Abstract
The histone acetyltransferase (HAT) p300/CBP has been shown to undergo autoacetylation on lysines in an apparent regulatory loop that stimulates HAT activity. Here we developed a strategy to introduce acetyl-Lys at up to six known modification sites in p300/CBP HAT using a combination of circular permutation and expressed protein ligation. We show that these semisynthetic, circularly permuted acetylated proteins retain high affinity for an acetyl-CoA substrate analog and that HAT activity correlates positively with degree of acetylation. This study provides novel evidence for control of p300/CBP HAT activity by site-specific autoacetylation and outlines a potentially general strategy to use expressed protein ligation and circular permutation to chemically interrogate internal regions of proteins.
Post-translational modifications (PTMs) by phosphorylation, acetylation, methylation, ubiquitylation, and glycosylation are major mechanisms for regulating protein function.1 Well-established in protein kinase signaling cascades, enzymatic activities can be activated or inhibited by reversible, site-specific covalent modification of protein sidechains.1-2 Several years ago, the transcriptional coactivator p300/CBP histone acetyltransferase (HAT) was shown to be densely autoacetylated on lysines in an apparent regulatory loop.3 Partial loop deletion or autoacetylation of up to 17 sites in p300 HAT leads to an increase in catalytic activity by 4-10-fold.3 In general, it is difficult to assess the specific functional contributions of individual Lys acetylation events using site-directed mutagenesis. Using classical mutagenesis, Lys is usually replaced with Arg and Gln, which are crude structural mimics of the unmodified and acetylated Lys. Recent advances in nonsense suppression mutagenesis to incorporate acetyl-Lys are promising4 but are difficult to apply to cases where multiple sites are modified. When post-translational modifications are near protein termini, expressed protein ligation (EPL) offers potential for installing multiple PTMs.5 However, in the case of p300/CBP, the ∼40 aa autoacetylation loop is in the middle of the catalytic domain (Figure 1) and is a difficult candidate for conventional EPL since multiple peptide ligation steps would be needed.
Figure 1.
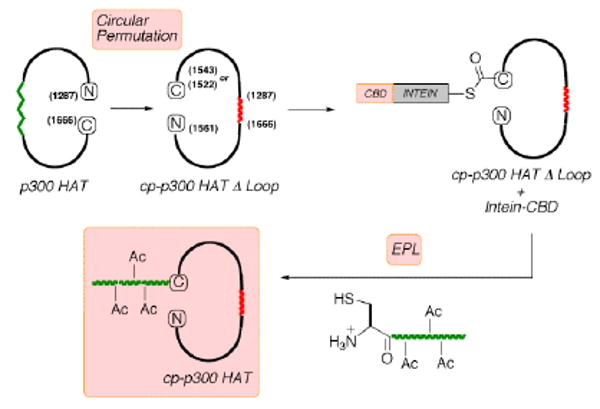
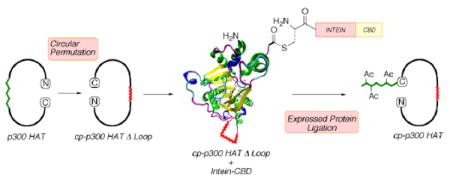
Approach to circularly permuted p300 HAT containing site-specific acetylations via expressed protein ligation (EPL).
We considered an alternative EPL approach to studying the autoacetylation loop in p300/CBP HAT that would involve generating a circularly permuted (cp) enzyme. In cp proteins, the natural N- and C-termini are fused and novel N- and C-termini are created in an alternative location (Figure 1).6 Examples where cp proteins have been generated successfully include green fluorescent protein, ribonuclease A, and beta-lactamase.6 If circular permutation were feasible for p300 HAT, designer regulatory loops could be installed containing stoichiometric, site-specific acetylations, and ligated to the C-terminus of the cp-p300 HAT construct (Figure 1). An X-ray crystal structure of the p300 HAT domain reveals that the natural N- and C-termini (aa 1287 and aa 1666) are close in space (ca. 15 Å apart),7 favorable for circular permutation since in principle, only a short linker might be required. Our strategy toward cp-p300 HAT involved genetically inserting a 7 aa flexible spacer (TGGGSGG) between the natural N- and C-termini and making residue 1543 the new C-terminus fused to an intein (Figures 1, S1). Ligation via the intein-generated C-terminal thioester would allow us to explore specific acetylations in an artificial 17 residue regulatory segment prepared with an N-Cys by solid phase peptide synthesis (Figure S2).
Since expression of wt p300 HAT in E. coli is greatly hampered by promiscuous acetyltransferase activity leading to host toxicity, we initially focused on the preparation of Y1467F cp-p300 HAT (cp-p300-F). Tyr1467 appears to serve as a general acid protonating the departing coenzyme A sulfur and Y1467F shows a 150-fold reduction in catalytic activity compared to wild type enzyme.7 Significant soluble expression (>1 mg/L) of cp-p300-F protein was confirmed so we performed EPL with three synthetic peptides (17-mers) containing 0, 3, or 6 acetyl-Lys residues. Prior mass spectrometry and mutagenesis studies had implicated several of these acetylation positions as early sites in autoacetylation reactions and potentially contributory to catalytic regulation.3 EPL proceeded smoothly and these semisynthetic cp-p300-F HAT proteins could be obtained in high purity by SDSPAGE and mass spectrometry (Figures 2 and S3).
Figure 2.
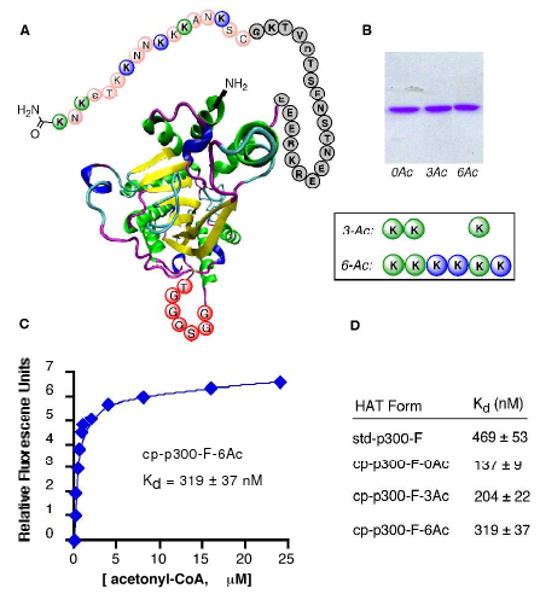
A. Schematic of cp-p300-F HAT (circularly permuted Y1467F) showing site-specific acetylations (blue, green) on the engineered loop (colored circles). The 7-aa flexible spacer is shown in red and the gray circles are the C-terminus of the recombinant moiety. B. SDSPAGE of cp-p300-F HATs. C. Representative fluorescence plot of acetonyl-CoA binding to cp-p300-F-6Ac. D. Dissociation constants of acetonyl-CoA with cp-p300-Fs.
To examine whether these cp-p300-F HAT proteins were still properly folded, we analyzed their affinities for acetonyl-CoA, a high affinity CoA analog for p300, using a fluorescence binding assay (Figures 2 and S4). In this way, it was shown that each of these semisynthetic proteins (unacetylated (0Ac), triacetylated (3Ac), and hexaacetylated (6Ac) cp-p300-F) retained high affinity for acetonyl-CoA binding (Kd = 140-320 nM), within 4-fold of that of standard Y1467F partial loop-deleted p300 HAT (Kd = 470 nM). These studies suggest that circular permutation of p300 does not dramatically alter the basic fold of the enzyme, although it may be relevant that hexa-acetylated form shows the closest Kd to that of the standard, loop-deleted enzyme.
Next, we chose to prepare catalytically active semisynthetic cp-p300s. Due to the aforementioned acetyltransferase cytotoxicity, the corresponding cp-HAT-intein fusion was co-expressed with the histone deacetylase sirtuin Hst2.3 Unfortunately, this failed to generate a significant amount of protein. However, we found that shortening the recombinant fragment to terminate at residue 1522 gave workable soluble protein yields (∼0.2 mg/L) with the intein system. The corresponding thioester successfully ligated to 38-mer synthetic peptides comprising residues 1523-1560 to generate the desired semisynthetic 0Ac, 3Ac, and 6Ac cp-p300 forms (Figure 3). While these proteins are likely to be autoacetylated on several Lys residues in the recombinant moiety, despite the presence of Hst2,3 these non-loop acetylation sites appear to be less important for regulation3 and in any case should be identical among the semisynthetic species which were all generated with the same parent thioester.
Figure 3.
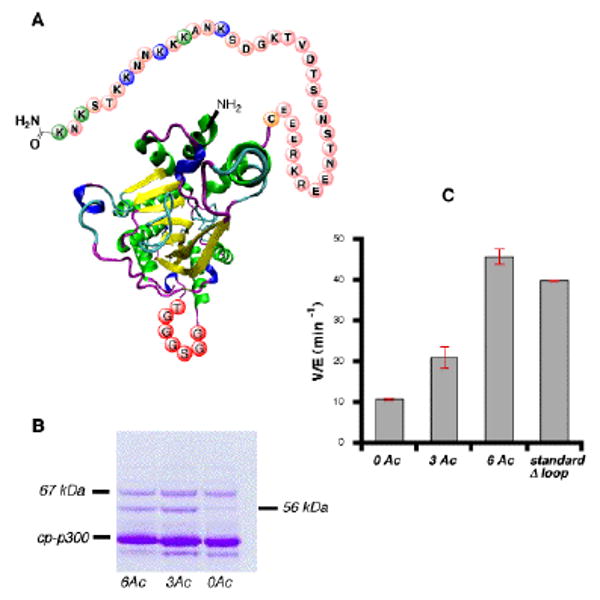
A. Schematic of catalytically active cp-p300 HAT. Color scheme as in Figure 2; 38-mer peptide used for loop. B. SDSPAGE of cp-p300 HATs; contaminants at 67 and 56 kDa are likely GroEL and DNAK, respectively. C. HAT activity of cp-p300s and standard, unacetylated p300 HAT containing partial loop deletion;3a [H4-15mer substrate] = 500 μM; [acetyl-CoA] = 20 μM. The error bars reflect duplicate measurements.
Acetyltransferase assays were performed with catalytically active 0Ac, 3Ac, and 6Ac cp-p300 using a synthetic histone H4 tail peptide substrate under conditions where autoacetylation is minimal, as previously described.3 These assays revealed that cp-p300-6Ac had a ∼5-fold greater activity compared with the cp-p300-0Ac form whereas the cp-p300-3Ac species showed intermediate activity. The HAT rate of cp-p300-6Ac proved to be similar to standard, loop deleted (constitutively active3a) p300 HAT under the same assay conditions (Figure 3). The enhanced rate of cp-p300-6Ac vs. cp-p300-0Ac is noteworthy since, unlike in prior studies,3 these cp-p300 proteins have well-defined acetylation modifications on specific sites rather than a more heterogeneous distribution, which is a natural consequence of promiscuous autoacetylation. These findings further suggest that functional aspects of the regulatory loop can be recapitulated in this circularly permuted system, even though the regulatory loop is only covalently anchored at one side in cp-p300 HAT.
In addition to providing new insights into p300 regulation, this study suggests a more general strategy to apply EPL to protein systems in which a flexible segment of interest lies in the middle of a large recombinant protein sequence. By judiciously selecting novel N- and C-termini for a protein, circular permutation coupled with EPL can offer the opportunity to install unnatural amino acids, biophysical probes, or PTMs and their mimics in functionally internal protein regions. Such integration of methods can thus extend the protein chemistry toolbox to previously difficult-to-approach biochemical problems.
Supplementary Material
Acknowledgments
We are grateful to R. Cotter and his group for assistance with mass spectrometry and R. Marmorstein and X. Liu for helpful discussions. We are thankful for support from the NIH.
Footnotes
Supporting Information Available: Experimental Methods and Supplementary Figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Walsh CT. Post-translational Modification of Proteins. Roberts and Co; Greenwood Village, CO: 2006. [Google Scholar]
- 2.Huse M, Kuriyan J. Cell. 2002;109:275–272. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 3.(a) Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, Levrero M, Sartorelli V, Cotter RJ, Cole PA. Nat Struct Mol Biol. 2004;11:308–315. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]; (b) Karanam B, Jiang L, Wang L, Kelleher NL, Cole PA. J Biol Chem. 2006;281:40292–40301. doi: 10.1074/jbc.M608813200. [DOI] [PubMed] [Google Scholar]; (c) Karanam B, Wang L, Wang D, Liu X, Marmorstein R, Cotter R, Cole PA. Biochemistry. 2007;46:8207–8216. doi: 10.1021/bi7000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Neumann H, Hazen JL, Weinstein J, Mehl RA, Chin JW. Nat Chem Biol. 2008;4:232–234. doi: 10.1038/nchembio.73. [DOI] [PubMed] [Google Scholar]; (b) Guo J, Melacon CE, Lee HS, Groff D, Schultz PG. Angew Chem Int Ed. 2008;47:6399–6401. doi: 10.1002/anie.200802336. [DOI] [PubMed] [Google Scholar]
- 5.(a) Muir TW, Sondhi D, Cole PA. Proc Natl Acad Sci USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Muralidharan V, Muir TW. Nat Methods. 2006;3:429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]; (c) Tarrant MK, Cole PA. Ann Rev Biochem. 2009;78:797–825. doi: 10.1146/annurev.biochem.78.070907.103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Heinemann U, Hahn M. Prog Biophys Mol Biol. 1995;64:121–143. doi: 10.1016/0079-6107(95)00013-5. [DOI] [PubMed] [Google Scholar]; (b) Gebhard LG, Risso VA, Santos J, Ferreyra RG, Noguera ME, Ermácora MR. J Mol Biol. 2006;358:280–8. doi: 10.1016/j.jmb.2006.01.095. [DOI] [PubMed] [Google Scholar]; (c) Baird GS, Zacharias DA, Tsien RY. Proc Natl Acad Sci USA. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Plainkum P, Fuchs SM, Wiyakrutta S, Raines RT. Nat Struct Biol. 2003;10:115–119. doi: 10.1038/nsb884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, Wang L, Zhao K, Thompson PR, Hwang Y, Marmorstein R, Cole PA. Nature. 2008;451:846–849. doi: 10.1038/nature06546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


