Abstract
Eukaryotic cells employ a suite of replication and mitotic checkpoints to ensure the accurate transmission of their DNA. In budding yeast, both the DNA damage checkpoint and the spindle assembly checkpoint (SAC) block cells prior to anaphase [1-5]. The presence of a single unrepaired double-strand break (DSB) activates ATR and ATM protein kinase homologs, Mec1 and Tel1, which then activate downstream effectors to trigger G2/M arrest and also phosphorylate histone H2A (creating γ-H2AX) in chromatin surrounding the DSB [6-8]. The SAC monitors proper attachment of spindle microtubules to the kinetochore formed at each centromere and the biorientation of sister centromeres toward opposite spindle pole bodies. Although these two checkpoints sense quite different perturbations, recent evidence has demonstrated both synergistic interactions and cross-talk between them [9-11]. Here we report that Mad2 and other SAC proteins play an unexpected role in prolonging G2/M arrest after induction of a single DSB. This function of the SAC depends not only on Mec1 and other components of the DNA damage checkpoint but also on the presence of the centromere located ≥ 90kb from the DNA damage. DNA damage induces epigenetic changes at the centromere, including the γ-H2AX modification, that appear to alter kinetochore function, thus triggering the canonical spindle assembly checkpoint. Thus, a single DSB triggers a response by both checkpoints to prevent the segregation of a damaged chromosome.
Results and Discussion
Mad2 prolongs arrest before adaptation after DNA damage
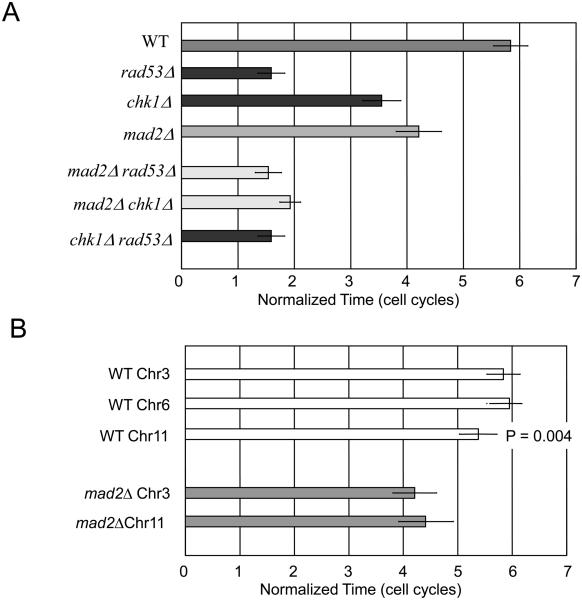
Cells with an unrepaired DSB exit checkpoint arrest and resume cell cycle progression either after repairing the damage (recovery) or even when damage persists (adaptation) (reviewed in[2]). We induced a single DSB at the MAT locus on chromosome 3 (Chr3) in a strain deleted for the homologous donor loci, so that the DSB cannot be repaired by homologous recombination [6, 12]. In this assay > 99% of cells remain arrested for about 15 h; this is equivalent to about 6 doubling times of isogenic cells lacking the HO endonuclease cut site (Fig. 1). Very similar arrest and adaptation is seen when a single DSB is induced on the left arm of Chr6 instead of at MAT [13]. In both cases the DSB is about 90-100 kb from its centromere. Cell cycle arrest is completely eliminated in cells lacking Mec1, but there is still a substantial delay in cells lacking Rad53 or Chk1 [13, 14]. A slightly shorter, though statistically significantly different, arrest response is seen with a DSB on Chr11, 294 kb from its centromere (Fig. 1B).
Figure 1.
mad2Δ shortens checkpoint-mediated cell cycle delay after induction of an unrepaired DSB. A) Induction of an irreparable DSB causes a delay in the cell cycle equivalent to 6 doubling times in wild type cells, which is fully eliminated by mec1Δ. mad2Δ reduces arrest in chk1Δ and is epistatic to rad53Δ. B) Response to a DSB in Chr6 is similar to that on Chr3, while a break on Chr11, where the DSB is 200 kb further from its centromere, provokes a significantly shorter Mad2-dependent arrest.
Unexpectedly, deletion of MAD2 markedly shortens the time that cells with a DSB remain arrested (Fig. 1). mad2Δ strains suffering a DSB on Chr3 or on Chr11 arrest only for about 10 h, 2/3 as long as in MAD2 cells. The SAC mutants mad1Δ and mad3Δ had comparable effects though bub1Δ and bub3Δ did not (data not shown). These results suggest that arrest of cells at G2/M may consist of two stages, the first enforced by Mec1, Rad53 and to a lesser extent by Chk1 and the second regulated by Mad1, Mad2, and Mad3.
Epistasis analysis suggests that Mad2's role may be executed through a Rad53-dependent pathway. The long residual cell cycle delay in chk1Δ cells is partially suppressed by deletion of any of the 5 mad and bub mutants tested, even bub1Δ and bub3Δ that did not affect arrest as single mutants (Suppl. Fig. 1). Arrest in mad2Δ chk1Δ is markedly reduced, but only to the length seen in rad53Δ cells. In contrast, deleting the SAC genes did not further shorten the arrest in rad53Δ (Fig. 1A and Suppl. Fig. 1). These results suggest that Mad and Bub proteins act to prolong constraints on cell cycle progression that are imposed by Rad53. Deleting Mad or Bub genes in rad53Δ chk1Δ or in rad9Δ does not further shorten the small, but statistically significant residual delay compared to mec1Δ (data not shown). Thus, Mad and Bub proteins play a significant role in prolonging G2/M arrest and may define one pathway by which Rad53 halts the cell cycle.
MAD2 is required for the permanent arrest of most adaptation-defective mutations
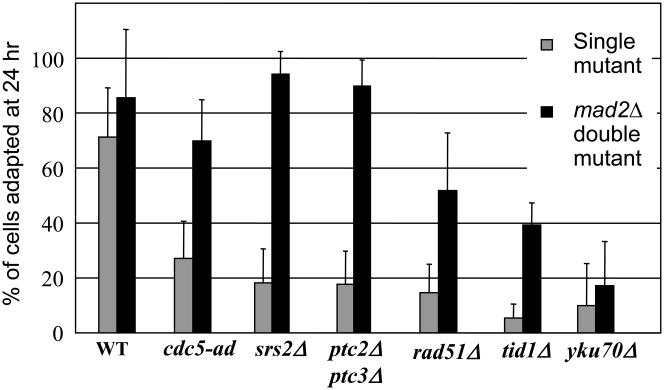
Several classes of mutants block checkpoint adaptation including: (1) those with increased DNA end processing, (2) those with defects in aspects of homologous recombination, and (3) those unable to turn off the checkpoint signaling pathway. As seen in Fig. 2, mad2Δ at least partially suppresses the permanent arrest of all adaptation-defective mutants tested except yku80Δ, but even the mad2Δ-suppressed mutants still arrest for at least 10 h. Thus Mad2 and presumably other SAC proteins play a central role in maintaining the permanent arrest of cells initially arrested by the DNA damage checkpoint. The consequence of deleting Mad2 is not as great as deleting Rad9, which eliminates nearly all DSB-mediated arrest of these mutants (data not shown).
Figure 2.
Deletion of MAD2 suppresses the permanent arrest phenotype of most adaptation-defective mutations. Unbudded (G1) cells were plated on YEP-Galactose and monitored at the times indicated.
Deletion of Mad2 also shortens the delay of a DSB on Chr11, located 294 kb from its centromere (Fig. 1B). It is noteworthy that, when Mad2 is present, cell cycle delay with a DSB on Chr11 is significantly less (p < 0.005) than that seen on either Chr3 or Chr6, where the DSB is 90-100 kb from the centromere. Thus the distance of the DSB to the centromere may play a role in maintaining the DNA damage response.
Deletion of the centromere mimics mad2Δ in shortening checkpoint arrest
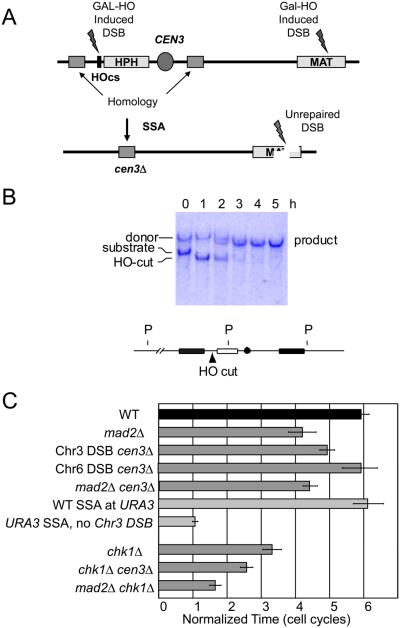
We tested whether centromere-derived signals can explain the role of Mad2 in the DSB response. A key question is whether Mad2's role reflects an alteration to the kinetochore in cis with the DSB. We have previously shown that 5′ to 3′ resection of the DSB continues for at least 24 hr at a rate of ~4 kb/h [15]; but the 90 kb distance of the DSB from CEN3 is too far to be degraded within 10 h, the time at which Mad2 is required to maintain G2/M arrest. However, the effect on the centromere could reflect a change in the topological constraint of the now-broken chromosome or it could be caused by a long-distance chromatin modification such as γ-H2AX extending from the DSB [8, 16]. To determine if the role of Mad2 depends on the presence of the centromere, we devised a way to delete CEN3 at the same time that HO is induced to create a DSB at MAT. As shown in Fig. 3, we simultaneously induced an irreparable DSB at MAT and another DSB at a site less than 1 kb from CEN3. The DSB at CEN3 is flanked by 2-kb identical DNA sequences, so 5′ to 3′ resection and repair of the DSB by single-strand annealing (SSA) lead to a complete deletion of CEN3. Galactose-mediated induction of HO endonuclease cleavage is essentially 100% efficient, as shown in the southern blot in Fig. 3B. By 3 h, nearly all cells have deleted CEN3.
Figure 3.
Deleting the CEN3 locus decreases cell cycle delay caused by the DNA DSB at MAT. A) An HO cut site was inserted to the left of CEN3. A 2 kb fragment from the left of the HO cutsite was inserted to the right of CEN3. Gal-HO induction creates DSBs at MAT and near CEN3. CEN3 is deleted as the nearby DSB is repaired by SSA between flanking homologous DNA segments. B) CEN3 is deleted within 3 h of HO induction, as seen in a southern blot probed with the sequences (black) flanking CEN3. C) Deleting CEN3 shortens the DNA DSB-dependent cell cycle arrest in WT cells but not in mad2Δ cells. An equivalent SSA event occurring at the URA3 locus does not affect cell cycle arrest. Deleting CEN3 does not affect arrest when the DSB is made on another chromosome. Deleting CEN3 shortens the checkpoint in a chk1Δ strain albeit not as much as deleting MAD2.
In the absence of a DSB at MAT, deleting CEN3 has no effect on the time it takes for cells to progress into the next cell cycle and bud a second time (Fig. 3). This is consistent with previous results that repairing a short-lived DSB by SSA does not delay cell cycle progression [17]. When there is a DSB at MAT, deleting CEN3 shortens the length of arrest caused by the remaining DSB at MAT, though not as profoundly as mad2Δ. Importantly, the mad2Δ CEN3 double deletion was indistinguishable from mad2Δ alone. Moreover, deleting CEN3 also significantly reduced the delay in a chk1Δ strain, as did mad2Δ. These data argue that deleting the centromere proximal to the DSB eliminates much of the Mad2-dependent response to the presence of a DSB.
If Mad2 responds to alterations at the kinetochore of the chromosome that suffered the DSB, then we would expect there to be no effect if we were to delete CEN3 in a strain where the DSB is induced on Chr 6. Indeed this is the case (Fig. 3). This experiment provides clear evidence that a significant portion of DSB-induced G2/M arrest is attributable to an alteration at the kinetochore of the broken chromosome, rather than by an indirect effect by increasing the pool of Cdc20 that could promote mitosis.
We note that there is a greater effect of deleting MAD2 than of deleting the CEN3. This difference in suppressing checkpoint-mediated arrest by deleting CEN3 versus mad2Δ could be explained by the fact that CEN3 deletion strain did – for a time – have two DSBs. Two unrepaired DSBs provoke permanent arrest, so that the checkpoint signal might have been initially stronger and possibly more difficult to turn off [12]. We therefore constructed a strain with an unrepairable DSB at MAT and a second DSB at the URA3 locus on Chr5, where it could be repaired by a comparable SSA event that did not involve centromere deletion [18]. There was no alteration in the time it took for adaptation to occur, and the SSA event by itself did not cause any visible arrest (Fig. 3).
Eliminating γ-H2AX shortens the checkpoint delay
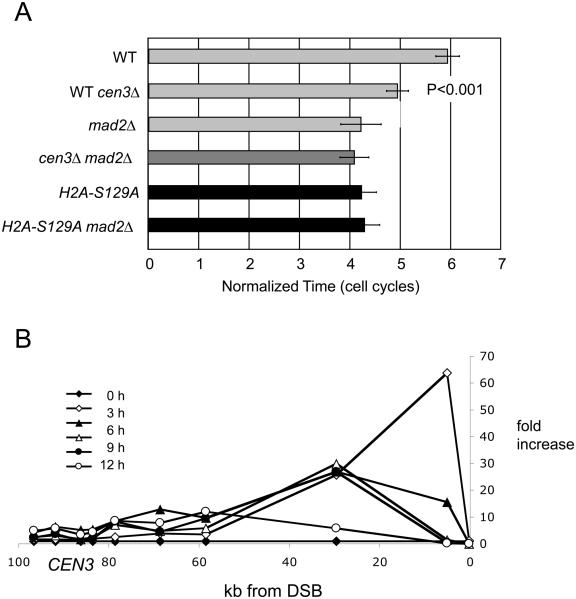
Shortly after a DSB is created, nearby histone H2A is phosphorylated (yielding γ-H2AX); this modification spreads prominently 40-50 kb on either side of the DSB with the strongest accumulation within ~20 kb of the DSB [16, 19]. As resection proceeds γ-H2AX is locally displaced from chromatin, but additional γ-H2AX is modified by Mec1 kinase ahead of the resection [16]. To ask if γ-H2AX modification affects the persistence of adaptation, we analyzed an isogenic strain in which the two copies of the H2A gene encode the non-phosphorylatable S129A allele. As seen in Fig. 4A, eliminating γ-H2AX shortens the length of arrest about as much as does mad2Δ. Importantly, the double mutant lacking both MAD2 and the phosphorylation site on H2A [hta1-S129A hta2-S129A] is no more defective than either defect alone. Cells carrying H2A-S129A also suppress adaptation-defective mutations, including tid1Δ, yku70Δ and ckb2Δ (data not shown). The H2A-S129A mutation markedly shortens the arrest seen in a chk1Δ mutant but, unlike mad2Δ, also reduces the duration of arrest in rad53Δ (Fig. 4). Just as Rad53 is likely to affect cell cycle arrest by several pathways, not all of which involve Mad2 [20], γ-H2AX may affect pathways other than those controlled by Rad53 (see below).
Figure 4.
γ-H2AX modification influences the persistence of the DNA damage checkpoint in a Mad2-dependent fashion. A) Mutating H2A-S129 shortens G2/M delay of a DSB at MAT. B) Spread of γ-H2AX leftward towards CEN3 from a DSB at MAT. ChIP using an anti-γ-H2AX antibody shows the spreading of γ-H2AX down the chromosome and its progressive loss from regions near the DSB. Peak accumulation at CEN3 of 10-fold was seen at 6 h, prior to the time cells adapt in mad2Δ.
To establish more directly that γ-H2AX modification could alter centromere function, we asked if γ-H2AX modification does in fact reach the centromere after resection has proceeded for many hours. Using an antibody specific for the γ-H2AX modification, we monitored sites around CEN3 as well as nearer the DSB at MAT. As shown in Fig. 4B, there is a 3-10 fold increase in γ-H2AX modification surrounding CEN3 after 6 h.
These data suggest that the Mad and Bub proteins, acting in their normal role as monitors of spindle function and centromere integrity, can sense a DSB created ≥ 90 kb away. Consequently, the G2/M arrest triggered by a single DSB that first activates the DNA damage checkpoint is significantly prolonged. Either contemporaneous deletion of the adjacent centromere or elimination of γ-H2AX formation largely, but not completely, abrogates the Mad2-dependent phase of cell cycle arrest. We conclude that the major way in which Mad2 responds to the DSB likely involves an epigenetic modification of the centromere in cis to the DSB. Centromere function depends not only on the core nucleosome containing Cse4 but on surrounding centric chromatin [21]. It is also possible that the accumulation of γ-H2AX on the chromosome could cause a rearrangement of the normally high level of cohesin binding around the centromere, as γ-H2AX enables damage-dependent recruitment of cohesins, which may include sliding from the centromeric region [19, 22]. An alternative model could be that the absence of Mad2 generally increases Cdc20 activity [23] that in turn could promote APC-mediated degradation of Pds1 and thus promote mitosis. However, this idea is hard to rationalize with the specific effect of completely deleting one centromere, which should in fact not perturb Mad2 monitoring of centromeres. However, release of Cdc20 could account for the larger effect of mad2Δ compared to centromere deletion (Fig. 1). We note that in a mec1Δ strain there is still Tel1-dependent γ-H2AX around the DSB site, but it fails to spread down the chromosome as resection proceeds because this requires Mec1 [16]; consequently the centromere region will not be modified and there is no Mad2-dependent delay of progression in a mec1Δ mutant.
Previous studies have also suggested that γ-H2AX plays an important role in Rad9-mediated steps in the DNA damage checkpoint response. Rad9 contains both BRCT domains that interact with γ-H2AX and a Tudor domain that binds to methylated histone H3-K79 [24, 25]. It is therefore possible that removing phosphorylation of histone H2A would produce a partial rad9 phenotype. This additional role can explain the shorter duration of arrest in H2A-S129A rad53Δ compared to rad53Δ. However, the fact that a mad2Δ H2A-S129A double mutant is not different from mad2Δ argues that the primary effect of eliminating γ-H2AX is acting through the Mad2 pathway.
To examine how partially limiting Rad9 response might affect DSB-induced arrest, we deleted the Dot1 methyltransferase that methylates H3-K79; this dot1Δ deletion has a more profound effect on DSB-induced arrest than H2A-S129A (Suppl. Fig. 2). Interestingly, a dot1Δ H2A-S129A double mutant shows significantly (p < 0.05) longer cell cycle delay than a rad9Δ mutation (Suppl. Fig. 2). This observation suggests there could be yet another histone modification to which Rad9 responds (we note that in Crb2 of S. pombe and in 53BP1 of mammals the methylated residue bound by the Tudor domain is H4-K20); moreover it appears that Rad9 may directly recognize single-stranded DNA [26].
To summarize, a persistent DSB establishes a spreading zone of ”chromosomal inflammation“, here marked by γ-H2AX. If this region is sufficiently close to a centromere the altered chromatin conformation can apparently trigger kinetochore dysfunction that is recognized by Mad2 and other components of the SAC. The SAC acts semi-redundantly with the DNA damage checkpoint to enforce long-term cell cycle arrest in the presence of chromosomal DSBs and thereby prevents the segregation of damaged chromosomes.
Experimental Procedures
Yeast strains, cell cycle arrest and adaptation assays, chromatin immunoprecipitation methods are presented in Supplemental Information.
Supplementary Material
Acknowledgements
This work was supported by NIH grant GM61766 to JEH. JCH was a postdoctoral fellow of the Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science (New York, N.Y. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 2.Harrison JC, Haber JE. Surviving the breakup: the DNA damage checkpoint. Annual review of genetics. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 3.Longhese MP, Foiani M, Muzi-Falconi M, Lucchini G, Plevani P. DNA damage checkpoint in budding yeast. The EMBO journal. 1998;17:5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lew DJ, Burke DJ. The spindle assembly and spindle position checkpoints. Annual review of genetics. 2003;37:251–282. doi: 10.1146/annurev.genet.37.042203.120656. [DOI] [PubMed] [Google Scholar]
- 5.Lim HH, Zhang T, Surana U. Regulation of centrosome separation in yeast and vertebrates: common threads. Trends in cell biology. 2009;19:325–333. doi: 10.1016/j.tcb.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Lee SE, Moore JK, Holmes A, Umezu K, Kolodner R, Haber JE. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 7.Toczyski DP, Galgoczy DJ, Hartwell LH. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell. 1997;90:1097–1106. doi: 10.1016/s0092-8674(00)80375-x. [DOI] [PubMed] [Google Scholar]
- 8.Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garber PM, Rine J. Overlapping roles of the spindle assembly and DNA damage checkpoints in the cell-cycle response to altered chromosomes in Saccharomyces cerevisiae. Genetics. 2002;161:521–534. doi: 10.1093/genetics/161.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemenson C, Marsolier-Kergoat MC. The spindle assembly checkpoint regulates the phosphorylation state of a subset of DNA checkpoint proteins in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:9149–9161. doi: 10.1128/MCB.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim EM, Burke DJ. DNA damage activates the SAC in an ATM/ATR-dependent manner, independently of the kinetochore. PLoS Genet. 2008;4:e1000015. doi: 10.1371/journal.pgen.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pellicioli A, Lee SE, Lucca C, Foiani M, Haber JE. Regulation of Saccharomyces Rad53 checkpoint kinase during adaptation from DNA damage-induced G2/M arrest. Mol Cell. 2001;7:293–300. doi: 10.1016/s1097-2765(01)00177-0. [DOI] [PubMed] [Google Scholar]
- 13.Dotiwala F, Haase J, Arbel-Eden A, Bloom K, Haber JE. The yeast DNA damage checkpoint proteins control a cytoplasmic response to DNA damage. Proc Natl Acad Sci U S A. 2007;104:11358–11363. doi: 10.1073/pnas.0609636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner R, Putnam CW, Weinert T. RAD53, DUN1 and PDS1 define two parallel G2/M checkpoint pathways in budding yeast. Embo J. 1999;18:3173–3185. doi: 10.1093/emboj/18.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–1017. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaze M, Pellicioli A, Lee S, Ira G, Liberi G, Arbel-Eden A, Foiani M, Haber J. Recovery from checkpoint-mediated arrest after repair of a double-strand break requires srs2 helicase. Molecular cell. 2002;10:373. doi: 10.1016/s1097-2765(02)00593-2. [DOI] [PubMed] [Google Scholar]
- 18.Sugawara N, Ira G, Haber JE. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol Cell Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strom L, Lindroos HB, Shirahige K, Sjogren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 20.Liang F, Wang Y. DNA damage checkpoints inhibit mitotic exit by two different mechanisms. Mol Cell Biol. 2007;27:5067–5078. doi: 10.1128/MCB.00095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson DJ, Vargas JD, Hsiao JP, Hetzer MW. Recruitment of functionally distinct membrane proteins to chromatin mediates nuclear envelope formation in vivo. J Cell Biol. 2009;186:183–191. doi: 10.1083/jcb.200901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 23.Yu H. Cdc20: a WD40 activator for a cell cycle degradation machine. Molecular cell. 2007;27:3–16. doi: 10.1016/j.molcel.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Toh GW, O'Shaughnessy AM, Jimeno S, Dobbie IM, Grenon M, Maffini S, O'Rorke A, Lowndes NF. Histone H2A phosphorylation and H3 methylation are required for a novel Rad9 DSB repair function following checkpoint activation. DNA Repair (Amst) 2006;5:693–703. doi: 10.1016/j.dnarep.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 25.Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lancelot N, Charier G, Couprie J, Duband-Goulet I, Alpha-Bazin B, Quemeneur E, Ma E, Marsolier-Kergoat MC, Ropars V, Charbonnier JB, et al. The checkpoint Saccharomyces cerevisiae Rad9 protein contains a tandem tudor domain that recognizes DNA. Nucleic acids research. 2007;35:5898–5912. doi: 10.1093/nar/gkm607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.