S, Barres BA. Distinct stages of myelination regulated by γ-secretase and astrocytes in a rapidly myelinating CNS coculture system. Neuron. 2008;60:555–569. doi: 10.1016/j.neuron.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner M. A matter of identity: Transcriptional control in oligodendrocytes. J Mol Neurosci. 2008;35:3–12. doi: 10.1007/s12031-007-9008-8. [DOI] [PubMed] [Google Scholar]
- Xin M, Yue T, Ma Z, Wu FF, Gow A, Lu QR. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J Neurosci. 2005;25:1354–1365. doi: 10.1523/JNEUROSCI.3034-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XW, Wynder C, Doughty ML, Heintz N. BAC-mediated gene-dosage analysis reveals a role for Zipro1 (Ru49/Zfp38) in progenitor cell proliferation in cerebellum and skin. Nat Genet. 1999;22:327–335. doi: 10.1038/11896. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]