Abstract
Hematopoietic development requires coordinated actions from a variety of transcription factors. The core binding factor (CBF), consisting of a Runx protein and the CBFβ protein, is a transcription factor complex that is essential for emergence of the hematopoietic stem cell (HSC) from an endothelial cell stage. The hematopoietic defects observed in either Runx1 or CBFβ knockout mice underscore the necessity of this complex for definitive hematopoiesis. Despite the requirement for CBF in establishing definitive hematopoiesis, Runx1 loss has minimal impact on maintaining the HSC state postnatally, while CBFβ may continue to be essential. Lineage commitment, on the other hand, is significantly affected upon CBF loss in the adult, indicating a primary role for this complex in modulating differentiation. Given the impact of normal CBF function in the hematopoietic system, the severe consequences of disrupting CBF activity, either through point mutations or generation of fusion genes, are obvious. The physiologic role of CBF in differentiation is subverted to an active process of self-renewal maintenance by the genetic aberrations, through several possible mechanisms, contributing to the development of hematopoietic malignancies including myelodysplastic syndrome and leukemia. The major impact of CBF on the hematopoietic system in both development and disease highlights the need for understanding the intricate functions of this complex and reiterate the necessity of continued efforts to identify potential points of therapeutic intervention for CBF-related diseases.
The core binding factor (CBF), composed of a Runx family member (Runx1, 2, or 3) and a CBFβ subunit, performs many crucial functions during hematopoiesis. The CBF complex regulates transcriptional activation of a multitude of hematopoietic target genes including interleukin 3, granulocyte–macrophage colony stimulating factor, neutrophil elastase, and granzyme B to name a few (Licht, 2001). CBF is also a critical transcriptional repressor, most notably in the repression of the CD4 gene during thymopoiesis and of the interleukin-4 gene in immune response (Taniuchi et al., 2002; Naoe et al., 2007). Genetic ablation of CBF has highlighted the significant role it plays in regulating multiple hematopoietic lineages. In this review we will focus on the function of CBF in the developing hematopoietic stem cell (HSC), outline its described roles during adult hematopoiesis, and discuss the consequences of CBF disruption on self-renewal.
Hematopoietic Development is Controlled by CBF
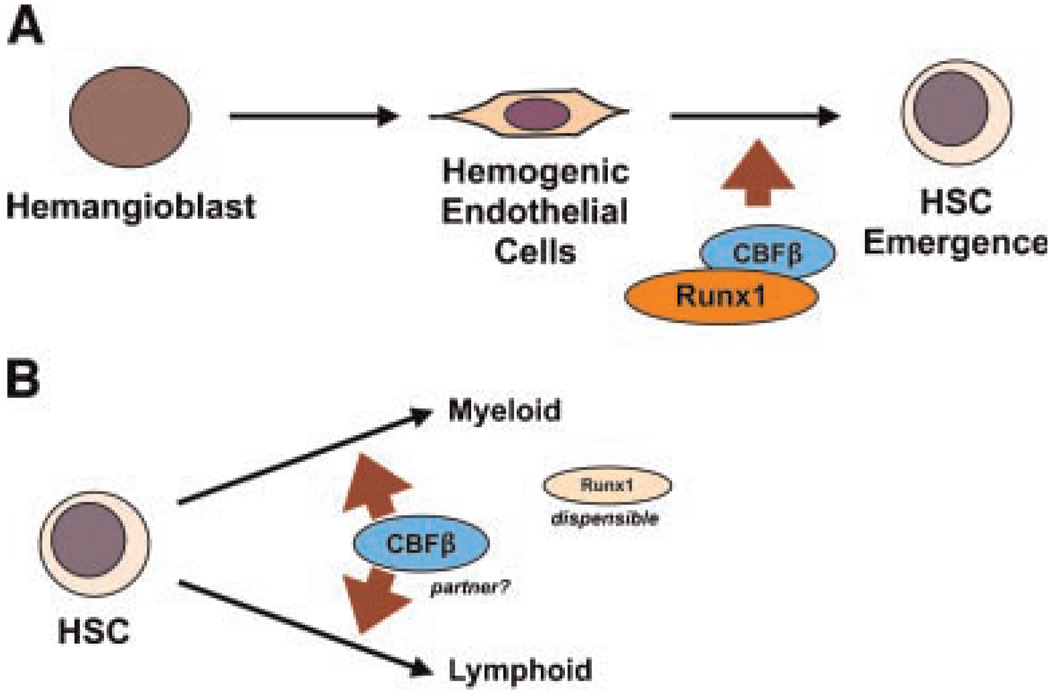
Fetal liver hematopoiesis does not occur in either the Runx1 or CBFβ knockout mice, and hemorrhaging within the central nervous system is evident, leading to death at E11.5–E12.5 (Okuda et al., 1996; Sasaki et al., 1996; Wang et al., 1996a,b). Colony forming assays using cells from the yolk sac and fetal liver revealed little to no erythroid or myeloid progenitor cell activity. Expression analyses have demonstrated Runx1 presence in endothelial cells in the dorsal aorta within the aorta/gonad/mesonephros (AGM) region and the umbilical and vitelline arteries (North et al., 1999), areas that are known to harbor hematopoietic stem and progenitor cells (HSPCs) in the developing embryo (Orkin and Zon, 2008). A recent study has described the transition from hemangioblast to hematopoietic cell formation through an essential endothelium stage (Lancrin et al., 2009). This seminal finding revealed a functional role for endothelial expression of Runx1 in directing definitive hematopoiesis, with no role for the complex in primitive hematopoiesis. Additionally, Chen et al. (2009) demonstrated that the Runx1 subunit is critical for the transition from endothelial cell to hematopoietic cell during development. Using the endothelial cell specific promoter, vascular endothelial cadherin (VEC), Chen et al. (2009) showed that most adult blood is derived from cells that developmentally expressed VEC. Conditional deletion of Runx1 with VEC-Cre resulted in severe hematopoietic deficiencies, similar to the Runx1 knockout mouse, and HSC formation was dependent on the presence of at least one allele of Runx1. In addition to endothelial to hematopoietic transition, Runx1 expression is critical for intra-aortic hematopoietic cluster formation, and knockout of either Runx1 or CBFβ prevents formation of these clusters. Surprisingly, however, Runx1 function was found to be dispensable for continued fetal hematopoiesis after the endothelial cell to HSC transition, as shown using a Vav1-Cre mouse that deletes Runx1 solely in hematopoietic-committed cells (Chen et al., 2009). Indeed, Runx1fl/fl mice expressing Vav1-Cre survive to adulthood, with a hematopoietic profile that resembles conditional Runx1 deletion in adult mice (Runx1fl/fl×Mx1-Cre mice postinduction, discussed in the following section). Restoration of Runx1 by a gene knockin approach demonstrated rescue of hematopoiesis in vivo, and this rescue required the transactivating but not the transrepression activity of Runx1 (Okuda et al., 2000; Goyama, 2004; Nishimura et al., 2004). Interestingly, CBFb−/− embryos failed to establish definitive hematopoiesis and died at E12.5. Restoration of CBFβ within the endothelium and hematopoietic progenitors of deficient mice (using the endothelial-specific Tek/Tie2 promoter to drive CBFβ expression) rescued fetal liver hematopoiesis at E12.5, with complications eventually resulting from incomplete bone formation as well as a lack of hematopoietic maturation in both myeloid and lymphoid (but not erythroid) lineages at E17.5, leading to death at birth (Miller et al., 2002). These findings suggest a requirement for sustained CBFβ expression in the more committed hematopoietic progenitors. Of note, as the article indicated, the failure in hematopoietic maturation is not secondary to defective bone formation and lack of bone marrow because Runx2-deficient mice, which display significant impairment in bone development, still retain intact definitive hematopoiesis. Thus, although Runx1 and CBFβ are both necessary for HSC formation at the initial stage of definitive hematopoiesis, only CBFβ is required for subsequent differentiation of the lymphoid and myeloid lineages. A Runx1-independent role for CBFβ in contributing to definitive hematopoiesis remains to be identified. Collectively, these data reveal a definitive requirement for the CBF transcription factor complex in the emergence of the self-renewing HSC but a differential role for Runx1 and CBFβ in hematopoietic development after the endothelial to hematopoietic cell transition (Fig. 1).
Fig. 1.
CBF complex in embryonic development. A: CBF complex promotes HSC emergence from hemogenic endothelial cells during embryonic development. B: After HSC emergence, CBFβ appears to remain a critical factor for progenitor cell production, but Runx1 is dispensable.
CBF in Adult Hematopoiesis
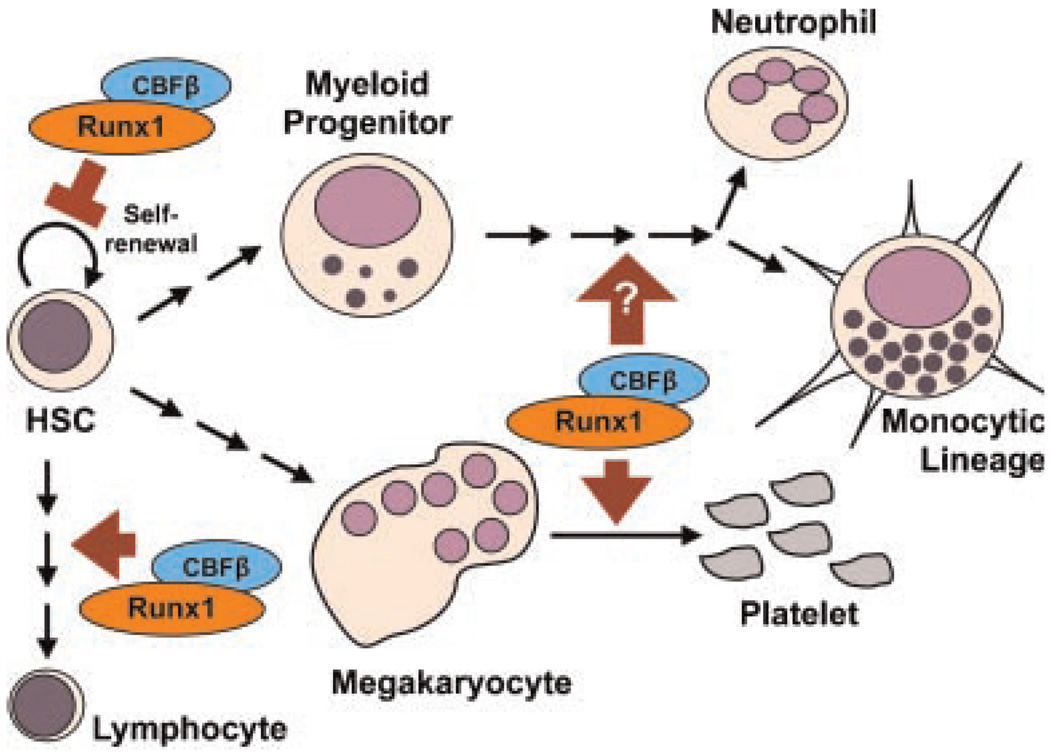
In addition to its role in fetal hematopoiesis, Runx1 was widely expected to play a critical role during adult hematopoiesis and HSC self-renewal, given its ubiquitous expression in adult hematopoietic lineages, including the adult HSC (Lorsbach et al., 2004; North et al., 2004). However, contrary to the dramatic effect upon CBF deletion (by VEC-Cre) in the emerging HSC, and similar to the phenotypes of Runx1 deletion by Vav1-Cre in embryonic HSC, Runx1 deletion in the adult HSC (by Mx1-Cre) presents with only a minor impact on HSC self-renewal (Ichikawa et al., 2004; Growney et al., 2005; Putz et al., 2006). One study demonstrated enhanced engrafting ability of the Runx1-deleted adult murine HSC using a competitive repopulating assay (Ichikawa et al., 2008), while another study using a similar Cre conditional approach concluded that loss of Runx1 had no effect on long-term HSC activity (Growney et al., 2005). In addition, Vav1-Cre-mediated Runx1 deletion results in an expansion of the Lin−Sca1+c-Kit+ population (Chen et al., 2009). Interestingly, haploinsufficiency of Runx1 results in fewer competitive repopulating HSCs when compared with control mice (Sun and Downing, 2004). These differences may be explained by the timing of Runx1 loss, by the possibility that heterozygosity for Runx1 through development may alter the properties of the adult HSC, or by technical differences in the experiments, but remain to be resolved. Conditional targeting of Runx1 is also associated with an expansion of myeloid progenitors, a phenotype possibly attributable to a partial block in myeloid differentiation or as a result of myeloid-biased differentiation of HSC (Ichikawa et al., 2004; Growney et al., 2005). In addition, extramedullary myeloid development and a myeloproliferative phenotype were observed upon Runx1 deletion, suggesting mobilization of excessive myeloid progenitors from the bone marrow cavity (Growney et al., 2005). B- and T-lymphoid differentiation was impaired upon Runx1 deletion, and a dramatic megakaryocytic maturation arrest was apparent in Runx1 deficient adult mice (Ichikawa et al., 2004; Growney et al., 2005). The megakaryocytic phenotype was closely recapitulated in CBFβ hypomorphic mouse embryos (expressing significantly reduced levels of CBFβ), and targeted deletion of CBFβ in T cells likewise demonstrated significant adverse effects on T-cell development (Naoe et al., 2007; Talebian et al., 2007). The CBFβ hypomorph experiments also detailed an exquisite sensitivity of the HSC to CBFβ dosage, with 15% of WT levels sufficient for emergence of the HSC while levels between 15% and 30% promoted an expansion of HSC and progenitor cells, reminiscent of the results upon conditional deletion of Runx1 (Talebian et al., 2007). Together, these results indicate a requirement for both CBF subunits in establishing and maintaining proper B- and T-cell function and a potential negative role for CBF in maintaining HSC quiescence. Given that maturation of some blood lineages are influenced by CBF loss and overexpression, a defined role in normal hematopoietic differentiation is well established. In contrast, maintenance of HSC self-renewal appears to be negatively correlated with the normal function of this hematopoietic transcription factor complex. Evidence obtained from embryonic and adult hematopoiesis indicates that CBF is indeed a potent differentiation promoter. Specifically, Runx1 promotes endothelial “differentiation” into HSC, as well as the subsequent myeloid, lymphoid, and megakaryocytic lineage differentiation. Interfering with CBF expression levels or function will ultimately lead to an expansion of stem and progenitor populations, a phenotype that resembles enhanced self-renewal (Fig. 2).
Fig. 2.
CBF complex is a potent differentiation modulator. CBF complex is required for myeloid, lymphoid, and megakaryocytic maturation in adult hematopoiesis.
CBF Complex Promotes Hematopoiesis Through Multiple Mechanisms
The above-mentioned findings indicate an essential role of the CBF complex as a master regulator in hematopoietic development, from emergence of HSCs to maintenance of multi-lineage differentiation. Recently, an elegant study using mouse genetic models has demonstrated that the ETS-family transcription factor, PU.1, is a major downstream target of Runx1 (Huang et al., 2008). This study showed that Runx1 binds to functionally important sites within the PU.1 − 14 kb upstream regulatory element and regulates PU.1 expression during embryonic and adult stages of hematopoietic development. Deletion of one allele of PU.1 enhanced certain aspects of the Runx1 knockout phenotype, including expansion of myeloid progenitors and a reduction in B220+ B cells, but reversed the deregulation of megakaryocyte differentiation and T-cell development observed in Runx1Δ/Δ mice, suggesting an ability of Runx1 to modulate PU.1 expression levels in a context dependent manner in different hematopoietic lineages. Ectopic expression of PU.1 in Runx1Δ/Δ mice partially rescued the granulocytic and B-cell phenotypes, demonstrating that PU.1 is a critical downstream target of Runx1 in adult hematopoiesis.
In addition to modulating PU.1 expression levels to regulate hematopoiesis, Runx1 is also capable of controlling hematopoietic cell fate by altering expression of the thrombopoietin (TPO) receptor, c-Mpl. Using mouse Lin−Sca1+c-Kit+ cells as well as a mouse embryonic stem (ES) cell model system, Satoh et al. (2008) demonstrated that Runx1 negatively regulates c-Mpl mRNA expression in HSPCs, but positively promotes c-Mpl expression in megakaryocytes. The differential regulation by Runx1 is due to recruitment of either a transcriptional repressor mSin3A (in HSPCs) or a transcriptional activator p300 (in megakaryocytes) to the c-Mpl promoter region, suggesting cell context-dependent function of Runx1. Moreover, because c-Mpl and its ligand, TPO, have been implicated in the maintenance of the HSC self-renewal phenotype, it is likely that, in addition to modulating the levels of PU.1, the CBF complex may also perturb HSC self-renewal and promote lineage differentiation by decreasing TPO/c-Mpl signaling (Alexander et al., 1996; Matsunaga et al., 1998; Yoshihara, 2007; Satoh et al., 2008).
Taken together, these examples support at the molecular level that the CBF complex is in fact a potent differentiation modulator. By regulating the expression of other transcription factors and surface receptors, CBF not only prevents HSC self-renewal, but may also specify lineage fate to ensure a balanced lineage development and maturation.
Disruption of CBF Impacts Self-Renewal
The CBF complex is commonly altered in human hematopoietic malignancies, including myelodysplastic syndrome (MDS) and leukemia. Various Runx1 mutations, in particular, have been identified in MDS and MDS-related acute myeloid leukemia (AML) (Harada and Harada, 2009). Several types of mutations have been observed, including in-frame mutations within the runt domain as well as C-terminal truncation mutants. Interestingly, each mutant class is thought to signal through distinct mechanisms based on the remaining intact functional domain(s), each resulting in a self-renewal phenotype (Harada and Harada, 2009). Expression of the D171N runt domain point mutant (N-terminal point mutation) in human CD34+ cells was capable of blocking differentiation and slightly enhanced the ability to self-renew (Watanabe-Okochi et al., 2008). However this mutant was unable to induce proliferation, suggesting additional mutations may be necessary to initiate disease. In contrast, expression of the C-terminal truncation mutant, S291fsX300, blocked differentiation, promoted self-renewal, and produced an increase in proliferation. These results highlight the diversity seen in MDS/AML patients harboring different Runx1 mutations and provide insight into the mechanisms involved in mutant Runx1-mediated disease development. Although the precise mechanism of action for these Runx1 mutants is unclear, it is unlikely to involve a straightforward dominant-negative effect on CBF activity or a sequestering of CBFβ protein as a predominant mode of action (Cammenga et al., 2007). The self-renewal phenotype associated with Runx1 heterozygous or null cells is significantly less dramatic than that seen with the Runx1 point mutations (Ichikawa et al., 2004; Cammenga et al., 2007). Thus, the Runx1 mutants show a gain of function and/or a disruption of the tightly regulated DNA-binding dependent and independent functions of CBF that promote the increased self-renewal phenotype.
In addition to Runx1 mutations in MDS/AML, both Runx1 and CBFβ are frequent targets of recurrent chromosomal translocations in human leukemia, comprising approximately 20% of AML and another 20% of acute lymphoblastic leukemia (ALL) cases (Look, 1997; Bernasconi, 2008). Translocation of chromosomes 8 and 21 [t(8;21)] and inversion of chromosome 16 [inv(16)] are two of the most frequent abnormalities associated with AML. The t(8;21) translocation leads to the N-terminal portion of the AML1 gene fused to nearly the entire eight twenty-one (ETO) gene, resulting in expression of the AML1-ETO protein. Inv(16) comprises a fusion of most of the CBFB gene and the 3′ portion of MYH11 (encoding part of the smooth muscle myosin heavy chain), which produces the CBFβ-MYH11 fusion gene. CBF requirement in normal hematopoiesis underlies the serious consequences that result upon disruption of this transcription factor complex in leukemia. Interestingly, knockin studies using one allele of either the AML1-ETO or the CBFB-MYH11 fusion showed striking similarities to the CBF knockout mice (Castilla et al., 1996; Yergeau et al., 1997; Okuda et al., 1998). Embryonic lethality occurred around E12.5–E13.5 as a result of hemorrhaging and a lack of definitive fetal liver hematopoiesis. Examination of colony formation at E10.5 and E11.5 revealed the absence of erythroid progenitors and a significant decrease in committed myeloid as well as multi-lineage progenitors in AML1-ETO expressing embryos, similar to CBF knockout mice (Okuda et al., 1996).
Nevertheless, despite demonstrating some striking similarities to knockout mice, the leukemia fusion knockin mice have phenotypes that strongly support a gain of function activity associated with a self-renewal phenotype. Analysis of E12.5–E13.5 mouse embryos expressing AML1-ETO demonstrated formation of large multi-lineage dysplastic colonies indicating an increased self-renewal capacity (Okuda et al., 1998), a characteristic not observed in CBF deficient embryos. Conditional expression of AML1-ETO in a mouse model did not result in a differentiation block in vivo; however, an increase in the replating ability of myeloid progenitors was observed (Higuchi et al., 2002). Furthermore, development of leukemia resulted upon exposure to theDNA alkylating agent N-ethyl-N-nitrosourea (ENU), highlighting the necessity of a cooperating event for CBF-mediated leukemogenesis. Additional mouse studies demonstrated the ability of AML1-ETO to generate a myeloproliferative phenotype with an increase in self-renewal when the stem cell compartment was targeted (de Guzman et al., 2002; Fenske et al., 2004). Similarly, conditional expression of CBFβ-MYH11 in murine bone marrow cells resulted in an increase in the HSC compartment (Kuo et al., 2006), whereas targeting of mouse thymocytes generated a block in T-cell development (Zhao et al., 2007). Interestingly, expression of CBFβ-MYH11 was able to induce spontaneous generation of AML, an outcome that is not realized upon expression of the AML1-ETO fusion (Higuchi et al., 2002). Introduction of AML1-ETO into normal human CD34+ hematopoietic cells presented a phenotype similar to that seen in mouse progenitor cells, namely, an enhanced self-renewal potential and an increase in long-term in vitro proliferative capacity (Mulloy et al., 2002, 2003; Tonks et al., 2003). Similar effects were seen in human CD34+ cells upon expression of CBFβ-MYH11, with a striking accumulation of eosinophils in the long-term cultures, a phenotype associated with the inv(16) in human patients but not seen in the mouse models of CBFβ-MYH11 (Wunderlich et al., 2006). The enhanced self-renewal capacity was evaluated in vivo in immunodeficient mice, implying an effect on a primitive progenitor cell by these two leukemia fusion proteins. Taken together, these studies demonstrate the profound effects of both leukemia fusion proteins on the self-renewal capacity of human and murine hematopoietic stem/progenitor cells and hint at the contribution these proteins make to the promotion of leukemia.
Mechanisms of CBF Fusion-Mediated Self-Renewal
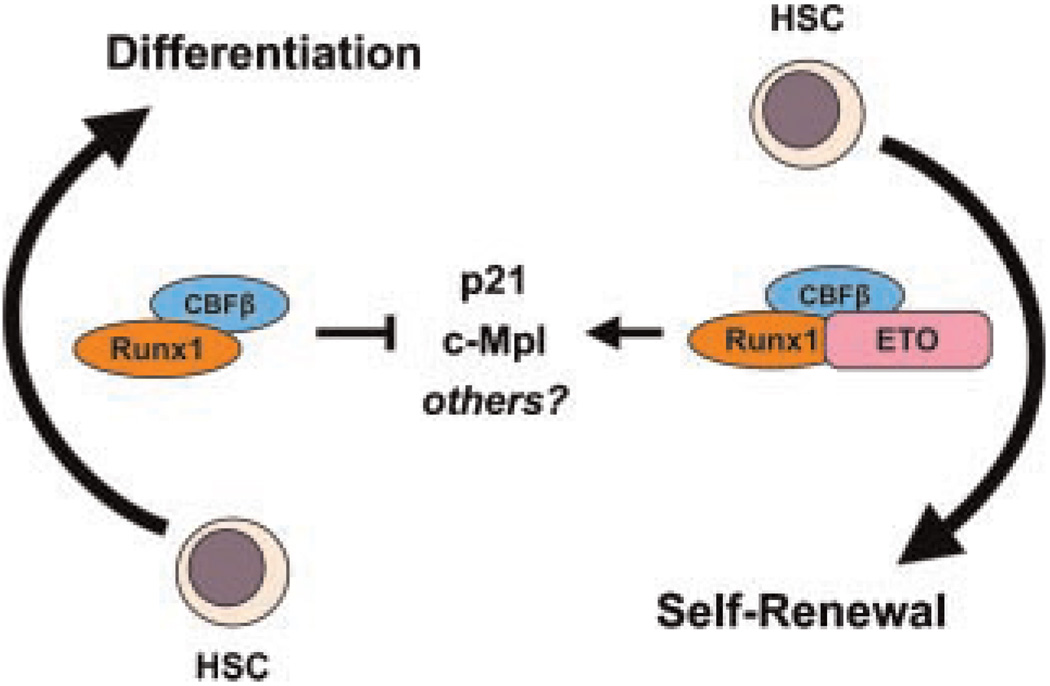
Normal CBF function involves the ability to modulate transcription of a variety of genes involved in directing hematopoietic differentiation. Thus, one can visualize the potential for significant deregulation of gene transcription in the presence of CBF fusion proteins and the resulting consequences that generate self-renewal signals. Using Drosophila genetics, Wildonger and Mann (2005) showed that AML1-ETO serves as a constitutive repressor of full-length AML1 and counteracts its ability to promote lineage differentiation. Notwithstanding this elegant demonstration in a powerful genetic model, numerous examples are evident from mammalian cell systems and mouse genetic models that argue for a more complex interaction between AML1 and AML1-ETO. For example, the promoter of the p21WAF1/CIP1 gene is directly repressed by AML1 in an in vitro reporter assay system (Lutterbach et al., 2000), while expression of the AML1-ETO fusion protein in various mammalian systems is consistently associated with an upregulation of p21WAF1/CIP1 (Yan et al., 2004; Peterson et al., 2007; Berg et al., 2008; Krejci et al., 2008), suggesting that AML1-ETO may abolish the regulation of AML1 on the promoter of p21WAF1/CIP1. Interestingly, evidence suggests that p21WAF1/CIP1 is critical for maintaining the self-renewal property of both normal HSCs and leukemia stem cells (Cheng et al., 2000; Viale et al., 2009), although more recent work has demonstrated a role for p21WAF1/CIP1 in HSC self-renewal under conditions of cellular stress but not necessarily during steady state hematopoiesis (van Os et al., 2007). Intriguingly, it has recently been demonstrated that expression of AML1-ETO induces significant cellular stress due to increased DNA damage and subsequent activation of p53 signaling (Krejci et al., 2008). Taken together, it is possible that AML1-ETO prevents the repressive action of AML1 on p21WAF1/CIP1 as a means to enhance self-renewal during an oncogene-induced stress response. To confound matters, AML1-ETO has also been shown to cooperate with loss of p21WAF1/CIP1 to promote leukemia formation in a mouse model using retroviral delivery of the AML1-ETO gene into mouse fetal liver hematopoietic progenitor cells in a p21WAF1/CIP1-null background (Peterson et al., 2007). How these different data converge on a common unifying mechanism of AML1-ETO action are unclear; it is likely that differences may reside in the unique approaches and model systems that have been used to address these questions. Similarly, the above-mentioned repressive effect ofAML1on c-Mpl gene expression is adversely regulated in the presence of AML1-ETO, providing another molecular example of the opposing effect of full-length AML1 and AML1-ETO fusion protein on HSC cell fate determination and tipping the balance from differentiation to self-renewal (Satoh et al., 2008) (Fig. 3).
Fig. 3.
Opposing effects of full-length Runx1 and AML1-ETO. Full-length Runx1 and Runx1-ETO individually partner with CBFβ to modulate levels of p21 and c-Mpl, both of which have been implicated in maintaining self-renewal phenotypes of HSC. Runx1 and AML1-ETO may also inversely regulate additional unidentified targets.
Additional studies have demonstrated novel potential mechanisms of AML1-ETO in promoting aberrant self-renewal and leukemogenesis. Recently, screening of a chemical library using an AML1-ETO zebrafish model established cyclooxygenase (Cox)-mediated production of prostaglandin E2 (PGE2) as a critical signaling mechanism for preventing differentiation of hemato poietic cells in AML1-ETO expressing zebrafish blood cells (Yeh et al., 2009). Suppression of the Cox enzymes promoted differentiation of hematopoietic cells expressing AML1-ETO. This finding fits nicely with the observation that Cox1/2 and PGE2 function as an essential component for normal HSC self-renewal (North et al., 2007). Together, these observations would suggest a role for Cox/PGE2 signaling in contributing to a self-renewal signal in AML. However, it remains to be determined whether this function is essential and/or aberrantly modulated in CBF leukemia.
Contribution of the CBF fusion partner must also be considered as a critical factor for understanding the aberrant function of the fusion protein. The AML1 fusion partner, ETO (MTG8), is a known transcriptional repressor that contains four Nervy Homology Regions (NHR1–4) named after the Drosophila ortholog Nervy (Davis et al., 2003). Functional studies have provided insight into the domains necessary for modulating the self-renewal phenotype. Loss of the NHR2 motif, responsible for oligomerization, significantly decreased the ability of AML1-ETO to inhibit differentiation of the human promonocytic U937 cell line and mouse primary hematopoietic cells, suggesting a role for the NHR2 domain in AML1-ETO-mediated self-renewal (Gelmetti et al., 1998; Zhang et al., 2001; Hug et al., 2002). An in vivo leukemia model of AML generated by an alternatively spliced form of the fusion gene, AML1-ETO9a (lacking the NHR3 and four regions), also relies on this domain to initiate leukemia (Yan et al., 2006, 2009). In addition to oligomerization, AML1-ETO requires DNA-binding ability to elicit its function. Point mutant disruption of DNA-binding, asshown in the context of AML1-ETO9a or full-length AML1-ETO, prevented leukemia development and self-renewal, respectively (Kwok et al., 2009; Roudaia et al., 2009; Yan et al., 2009). In contrast, the contribution of CBFβ to AML1-ETO associated self-renewal and leukemogenesis remains somewhat controversial. Two recent studies using different point mutants to disrupt the CBFβ interaction with AML1-ETO reached opposite conclusions. The Y113A/T161A CBFβ-binding mutant of AML1-ETO showed impaired replating ability and loss of leukemia generation (Roudaia et al., 2009); however, a second study suggested that AML1-ETO dependent self-renewal is independent of CBFβ function using the M106V and A107T mutants (Kwok et al., 2009). In addition, Kwok et al. showed that knockdown of CBFβ using RNAi did not affect the self-renewal ability of AML1-ETO in vitro. Clearly, key points must be addressed to clarify the disparate findings.
Corepressor Involvement in Self-Renewal
Functionally, ETO is a transcriptional repressor that contains binding sites for corepressors including nuclear corepressor (N-CoR), SMRT, and mSin3A (Wang et al., 1998; Amann et al., 2001). Corepressor interaction primarily occurs through the NHR2 and NHR4 domains of ETO (Gelmetti et al., 1998; Lutterbach et al., 1998). Interestingly, studies have revealed that AML1-ETO activity is not dependent on corepressor interaction with the NHR2 domain; however, oligomerization elicited through this domain is necessary to promote AML1-ETO-mediated self-renewal (Liu et al., 2006). These findings do not negate the potentially critical role of corepressor complexes in AML1-ETO function but do demonstrate the obligatory role of oligomerization. Indeed, suppression of SMRT corepressor interaction with the NHR4 domain, achieved through point mutations within this region, resulted in impaired function compared to wild-type AML1-ETO (Liu et al., 2007). When coupled with disruption of the NHR2 domain, loss of corepressor interaction with NHR4 mitigated the block in differentiation caused by AML1-ETO (Hug et al., 2002; Liu et al., 2007). Together, these data highlight the importance of NHR2 oligomerization and NHR4 corepressor interaction as key mechanisms in regulating AML1-ETO induced self-renewal. Future studies will be required to determine the specific targets and pathways affected by these domains.
In conjunction with corepressor action, ETO has been shown to repress gene transcription of E protein transcription factors, such as HEB (HeLa E-box-binding protein) and E2A, involved in promoting differentiation and proliferation (Massari and Murre, 2000; Zhang et al., 2004). Recent work has demonstrated the colocalization of AML1-ETO with AML1 and HEB on specific AML1 target genes, and the ability of AML1-ETO to re-direct AML1 DNA binding (Gardini et al., 2008; Okumura et al., 2008). These data give insight into the potential actions of AML1-ETO in disrupting normal AML1 function and suggest one possible mechanism for promoting aberrant self-renewal. The interaction between AML1-ETO and HEB is mediated through the NHR1 domain, and AML1-ETO binding inhibits E protein activity by blocking recruitment of the transcriptional coactivator p300/CBP (Zhang et al., 2004). These data implicate the NHR1 region as a critical domain through which AML1-ETO acts to aberrantly repress transcription and potentially promote leukemogenesis. However, recent evidence suggests that theNHR1 domain may be dispensable for E protein repression by AML1-ETO (Park et al., 2009) and for leukemia induction in the context of the AML1-ETO-9a protein (Yan et al., 2009). In addition, disrupting HEB interaction through point mutations within the NHR1 domain had little to no effect on AML1-ETO function (Park et al., 2009). Thus, the role of E protein repression in AML1-ETO induced leukemogenesis may be limited.
Similar to AML1-ETO, CBFβ-MYH11 has exhibited several mechanisms by which it is able to promote self-renewal and leukemia development. The CBFβ-MYH11 fusion retains the ability to bind AML1 with increased affinity as compared to CBFβ and thus is able to sequester Runx proteins away from target genes (Wijmenga et al., 1996; Adya et al., 1998; Kanno et al., 1998; Huang et al., 2004). This binding ability was shown to mediate hyperdimerization with the AML1 Runt domain through a region near the junction of CBFβ and MYH11 (Huang et al., 2004). Recently, CBFβ-MYH11 was also shown to bind and sequester HIPK2, preventing the critical phosphorylation of AML1 and consequently p300 (Wee et al., 2008). Impairment of AML1 and p300 may be one mechanism utilized by CBFβ-MYH11 to circumvent differentiation and promote self-renewal. Active transcriptional repression has also been exhibited by CBFβ-MYH11 and is mediated through the C-terminal portion of MYH11. Similar to AML1-ETO, CBFβ-MYH11 contains a corepressor-binding domain (Durst et al., 2003). The assembly complex domain (ACD) within the C-terminal portion of CBFβ-MYH11 is necessary for association with the mSin3A and HDAC8 corepressors, and through these interactions the fusion is proposed to generate aberrant transcriptional repression of CBF target genes by interacting with AML1 on DNA. The ACD is also required for multimerization of MYH11, a function that was demonstrated through the actions of CBFβ-MYH11 to confer nuclear retention of the fusion and mediate CBF complex DNA binding (Kummalue et al., 2002). Together, these data demonstrate the range of function produced by the CBF fusion proteins and portray the complexity behind identifying a target-worthy protein and/or pathway in CBF dependent malignancies.
Conclusion
It is clear that the CBF complex plays a critical role in hematopoiesis and HSC development. Many advances have been made to uncover detailed evidence identifying the molecular mechanisms behind CBF function in these processes. The importance of CBF for embryonic development emphasizes the potential consequences resulting from disruption of normal complex activities and demonstrates the consequential impact that this has on regulating aberrant self-renewal of the HSC. Continued exploration of the molecular mechanisms regulating CBF activities in normal and disease states is necessary to address the pressing issue of effective and lasting treatment options for CBF-associated malignancies.
Acknowledgments
The authors would like to thank Dr. Benjamin Mizukawa, Dr. Gang Huang, and Mark Wunderlich for critical reading of the manuscript and helpful suggestions.
Contract grant sponsor: NIH NCI;
Contract grant number: CA118319.
Literature Cited
- Adya N, Stacy T, Speck NA, Liu PP. The leukemic protein core binding factor beta (CBFbeta)-smooth-muscle myosin heavy chain sequesters CBFalpha2 into cytoskeletal filaments and aggregates. Mol Cell Biol. 1998;18:7432–7443. doi: 10.1128/mcb.18.12.7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander WS, Roberts AW, Nicola NA, Li R, Metcalf D. Deficiencies in progenitor cells of multiple hematopoietic lineages and defective megakaryocytopoiesis in mice lacking the thrombopoietic receptor c-Mpl. Blood. 1996;87:2162–2170. [PubMed] [Google Scholar]
- Amann JM, Nip J, Strom DK, Lutterbach B, Harada H, Lenny N, Downing JR, Meyers S, Hiebert SW. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol Cell Biol. 2001;21:6470–6483. doi: 10.1128/MCB.21.19.6470-6483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg T, Fliegauf M, Burger J, Staege MS, Liu S, Martinez N, Heidenreich O, Burdach S, Haferlach T, Werner MH, Lubbert M. Transcriptional upregulation of p21/WAF/ Cip1 in myeloid leukemic blasts expressing AML1-ETO. Haematologica. 2008;93:1728–1733. doi: 10.3324/haematol.13044. [DOI] [PubMed] [Google Scholar]
- Bernasconi P. Molecular pathways in myelodysplastic syndromes and acute myeloid leukemia: Relationships and distinctions—A review. Br J Haematol. 2008;142:695–708. doi: 10.1111/j.1365-2141.2008.07245.x. [DOI] [PubMed] [Google Scholar]
- Cammenga J, Niebuhr B, Horn S, Bergholz U, Putz G, Buchholz F, Lohler J, Stocking C. RUNX1 DNA-binding mutants, associated with minimally differentiated acute myelogenous leukemia, disrupt myeloid differentiation. Cancer Res. 2007;67:537–545. doi: 10.1158/0008-5472.CAN-06-1903. [DOI] [PubMed] [Google Scholar]
- Castilla LH, Wijmenga C, Wang Q, Stacy T, Speck NA, Eckhaus M, Marín-Padilla M, Collins FS, Wynshaw-Boris A, Liu PP. Failure of embryonic hematopoiesis and lethal hemorrhages in mouse embryos heterozygous for a knocked-in leukemia gene CBFB-MYH11. Cell. 1996;87:687–696. doi: 10.1016/s0092-8674(00)81388-4. [DOI] [PubMed] [Google Scholar]
- Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck N. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, Scadden DT. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- Davis JN, McGhee L, Meyers S. The ETO (MTG8) gene family. Gene. 2003;303:1–10. doi: 10.1016/s0378-1119(02)01172-1. [DOI] [PubMed] [Google Scholar]
- de Guzman CG, Warren AJ, Zhang Z, Gartland L, Erickson P, Drabkin H, Hiebert SW, Klug CA. Hematopoietic stem cell expansion and distinct myeloid developmental abnormalities in a murine model of the AML1-ETO translocation. Mol Cell Biol. 2002;22:5506–5517. doi: 10.1128/MCB.22.15.5506-5517.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durst KL, Lutterbach B, Kummalue T, Friedman AD, Hiebert SW. The inv(16) fusion protein associates with corepressors via a smooth muscle myosin heavy-chain domain. Mol Cell Biol. 2003;23:607–619. doi: 10.1128/MCB.23.2.607-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenske TS, Pengue G, Mathews V, Hanson PT, Hamm SE, Riaz N, Graubert TA. Stem cell expression of the AML1/ETO fusion protein induces a myeloproliferative disorder in mice. Proc Natl Acad Sci USA. 2004;101:15184–15189. doi: 10.1073/pnas.0400751101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardini A, Cesaroni M, Luzi L, Okumura A, Biggs J, Minardi S, Venturini E, Zhang D, Pelicci P, Alcalay M, Cheung V. AML1/ETO oncoprotein is directed to AML1 binding regions and co-localizes with AML1 and HEB on its targets. PLoS Genet 4:e1000275. 2008 doi: 10.1371/journal.pgen.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci PG, Lazar MA. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18:7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyama S. The transcriptionally active form of AML1 is required for hematopoietic rescue of the AML1-deficient embryonic para-aortic splanchnopleural (P-Sp) region. Blood. 2004;104:3558–3564. doi: 10.1182/blood-2004-04-1535. [DOI] [PubMed] [Google Scholar]
- Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, Curley DP, Kutok JL, Akashi K, Williams IR, Speck NA, Gilliland DG. Loss of Runx1 perturbs adult hematopoiesis and is associated with amyeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y, Harada H. Molecular pathways mediating MDS/AML with focus on AML1/ RUNX1 point mutations. J Cell Physiol. 2009;220:16–20. doi: 10.1002/jcp.21769. [DOI] [PubMed] [Google Scholar]
- Higuchi M, O’Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 2002;1:63–74. doi: 10.1016/s1535-6108(02)00016-8. [DOI] [PubMed] [Google Scholar]
- Huang G, Shigesada K, Wee HJ, Liu PP, Osato M, Ito Y. Molecular basis for a dominant inactivation of RUNX1/AML1 by the leukemogenic inversion 16 chimera. Blood. 2004;103:3200–3207. doi: 10.1182/blood-2003-07-2188. [DOI] [PubMed] [Google Scholar]
- Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, Koschmieder S, Okuno Y, Dayaram T, Growney JD, Shivdasani RA, Gilliland DG, Speck NA, Nimer SD, Tenen DG. PU.1 is a major downstream target of AML1 (RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [DOI] [PubMed] [Google Scholar]
- Hug BA, Lee SY, Kinsler EL, Zhang J, Lazar MA. Cooperative function of Aml1-ETO corepressor recruitment domains in the expansion of primary bone marrow cells. Cancer Res. 2002;62:2906–2912. [PubMed] [Google Scholar]
- Ichikawa M, Asai T, Saito T, Yamamoto G, Seo S, Yamazaki I, Yamagata T, Mitani K, Chiba S, Hirai H, Ogawa S, Kurokawa M. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- Ichikawa M, Goyama S, Asai T, Kawazu M, Nakagawa M, Takeshita M, Chiba S, Ogawa S, Kurokawa M. AML1/Runx1 negatively regulates quiescent hematopoietic stem cells in adult hematopoiesis. J Immunol. 2008;180:4402–4408. doi: 10.4049/jimmunol.180.7.4402. [DOI] [PubMed] [Google Scholar]
- Kanno Y, Kanno T, Sakakura C, Bae SC, Ito Y. Cytoplasmic sequestration of the polyomavirus enhancer binding protein 2 (PEBP2)/core binding factor alpha (CBFalpha) subunit by the leukemia-related PEBP2/CBFbeta-SMMHC fusion protein inhibits PEBP2/ CBF-mediated transactivation. Mol Cell Biol. 1998;18:4252–4261. doi: 10.1128/mcb.18.7.4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci O, Wunderlich M, Geiger H, Chou FS, Schleimer D, Jansen M, Andreassen PR, Mulloy JC. p53 signaling in response to increased DNA damage sensitizes AML1-ETO cells to stress-induced death. Blood. 2008;111:2190–2199. doi: 10.1182/blood-2007-06-093682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummalue T, Lou J, Friedman AD. Multimerization via its myosin domain facilitates nuclear localization and inhibition of core binding factor (CBF) activities by the CBFbeta-smooth muscle myosin heavy chain myeloid leukemia oncoprotein. Mol Cell Biol. 2002;22:8278–8291. doi: 10.1128/MCB.22.23.8278-8291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo YH, Landrette SF, Heilman SA, Perrat PN, Garrett L, Liu PP, Le Beau MM, Kogan S, Castilla LH. Cbf beta-SMMHC induces distinct abnormalmyeloid progenitors able to develop acute myeloid leukemia. Cancer Cell. 2006;9:57–68. doi: 10.1016/j.ccr.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Kwok C, Zeisig BB, Qiu J, Dong S, So CW. Transforming activity of AML1-ETO is independent of CBFbeta and ETO interaction but requires formation of homo-oligomeric complexes. Proc Natl Acad Sci USA. 2009;106:2853–2858. doi: 10.1073/pnas.0810558106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457:892–895. doi: 10.1038/nature07679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licht JD. AML1 and the AML1-ETO fusion protein in the pathogenesis of t(8;21) AML. Oncogene. 2001;20:5660–5679. doi: 10.1038/sj.onc.1204593. [DOI] [PubMed] [Google Scholar]
- Liu Y, Cheney MD, Gaudet JJ, Chruszcz M, Lukasik SM, Sugiyama D, Lary J, Cole J, Dauter Z, Minor W, Speck NA, Bushweller JH. The tetramer structure of theNervy homology two domain, NHR2, is critical for AML1/ETO’s activity. Cancer Cell. 2006;9:249–260. doi: 10.1016/j.ccr.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Liu Y, Chen W, Gaudet J, Cheney M, Roudaia L, Cierpicki T, Klet R, Hartman K, Laue T, Speck N. Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO’s activity. Cancer Cell. 2007;11:483–497. doi: 10.1016/j.ccr.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Look A. Oncogenic transcription factors in the human acute leukemias. Science. 1997;278:1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- Lorsbach RB, Moore J, Ang SO, Sun W, Lenny N, Downing JR. Role of RUNX1 in adult hematopoiesis: Analysis of RUNX1-IRES-GFP knock-in mice reveals differential lineage expression. Blood. 2004;103:2522–2529. doi: 10.1182/blood-2003-07-2439. [DOI] [PubMed] [Google Scholar]
- Lutterbach B, Westendorf JJ, Linggi B, Patten A, Moniwa M, Davie JR, Huynh KD, Bardwell VJ, Lavinsky RM, Rosenfeld MG, Glass C, Seto E, Hiebert SW. ETO, a target of t(8;21) in acute leukemia, interacts with theN-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18:7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutterbach B, Westendorf JJ, Linggi B, Isaac S, Seto E, Hiebert SW. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J Biol Chem. 2000;275:651–656. doi: 10.1074/jbc.275.1.651. [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C. Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga T, Kato T, Miyazaki H, Ogawa M. Thrombopoietin promotes the survival of murine hematopoietic long-term reconstituting cells: Comparison with the effects of FLT3/FLK-2 ligand and interleukin-6. Blood. 1998;92:452–461. [PubMed] [Google Scholar]
- Miller J, Horner A, Stacy T, Lowrey C, Lian JB, Stein G, Nuckolls GH, Speck NA. The core-binding factor beta subunit is required for bone formation and hematopoietic maturation. Nat Genet. 2002;32:645–649. doi: 10.1038/ng1049. [DOI] [PubMed] [Google Scholar]
- Mulloy JC, Cammenga J, MacKenzie KL, Berguido FJ, Moore MA, Nimer SD. The AML1-ETO fusion protein promotes the expansion of human hematopoietic stem cells. Blood. 2002;99:15–23. doi: 10.1182/blood.v99.1.15. [DOI] [PubMed] [Google Scholar]
- Mulloy JC, Cammenga J, Berguido FJ, Wu K, Zhou P, Comenzo RL, Jhanwar S, Moore MA, Nimer SD. Maintaining the self-renewal and differentiation potential of human CD34+ hematopoietic cells using a single genetic element. Blood. 2003;102:4369–4376. doi: 10.1182/blood-2003-05-1762. [DOI] [PubMed] [Google Scholar]
- Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, Littman DR, Taniuchi I. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J Exp Med. 2007;204:1749–1755. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M, Fukushima-Nakase Y, Fujita Y, Nakao M, Toda S, Kitamura N, Abe T, Okuda T. VWRPY motif-dependent and -independent roles of AML1/Runx1 transcription factor in murine hematopoietic development. Blood. 2004;103:562–570. doi: 10.1182/blood-2003-06-2109. [DOI] [PubMed] [Google Scholar]
- North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, Marín-Padilla M, Speck NA. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development. 1999;126:2563–2575. doi: 10.1242/dev.126.11.2563. [DOI] [PubMed] [Google Scholar]
- North TE, Stacy T, Matheny CJ, Speck NA, de Bruijn MF. Runx1 is expressed in adult mouse hematopoietic stem cells and differentiating myeloid and lymphoid cells, but not in maturing erythroid cells. Stem Cells. 2004;22:158–168. doi: 10.1634/stemcells.22-2-158. [DOI] [PubMed] [Google Scholar]
- North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang IH, Grosser T, Fitzgerald GA, Daley GQ, Orkin SH, Zon LI. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- Okuda T, Cai Z, Yang S, Lenny N, Lyu CJ, van Deursen JM, Harada H, Downing JR. Expression of a knocked-in AML1-ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood. 1998;91:3134–3143. [PubMed] [Google Scholar]
- Okuda T, Takeda K, Fujita Y, Nishimura M, Yagyu S, Yoshida M, Akira S, Downing JR, Abe T. Biological characteristics of the leukemia-associated transcriptional factor AML1 disclosed by hematopoietic rescue of AML1-deficient embryonic stem cells by using a knock-in strategy. Mol Cell Biol. 2000;20:319–328. doi: 10.1128/mcb.20.1.319-328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura A, Peterson LF, Okumura F, Boyapati A, Zhang D. t(8;21)(q22;q22) Fusion proteins preferentially bind to duplicated AML1/RUNX1 DNA-binding sequences to differentially regulate gene expression. Blood. 2008;112:1392–1401. doi: 10.1182/blood-2007-11-124735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S, Zon L. Hematopoiesis: An evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Chen W, Cierpicki T, Tonelli M, Cai X, Speck NA, Bushweller JH. Structure of the AML1-ETO eTAFH domain-HEB peptide complex and its contribution to AML1-ETO activity. Blood. 2009;113:3558–3567. doi: 10.1182/blood-2008-06-161307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson L, Yan M, Zhang D. The p21Waf1 pathway is involved in blocking leukemogenesis by the t(8;21) fusion protein AML1-ETO. Blood. 2007;109:4392–4398. doi: 10.1182/blood-2006-03-012575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putz G, Rosner A, Nuesslein I, Schmitz N, Buchholz F. AML1 deletion in adult mice causes splenomegaly and lymphomas. Oncogene. 2006;25:929–939. doi: 10.1038/sj.onc.1209136. [DOI] [PubMed] [Google Scholar]
- Roudaia L, Cheney MD, Manuylova E, Chen W, Morrow M, Park S, Lee CT, Kaur P, Williams O, Bushweller JH, Speck NA. CBFbeta is critical for AML1-ETO and TEL-AML1 activity. Blood. 2009;113:3070–3079. doi: 10.1182/blood-2008-03-147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Yagi H, Bronson RT, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci USA. 1996;93:12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh Y, Matsumura I, Tanaka H, Ezoe S, Fukushima K, Tokunaga M, Yasumi M, Shibayama H, Mizuki M, Era T, Okuda T, Kanakura Y. AML1/RUNX1 Works as a Negative Regulator of c-Mpl in Hematopoietic Stem Cells. J Biol Chem. 2008;283:30045–30056. doi: 10.1074/jbc.M804768200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Downing JR. Haploinsufficiency of AML1 results in a decrease in the number of LTR-HSCs while simultaneously inducing an increase in more mature progenitors. Blood. 2004;104:3565–3572. doi: 10.1182/blood-2003-12-4349. [DOI] [PubMed] [Google Scholar]
- Talebian L, Li Z, Guo Y, Gaudet J, Speck ME, Sugiyama D, Kaur P, Pear WS, Maillard I, Speck NA. T-lymphoid, megakaryocyte, and granulocyte development are sensitive to decreases in CBFbeta dosage. Blood. 2007;109:11–21. doi: 10.1182/blood-2006-05-021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- Tonks A, Pearn L, Tonks AJ, Pearce L, Hoy T, Phillips S, Fisher J, Downing JR, Burnett AK, Darley RL. The AML1-ETO fusion gene promotes extensive self-renewal of human primary erythroid cells. Blood. 2003;101:624–632. doi: 10.1182/blood-2002-06-1732. [DOI] [PubMed] [Google Scholar]
- van Os R, Kamminga LM, Ausema A, Bystrykh LV, Draijer DP, van Pelt K, Dontje B, de Haan G. A limited role for p21Cip1/Waf1 in maintaining normal hematopoietic stem cell functioning. Stem Cells. 2007;25:836–843. doi: 10.1634/stemcells.2006-0631. [DOI] [PubMed] [Google Scholar]
- Viale A, De Franco F, Orleth A, Cambiaghi V, Giuliani V, Bossi D, Ronchini C, Ronzoni S, Muradore I, Monestiroli S, Gobbi A, Alcalay M, Minucci S, Pelicci P. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature. 2009;457:51–56. doi: 10.1038/nature07618. [DOI] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Binder M, Marín-Padilla M, Sharpe AH, Speck NA. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996a;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Miller JD, Lewis AF, Gu TL, Huang X, Bushweller JH, Bories JC, Alt FW, Ryan G, Liu PP, Wynshaw-Boris A, Binder M, Marín-Padilla M, Sharpe AH, Speck NA. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996b;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/ HDAC1 complex. Proc Natl Acad Sci USA. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe-Okochi N, Kitaura J, Ono R, Harada H, Harada Y, Komeno Y, Nakajima H, Nosaka T, Inaba T, Kitamura T. AML1 mutations induced MDS and MDS/AML in a mouse BMT model. Blood. 2008;111:4297–4308. doi: 10.1182/blood-2007-01-068346. [DOI] [PubMed] [Google Scholar]
- Wee HJ, Voon DC, Bae SC, Ito Y. PEBP2-beta/CBF-beta-dependent phosphorylationof RUNX1 and p300 by HIPK2: Implications for leukemogenesis. Blood. 2008;112:3777–3787. doi: 10.1182/blood-2008-01-134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijmenga C, Gregory PE, Hajra A, Schröck E, Ried T, Eils R, Liu PP, Collins FS. Core binding factor beta-smooth muscle myosin heavy chain chimeric protein involved in acute myeloid leukemia forms unusual nuclear rod-like structures in transformed NIH 3T3 cells. Proc Natl Acad Sci USA. 1996;93:1630–1635. doi: 10.1073/pnas.93.4.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildonger J, Mann RS. The t(8;21) translocation converts AML1 into a constitutive transcriptional repressor. Development. 2005;132:2263–2272. doi: 10.1242/dev.01824. [DOI] [PubMed] [Google Scholar]
- Wunderlich M, Krejci O, Wei J, Mulloy JC. Human CD34+ cells expressing the inv(16) fusion protein exhibit a myelomonocytic phenotype with greatly enhanced proliferative ability. Blood. 2006;108:1690–1697. doi: 10.1182/blood-2005-12-012773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Burel SA, Peterson LF, Kanbe E, Iwasaki H, Boyapati A, Hines R, Akashi K, Zhang DE. Deletion of an AML1-ETO C-terminal NcoR/SMRT-interacting region strongly induces leukemia development. Proc Natl Acad Sci USA. 2004;101:17186–17191. doi: 10.1073/pnas.0406702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M, Kanbe E, Peterson L, Boyapati A, Miao Y, Wang Y, Chen I, Chen Z, Rowley J, Willman C, Zhang D. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med. 2006;12:945–949. doi: 10.1038/nm1443. [DOI] [PubMed] [Google Scholar]
- Yan M, Ahn EY, Hiebert SW, Zhang D. RUNX1/AML1 DNA-binding domain and ETO/MTG8 NHR2-dimerization domain are critical to AML1-ETO9a leukemogenesis. Blood. 2009;113:883–886. doi: 10.1182/blood-2008-04-153742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh JR, Munson KM, Elagib KE, Goldfarb AN, Sweetser DA, Peterson RT. Discovering chemical modifiers of oncogene-regulated hematopoietic differentiation. Nat Chem Biol. 2009;5:236–243. doi: 10.1038/nchembio.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau DA, Hetherington CJ, Wang Q, Zhang P, Sharpe AH, Binder M, Marín-Padilla M, Tenen DG, Speck NA, Zhang DE. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]
- Yoshihara H. Thrombopoietin/MPL signaling regulates hematopoietic stem cell quiescence and interaction with the osteoblastic niche. Cell Stem Cell. 2007;1:685–697. doi: 10.1016/j.stem.2007.10.020. [DOI] [PubMed] [Google Scholar]
- Zhang J, Hug BA, Huang EY, Chen CW, Gelmetti V, Maccarana M, Minucci S, Pelicci PG, Lazar MA. Oligomerization of ETO is obligatory for corepressor interaction. Mol Cell Biol. 2001;21:156–163. doi: 10.1128/MCB.21.1.156-163.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Yamamura S, Chait BT, Roeder RG. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science. 2004;305:1286–1289. doi: 10.1126/science.1097937. [DOI] [PubMed] [Google Scholar]
- Zhao L, Cannons JL, Anderson S, Kirby M, Xu L, Castilla LH, Schwartzberg PL, Bosselut R, Liu P. CBFB-MYH11 hinders early T-cell development and induces massive cell death in the thymus. Blood. 2007;109:3432–3440. doi: 10.1182/blood-2006-10-051508. [DOI] [PMC free article] [PubMed] [Google Scholar]