Abstract
Previously, we reported a method for the attachment of living cells to surfaces through the hybridization of synthetic DNA strands attached to their plasma membrane. The oligonucleotides were introduced using metabolic carbohydrate engineering, which allowed reactive tailoring of the cell surface glycans for chemoselective bioconjugation. While this method is highly effective for cultured mammalian cells, we report here a significant improvement of this technique that allows the direct modification of cell surfaces with NHS-DNA conjugates. This method is rapid and efficient, allowing virtually any mammalian cell to be patterned on surfaces bearing complementary DNA in under 1 h. We demonstrate this technique using several types of cells that are generally incompatible with integrin-targeting approaches, including red blood cells and primary T-cells. Cardiac myoblasts were also captured. The immobilization procedure itself was found not to activate primary T-cells, in contrast to previously reported antibody- and lectin-based methods. Myoblast cells were patterned with high efficiency and remained undifferentiated after surface attachment. Upon changing to differentiation media, myotubes formed in the center of the patterned areas with an excellent degree of edge alignment. The availability of this new protocol greatly expands the applicability of the DNA-based attachment strategy for the generation of artificial tissues and the incorporation of living cells into device settings.
Introduction
The ability to pattern cells on surfaces provides a new platform for the study of cell biology,1–4 the control of stem cell differentiation, 5 and the engineering of new tissues.6 Typically, cell-based arrays are formed by printing surfaces of interest with “RGD” peptides that are designed to bind to integrins on the cell surface.7 While this approach has been widely adopted for the immobilization of many cell types, it cannot be used to capture nonadherent cell lines (such as leukocytes) or to bind multiple cell types to unique array features. It also can cause undesired changes in cell differentiation or behavior because it engages the very surface receptors that are involved in controlling these processes.8,9 As an alternative method that can circumvent these limitations, we have reported the capture of live cells through the hybridization of synthetic DNA strands covalently linked to their plasma membranes to surfaces printed with complementary sequences.10–12 In addition to allowing multiple cell types to be patterned on a single substrate, this method offers the important advantages of substrate reuse and tunability. Most importantly, we have used this approach to capture nonadherent cells in addition to adherent ones, and we have shown that the cells experience minimal changes in behavior as a result of immobilization through this receptor-independent process. In previous reports, we have shown the utility of this method for the formation of complex cell patterns,13 the capture of single cells for reverse transcription polymerase chain reaction (RT-PCR) analysis,14 and the attachment of living cells to AFM tips for force measurement.15
The DNA strands used in previous studies were installed into cell surface glycans through a two-step process. First, the cells were fed with an azide-containing mannose derivative for 1–3 days. This sugar was subsequently metabolized and incorporated into sialic-acid-containing cell surface glycans.16 The DNA was then targeted to the azide functionality by the Staudinger ligation.10 While effective, this protocol is most appropriate for cultured mammalian cell lines, as it requires multiple days of exposure to install a sufficient number of azide groups. To expand the generality of this DNA-based adhesion method, we report here an improved method for the direct installation of DNA strands onto virtually any cell surface. This procedure can be carried out in less than 1 h, and it leads to equivalent levels of cell surface functionalization with any oligonucleotide sequence of interest. We demonstrate the use of this new labeling method for the capture of red blood cells, primary T-cells, and myoblasts. This new technique greatly expands the scope of the DNA-based adhesion strategy and is sufficiently straightforward to be used in laboratories that do not specialize in organic synthesis.
Materials and Methods
General Experimental Procedures
All cell culture reagents were obtained from Gibco/Invitrogen Corp. (Carlsbad, CA) unless otherwise noted. Cell culture was conducted using standard techniques. Jurkat cells were grown in T-25 culture flasks (Corning) in RPMI Medium 1640 supplemented with 10% (v/v) fetal bovine serum (FBS, HyClone) and 1% penicillin/streptomycin (P/S, Sigma). MCF-7 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 1% nonessential amino acids and 10% fetal bovine serum, plus 1% penicillin/streptomycin. MDA-MB-231 cells were grown under the same conditions as the MCF-7 cells but without nonessential amino acids.
Fluorescence micrographs were acquired using an Axiovert 200 M inverted microscope (ZEISS) with fluorescence filter sets for DAPI/Hoechst, fluorescein/fluo-3, and rhodamine. Ultraviolet absorption of the different oligonucleotides was determined at 260 nm on a UVIKON 933 double beam UV/vis spectrophotometer (Kontron Instruments, United Kingdom).
Synthesis of NHS-DNA Conjugates
For cell adhesion studies, three complementary oligonucleotide pairs were designed such that they were identical in overall base composition, but differed in their position along the sequence. Each sequence pair was also calculated to possess comparable melting temperatures (55 °C) and minimal secondary structures. The sequence identities were as follows:
C1: 5′-GTA ACG ATC CAG CTG TCA CT-3′
M1: 5′-AGT GAC AGC TGG ATC GTT AC-3′
C2: 5′-TCA TAC GAC TCA CTC TAG GG-3′
M2: 5′-CCC TAG AGT GAG TCG TAT GA-3′
C3: 5′-ACT GAC TGA CTG ACT GAC TG-3′
M3: 5′-CAG TCA GTC AGT CAG TCA GT-3′
The oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA) with thiol groups installed at the 5′-end. Aqueous samples (2 mg in 80 µL) were combined with 320 µL of 10 mM tris(2-carboxyethyl)phosphine (TCEP) and 400 µL of 1× TE buffer (10 mM Tris with 1 mM EDTA, brought to pH 7.5 with HCl) and stored frozen at −20 °C until use.
Succinimidyl-[(N-maleimidopropionamido)-hexaethyleneglycol] ester (NHS-PEO6-maleimide) was purchased from Pierce. A stock solution was prepared by dissolving 5 mg of NHS-PEO6-maleimide in 1 mL of dimethyl sulfoxide (DMSO, Sigma). Aliquots of this solution (20 µL each) were stored at −20 °C until use.
DNA modification was achieved by passing a thawed solution of 5′-thiol single stranded DNA (ssDNA) (30 µL, 0.39 mM) through a NAP-5 size-exclusion column (GE Healthcare). The eluent was then exposed to 20 µL of the NHS-PEO6-maleimide solution at room temperature for 10 min. The reaction was then purified by passing it through a second NAP-5 column that was pre-equilibrated with phosphate buffered saline (PBS) solution (pH 7.2). The concentration of DNA in the column eluent was verified using UV-vis spectroscopy. The resulting solution was then applied to samples of live cells (see below).
To confirm the nature of the modification chemistry, models of the oligonucleotide conjugates were prepared and characterized. To do this, 0.5 mL of dimethylformamide (DMF) was saturated with 6-amino-N-(4-aminophenethyl)hexanamide and added to 1 mL of the reaction solution obtained after NAP-5 purification. After 30 min of incubation at room temperature, the oligonucleotide conjugates were analyzed using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). Observed masses were within 0.090% of expected values.
Modification of Live Cells and Quantification of Attached DNA Molecules
Immediately prior to modification, a sample of 5 × 106 Jurkat cells was washed with PBS buffer three times to remove any proteins from the culture medium. The cells were then exposed to solutions of NHS-DNA (C3 sequence, 3–54 µM final concentrations) for 30 min at room temperature. After isolation via centrifugation, the cells were returned to the culture medium or labeled with fluorescent DNA complements as described in the next paragraph.
In order to quantify the number of surface DNA molecules, portions of the modified cells were incubated with 10 µL solutions of fluorescein isothiocyanate (FITC)-labeled complementary DNA strands at 0 °C for 30 min. The cells were then washed with PBS solution and resuspended in PBS containing 1% FBS prior to analysis. The cells were then analyzed by flow cytometry. Fluorescence measurements were calibrated using fluorescent beads of known fluorophore density (QuantumTM FITC MESF, Bangs Laboratories, Inc.). Fluorescence measurements were compared to those corresponding to control cells lacking DNA modification and cells that had been reacted with mismatch DNA sequences.
General Protocol for the Attachment of DNA Strands to Cells and for Their Immobilization onto DNA-Printed Surfaces
Immediately prior to modification, a sample of 5 × 106 Jurkat cells was washed with PBS buffer three times to remove any proteins from the culture medium. The cell suspension was then reacted with 1 mL of NHS-DNA (11.7 µM) solution synthesized and purified from 30 µL of 5′-thiol ssDNA (C2 sequence). The mixture was allowed to react at room temperature for 30 min and was then washed three times with PBS containing 1% FBS. The cells were then resuspended in 0.5 mL of PBS containing 1% FBS.
To print the glass surfaces, a 20 µM solution of 5′ amine functionalized ssDNA in 3× saline sodium citrate buffer (SSC: 45 mM sodium citrate, 450 mM NaCl, pH 7.0) was used for sample preparation. DNA solutions were deposited onto aldehyde-functionalized glass slides (SCHOTT Nexterion, Louisville, KY) by manual pipetting or by using a robotic microarray printing system at the UC Berkeley Functional Genomic Laboratory. Spotted DNA was immobilized, and the slides were passivated according to the manufacturer′s protocol. After printing, the slides were dried under a stream of N2 and stored in the dark under a dry atmosphere. Patterned slides were typically used within 1 month.
Micropatterning of the glass slides was achieved using photo-lithography in conjunction with an aluminum lift-off technique,17 and will be described in full detail elsewhere.
For studies with one cell type, all cells were labeled with the C2 sequence. Slides were patterned with complementary DNA sequence M2 unless otherwise noted. Solutions of DNA-modified cells were introduced onto each surface and incubated for 3–5 min without agitation. The devices were then washed twice with PBS containing 1%FBS. Replicate data sets were collected by selecting three device regions at random before washing. Each location was photographed, washed, and then visualized again.
Evaluation of Cell Viability
Jurkat cells coated with the C2 strand were seeded in a 1 mL Petri dish with normal growth media, and M2 strand DNA was added into the solution to a concentration of 2 µM. Unmodified Jurkat cells were cultured under identical conditions as a control Cells were counted in each of the four samples using a hemocytometer at 24, 48, and 72 h Cell viability was monitored by adding trypan blue.
The extent of apoptosis of surface bound cells was determined by annexin V/propidium iodide staining (BD Biosciences). After immobilization on the slides by DNA, the cells were incubated in normal media at 37 °C for 24 h. A sample of unbound Jurkat cells (lacking surface DNA strands) was incubated under the same conditions as a control. A solution consisting of 900 µL of 1× binding buffer, 30 µL of the annexin V-FITC stock solution, and 30 µL of the PI stock solution was prepared. After 24 h, 100 µL of this solution was applied to the slides for 15min at room temperature. The cells were imaged by fluorescence microscopy, and the number of nonviable cells were counted after 1 h.
Immobilization of Adherent Cell Lines on Patterned Surfaces
Two breast cancer cell lines, MCF-7 and MDA-MB-231, were obtained from the American Type Culture Collection (ATCC). The cells were detached from culture plates with 1 mM ethylenediaminetetraacetate (EDTA) without any trypsin, and the cell solutions were washed with PBS three times. A 5 mL portion of the cell solution (1 × 106 cell/mL) was reacted with 1 mL of NHS-DNA solution synthesized from 30 µL of 5′-thiol ssDNA (C2 sequence) as described above. The mixture was allowed to react at room temperature for 30 min and was then washed three times with PBS containing 1%FBS. The cells were then resuspended in 0.5 mL of PBS containing 1%FBS. The cell solution was introduced onto glass slides patterned with complementary DNA sequence M2, and the samples were incubated for 5min.The slides were then washed two times with PBS containing 1% FBS. After immobilization onto slides via DNA hybridization, the cells were incubated in their normal media and observed for 36 h. Replicate data sets were collected by photographing three different surface regions at 12 h intervals.
Confirmation of the Sequence-Specificity of Cell Immobilization
DNA-modified Jurkat cells and MDA cells were prepared by incubating each cell population with NHS-DNA (sequence C2 or C1, respectively) in PBS for 30 min as described above To facilitate visual differentiation of the cells, the cytosol of each population was labeled with either CellTracker Blue or CellTracker Green live cell stains. After rinsing, equal amounts of each population were mixed, introduced onto microspotted DNA microarrays bearing either sequence M2 or M1 (constructed as above), and incubated for 5 min. The microarray was then washed twice with PBS containing 1% FBS and observed under a fluorescence microscope.
Immobilization of Human Red Blood Cells
Fresh samples of red blood cells were obtained froma blood sample of a healthy human and stored in1%citric acid solution at room temperature. Cells were used within 1 h. The cell solution was washed three times with PBS and was then incubated in the NHS-ssDNA solution for 30min to allow modification of cell surfaces. The cell suspension was then washed three times with 1% FBS/PBS solution before being applied to glass slides bearing the complementary ssDNA strands. After cell attachment, the glass slides were washed with 1% FBS/PBS to remove any unbound cells and viewed under an optical microscope. Cells were incubated in 1% FBS/PBS after immobilization, and their viability was examined after 3 h using trypan blue staining.
Patterning of Primary CD4+ T-Cells and IL-2 Production Assay
Primary CD4+ T-cells (obtained in collaboration with Jay T. Groves’ laboratory, UC Berkeley) were harvested from mice and grown under reported conditions18 before use. The primary T-cells were then modified using the NHS-DNA protocol and exposed to different DNA patterns printed by spotting or by using photolithography, as described above. The glass slides with DNA-immobilized cells were washed with 1% FBS/PBS to remove any unbound cells and viewed under a microscope.
The IL-2 production of primary T-cells immobilized with DNA duplexes was examined via enzyme-linked immunoadsorbent assay (ELISA). A population of 2 × 105 primary T-cells was modified with DNA strands and immobilized on a series of slides (1 cm2) bearing the complementary sequence. These samples were then divided into three portions. The first sample was incubated in normal T-cell growth media without any additional reagents. The second sample was treated with PHA (1 µg/mL) and phorbol 12-myristate 13-acetate (PMA, 50 ng/mL). The third sample was treated with concanavalin A (ConA, 1 µg/mL), PMA (50 ng/mL), and cyclosporin A (CSA, µg/mL). Analogous samples of free T-cells with no surface DNA were prepared as controls. All the cell samples were incubated at 37 °C for 20 h and then centrifuged. Portions of the culture media (1 mL) were withdrawn from each population of cells and tested for IL-2 production using a Mouse Interleukin-2 ELISA test kit (Thermo Scientific).
Patterning of Primary Myoblasts
Primary myoblasts (obtained in collaboration with Randall Lee’s laboratory, UCSF) were harvested from mice and purified according to a published protocol.19 Normal cell growth was achieved in Ham′s F-10 media (Invitrogen) with 10% (v/v) fetal bovine serum (FBS, HyClone), 1%bGF (Invitrogen), and 1% penicillin/streptomycin (P/S, Sigma). Immediately before surface modification, the cells were detached with 1 mM EDTA without any trypsin. The resulting cell suspensions were rinsed with PBS three times. A 5 mL portion of the cell solution (1 × 106 cell/mL) was reacted with 1 mL of NHS-DNA solution. The mixture was allowed to react at room temperature for 30 min, and the cells were then washed three times with PBS containing 1% FBS. Surfaces were patterned with the complementary DNA sequence through spotting or photolithography and then incubated with PBS containing 1% FBS at room temperature for 1 h. The cell solution was introduced onto the slides and incubated for 5 min. The devices were then washed three times with PBS containing 1% FBS. After immobilization, the cells were incubated in growth media or fusion media (DMEM (Invitrogen) containing 5% horse serum and 1% penicillin/streptomycin (P/S, Sigma)) at 37 °C for 14 days. The unbound myoblasts were cultured under identical conditions as a control. Images and movies of all cell samples were recorded every 24 h.
Results
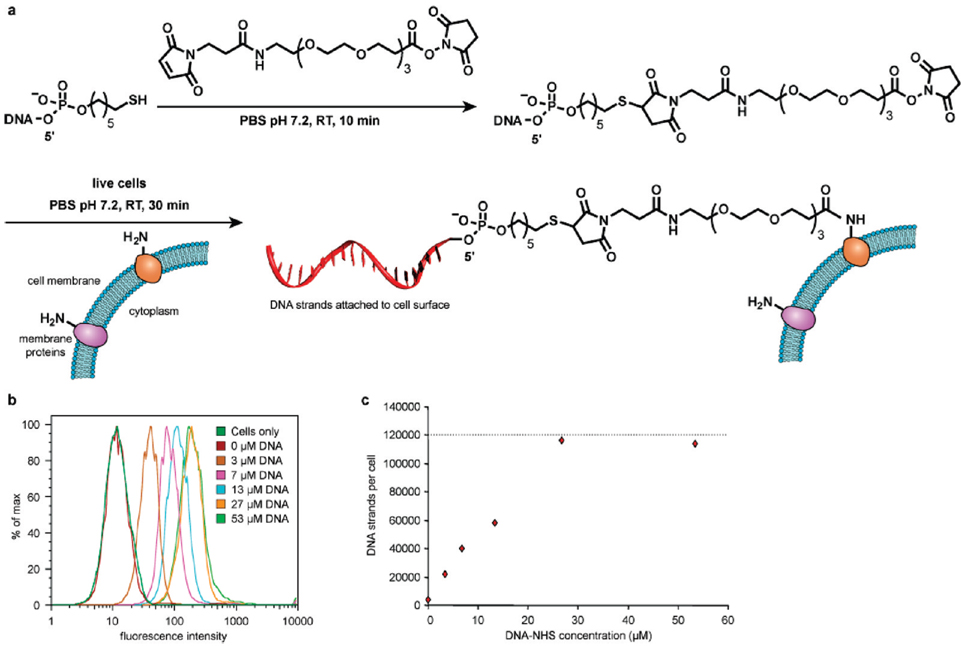
Our general strategy for the direct modification of cell surfaces with DNA strands targeted amino groups present on membrane proteins (Figure 1a). To prepare an appropriately reactive DNA conjugate, commercially available 5′-thiol modified DNA was conjugated to an NHS-PEG-maleimide cross-linker at pH7.2 for 10 min, yielding an NHS-DNA conjugate. This reagent was immediately incubated with live cells in PBS at room temperature for 30 min, and the cells were then applied to patterned surfaces for study. The NHS-DNA conjugates were characterized using MALDI-TOF MS, and model studies conducted by exposing the conjugate to a small molecule amine confirmed the formation of the expected amide product (Supporting Information Figure S1). Based on the mass spectrometry results, there was no thiol-substituted DNA remaining in the final purified product. The concentration of the NHS-DNA product was determined using UV absorption.
Figure 1.
Covalent attachment of ssDNA to cell surfaces. (a) Thiolated single-stranded DNA was first reacted with NHS-PEG-maleimide in PBS at room temperature to form the NHS-DNA conjugate. This solution was then incubated with suspensions of live cells in PBS at room temperature for 30 min. After attachment of the DNA strands, the cells were returned to culture media. (b,c) Jurkat cells were exposed to NHS-DNA solutions of varying concentrations as described in (a). The fluorescent strand complement was then added, and the level of cell modification was quantified using flow cytometry. Up to 120 000 DNA strands could be installed on each cell.
To verify the ability of the DNA-NHS conjugates to react with lysines on the cell surfaces, human T-cell lymphocytes (Jurkat cell line) were used as a model because they are non-adherent and thus any observed binding could be attributed to DNA hybridization. Jurkat cells were first washed with PBS buffer to remove serum proteins that would compete for amine labeling. The cells were then reacted with the NHS-DNA conjugates for 30 min at room temperature to conjugate ssDNA to the cell surface. In order to quantify the amount of DNA on their surfaces, the modified Jurkat cells were incubated with complementary ssDNA strands labeled with FITC. Flow cytometry was then used to quantify the number of DNA molecules by comparing the signal to that obtained with fluorescent beads of known fluorophore density (Bangs Laboratories, Inc., Fishers, IN).20 By varying the concentration of the NHS-ssDNA conjugate from 3 to 54 µM, up to 120 000 DNA strands could be introduced on each cell (Figure 1b,c).Control experiments carried out with unmodified cells and cells modified with a mismatched DNA sequence showed the same fluorescence intensity as native Jurkat cells with no added fluorophore.
Glass slides were prepared for cell adhesion studies as previously described.13 Upon exposure to Jurkat cells labeled with NHS-DNA, rapid cell capture was observed. The capture efficiency was comparable to that observed in previous studies, with close-packed arrays forming in as little as 3 min. No cells adhered to regions of the slide that lacked DNA or to spots prepared with the incorrect sequence (Figure 2a).
Figure 2.
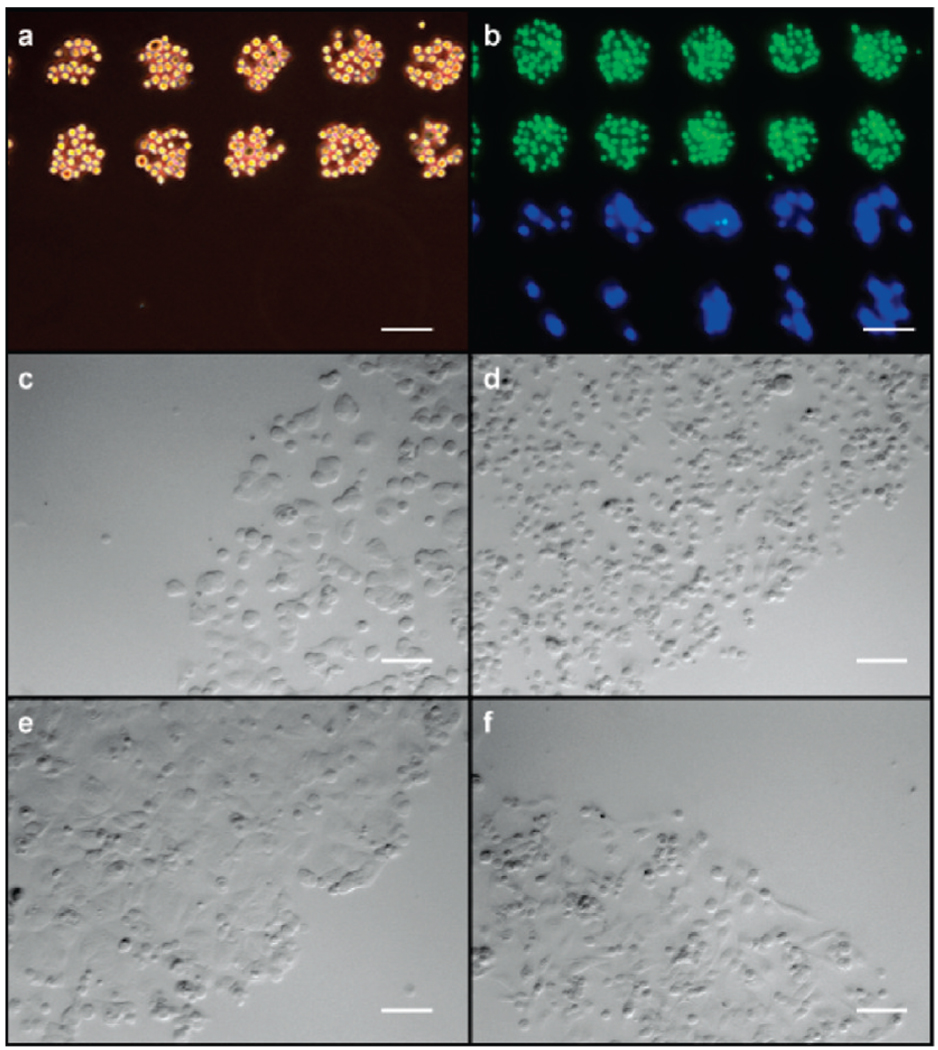
Immobilization of cells in a DNA sequence-specific manner. (a) Cells bearing DNA sequence C2 bound to complementary (sequence M2) spots on a DNA microarray (spot size = 60 µm). Neighboring spots with noncomplementary sequenceM1 remained unoccupied. (b) The same microarray substrate was exposed to a mixed suspension of Jurkat and MDA cells bearing sequences C2 and C1, respectively. Jurkat cells were stained with CellTracker Green, and MDA cells were stained with CellTracker Blue. (c) MCF-7 and (d) MDA cells also bound to DNA-coated surfaces in a rapid, stable, and sequence-specific manner. Clear delineation between the DNA-coated and uncoated regions was observed. Phase contrast images are shown after 2 h of incubation. (e) MCF-7 and (f) MDA cells had spread and proliferated after 36 h but were still confined to the DNA-printed area. In each panel, the scale bar represents 100 µm.
The effects of the synthetic DNA molecules on cell viability were assessed using two different methods. First, suspensions of DNA-coated cells were supplemented with the complementary DNA sequence and the growth curves were monitored over a 3 day period (Supporting Information Figure S2a).No changes in cell growth were observed relative to unmodified cells. In a second assay, the viability of DNA-bound cells was determined after 24 h using FITC-labeled annexin V and propidium iodide solutions.15 For the DNA-immobilized cells, the low percentages of apoptotic and necrotic cells were similar to those obtained for unmodified cells maintained in the same culture media (Supporting Information Figure S2b).
To test the compatibility of the platform with adherent cells, two breast cancer cell lines (MCF-7 and MDA-MB-231) were investigated. Cultured MCF-7 and MDA cells were first treated with EDTA to detach them from the culture plate surface, and they were then modified with NHS-DNA as described above. Trypsin was not used to detach them because it was found to prevent DNA conjugation. This is presumably because it reduces the overall number of available amino groups on the cell surfaces. The DNA-modified MCF-7 and MDA cells were then applied to glass slides spotted with complementary sequences, and the cells were returned to culture conditions (growth media at 37 °C with an atmosphere of 5% CO2). Phase contrast images were acquired 2 and 36 h after washing (Figure 2c–f). At early time points (2 h), the MCF-7 and MDA cells appear morphologically identical to unmodified cells cultured on treated polystyrene plates. However, after 36 h, the MCF-7 and MDA cells had adhered and spread at the site of their initial binding. Their shapes and morphology were identical to the native cells cultured on Petri dishes. The initial cellular patterns remained, validating both the utility of this system for adherent cells and the ability to interrogate such cell populations grown in defined 2D arrays over sustained periods. Cell proliferation was observed, confirming the viability of the adhered cells (Supporting Information Figure S3).
With the successful DNA-programmed capture of both Jurkat and breast cancer cells, we sought to demonstrate the parallel self-assembly of patterns of these cells from a mixed population. To this end, a spotted microarray of DNA was produced with alternating sequences M1 and M2. MDA and Jurkat cells were then labeled with complementary sequences C1 and C2, respectively. The labeled cells were mixed in equal proportion, and the mixture was incubated on the array for 30 min. The array was then rinsed and visualized, revealing the ordered multitype array shown in Figure 2b. Visual inspection of cell morphology suggested that bound cells remained viable.
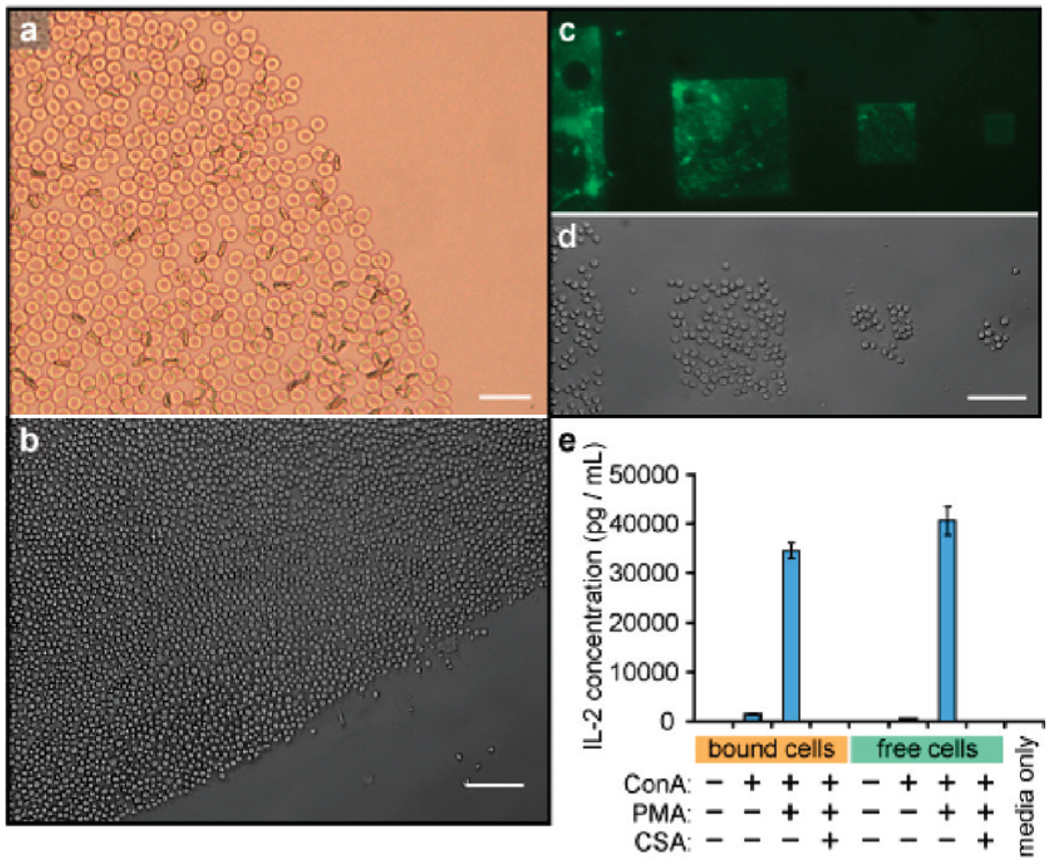
We next applied this method to the capture of primary cells taken directly from a living organism. These cells are often difficult to culture for long periods and thus are incompatible with many surface patterning techniques. As a first example, this technique was applied to capture red blood cells obtained from a healthy human donor. DNA strands were introduced onto the cell surfaces as described above, and the cells were then immobilized on complementary DNA spots (Figure 3a). Although red blood cells are inherently nonviable, assays using annexin-V and trypan blue indicated that the cell membranes remained intact after surface binding.21
Figure 3.
Direct DNA modification and capture of primary cells. (a) Human red blood cells were bound in the same manner as Jurkat cells on a DNA spot, and appeared to be morphologically identical immediately after binding. Trypan blue staining indicated that the membranes remained intact. The scale bar represents 25 µm. (b) DNA-coated mouse CD4+ helper T-cells were bound by spots coated with complementary DNA. After 3 min of exposure, a clear boundary can be seen between the printed and unprinted regions of the slide. (c) Microscale DNA patterns made by photolithography and microfabrication. Fluorescein conjugated ssDNA strands were patterned on the substrate to allow visualization. (d) Mouse primary T-cells were captured on the same DNA patterns. (e) IL-2 production of DNA-immobilized T-cells and free T-cells, as determined by ELISA. ConA = concanavalin A; PMA = phorbolmeristyl acetate;CSA = cyclosporin A. In (b) and (d), the scale bars represent 100 µm.
As a second target, primary CD4+ T-cells were harvested from mice and grown using literature protocols.18 After surface modification with NHS-DNA, the cells were successfully immobilized on glass surfaces bearing patterns of complementary DNA generated using photolithography (Figure 3c,d). Successful cell capture was observed for a variety of different patterns (Figure 3b–d). Both the DNA-immobilized T-cells and free T-cells were then cultured in normal growth media for 20 h, and the interleukin-2 (IL-2) production levels of the cell samples were examined using ELISA. Both DNA-bound T-cells and free T-cells produced a very low level of IL-2 in normal media, indicating that the adhesion process did not activate T-cell signaling pathways. When both populations were treated with phorbol 12-myristate 13-acetate (PMA) and concanavalin A (ConA) to costimulate IL-2 production, similar increases in IL-2 production were observed. This increase could be suppressed by adding cyclosporin (CSA) in both cases (Figure 3e).22
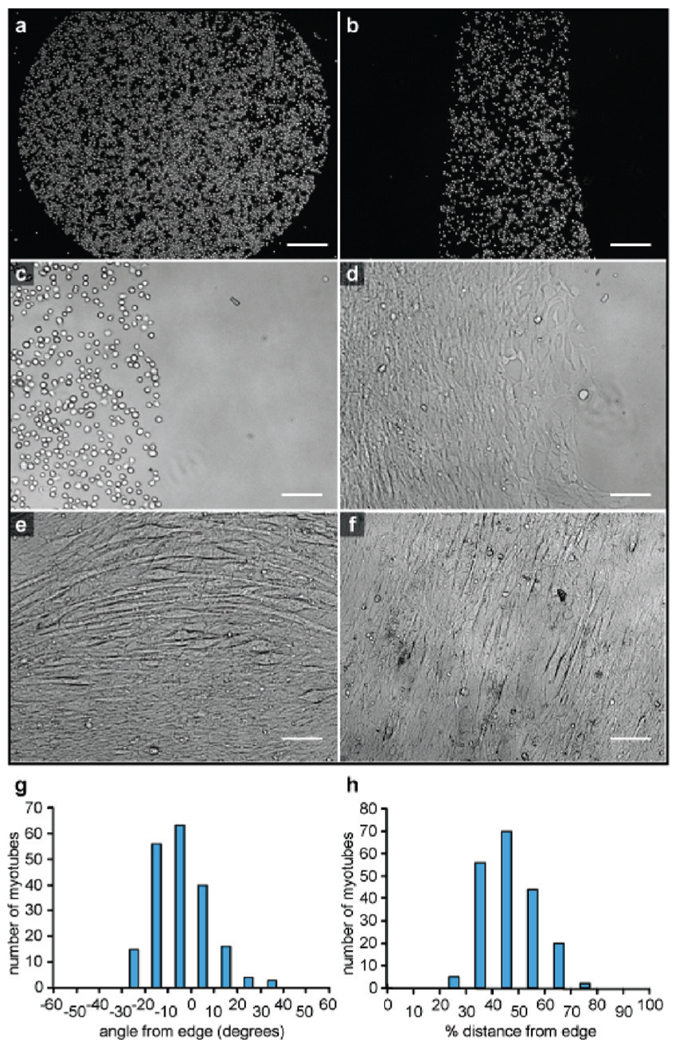
Finally, we used the same NHS-DNA labeling method to pattern primary myoblast cells on DNA-coated surfaces (Figure 4a–c).After initially binding to the oligonucleotide-coated regions, the cells were able to spread but retained the original pattern. They were maintained in the undifferentiated state for a period of 1 day. Following a switch to fusion media, differentiation into skeletal and heart muscle cells occurred. After incubation for 5 days, myotube formation could be observed in some areas (Figure 4d–f), and after 5 to 7 days the myotubes began to contract spontaneously (Supporting Information Movie 1). Similar myoblast growth and myotube formation were observed on collagen-coated surfaces (Supporting Information Figure S4 and Figure S5)19 but with less control over the patterns that were formed.
Figure 4.
Capture and differentiation of primary myoblast cells. (a,b) DNA patterns on glass slides dictate the areas in which cells are bound. (c) Myoblast cells show no signs of differentiation immediately after capture (shown) or after 1 day when kept in growth media. (d) Myotubes form upon addition of differentiation media. The photo was taken 5 days after the switch was made. (e) After 6 days of incubation in differentiation media, circularly patterned myoblasts form arced myotubes that are aligned with the edge. (f) After 6 days, myocytes in rectangular arrangements form myotubes that are aligned with the long axis of the patterns. For the linear patterns, the majority of the myocytes (g) align to within 20° of the pattern boundary angle and (h) are found halfway between the edges. The scale bars represent 250 µmin (a) and (b) and 100 µm in (c–f).
By patterning myoblasts in different 2D arrangements, we were able to study the influence of cell patterns on myotube formation. We noted that myotube formation tracked with the nearest edge of the DNA patterns, leading either to arced tubes for myocytes patterned in circles (Figure 4e) or aligned linear bundles for rectangular arrangements (Figure 4f). After differentiation, myotube alignments were analyzed using ImageJ (Supporting Information Figure S6). The majority of the myotubes were found in the middle of the patterned areas, and for linear patterns they were predominantly aligned to within 20° of the edge angles (Figure 4e,f).
Discussion
The most significant finding of these studies is the fact that NHS-DNA can modify a wide range of different cells in under 1 h. The required reagents can be prepared from commercially available materials in two steps with minimal purification and are therefore accessible to almost any laboratory. Unlike our previous labeling method, which required the modification of azido sialic acid residues on the cell surfaces, this new technique does not require prolonged cell culture and is thus more suitable for modifying primary cells. It is also compatible with cells that are not actively adding new glycoproteins to their membranes, such as erythrocytes, which would be expected to show little-to-no incorporation of artificial functional groups into sialic acid residues. As morphological changes in red blood cells are associated with sickle-cell anemia and spherocytosis,23 the ability to immobilize them into arrays could facilitate diagnosis and render them compatible with laboratory-on-a-chip technologies. While it has been previously shown that red blood cells can be immobilized using phytohemagglutinin,24 our DNA-based method leads to more densely packed arrays in significantly less time and could potentially be used to isolate single cells.
The results of the IL-2 production assay indicate that DNA-bound T-cells are not activated by virtue of surface attachment, a result that is difficult to achieve using lectins or antibody-based methods.25–28 However, upon the addition of ConA and PMA, robust activation was observed with similar levels of IL-2 production to that of free cells in solution. In combination with previous studies of cell viability after DNA-based adhesion,15 we take these results to indicate that minimal perturbation of the cell function occurs as a result of the DNA attachment process. While any adhesion method would be expected to have some influence on cellular behavior, this method stands as the least perturbing method available for the anchoring of cells to surfaces, and it is readily compatible with DNA printing methodologies.
The ability to create defined patterns comprising multiple cell types could provide a powerful tool for the study of cell-to-cell communication and the generation of tissue models. In the latter context, we have begun to pattern cells that exhibit collective behavior, with the long-term goal of learning how spatial arrangements influence differentiation and growth. We chose myoblasts for this purpose, as they have the ability to differentiate into cardiac muscle cells or into skeletal muscle cells.29–31 Using the NHS-DNA modification technique, we found that these cells can be patterned on surfaces with similarly high efficiency. Upon addition of differentiation media, many of the cells formed myotubes that were capable of spontaneous contraction.
Our preliminary observations in these experiments have shown that the underlying pattern can influence myotube formation and alignment significantly. The highest density of myotube formation was observed in the center of the patterned regions. There is also a clear alignment of the tubes with the shape of the patterned region, with the majority lying within 20° of the pattern edge. This occurs for both rectangular and circular patterns. Although other groups have previously reported that myotubes can be aligned using electric fields,32 spun nylon 6/6 fibrils,33 micropatterned glass,34 16 µm wide laminin strips,35 and proteins deposited using inkjet printing techniques,36 this is one of the first demonstrations of the spontaneous alignment of myotubes through long-range patterning effects. Our ability to prepare complex spatial arrangements of the cells provides an effective tool with which to study this behavior, and is currently being used to determine the sensing mechanism that generates these ordered patterns.
Conclusion
We have developed an efficient new method that allows the direct modification of the cell surface with DNA strands using NHS-DNA conjugates. The NHS-DNA conjugates react with lysine residues of cell surface proteins to conjugate single-stranded DNA to the cell surface. This method allows us to capture many new types of cells, including primary cells, at specific surface locations in a sequence-dependent fashion. This strategy allows complex networks of living cells to be created through self-assembly. It should be noted that virtually all of the state-of-the-art methods for printing biomolecules on surfaces could be used to prepare the DNA patterns, and thus, we anticipate that the adhesion method reported herein could be adapted readily to a number of experimental formats. In combination with laboratory-on-a-chip RT-PCR analysis,12 it also provides a valuable tool for the study of cell behavior and differentiation mechanisms. Currently, we are using this method to understand the effects of spatial patterning on the behavior of myocytes in more detail. We are also combining our method with microfabrication techniques to integrate living cells with sensing and stimulatory electrodes.
Supplementary Material
Acknowledgment
This work was supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC03-76SF00098. This investigation was partially funded by a grant to Z.J.G. by the Jane Coffin Childs Memorial Fund for Medical Research. The authors wish to thank Prof. Randall Lee (UCSF Department of Cardiology) for myoblast cells and for many helpful discussions. Primary T-cells harvested from mice were obtained in collaboration with Nina Hartman and the laboratory of Prof. Jay Groves (UC Berkeley Chemistry).
Footnotes
Supporting Information Available: Supplemental figures, movie, and characterization data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Falconnet D, Csucs G, Grandin HM, Textor M. Biomaterials. 2006;27:3044–3063. doi: 10.1016/j.biomaterials.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 2.Lee H, Purdon AM, Westervelt RM. Appl. Phys. Lett. 2004;85:1063–1065. [Google Scholar]
- 3.Li Y, Yuan B, Ji H, Han D, Chen SQ, Tian F, Jiang XY. Angew. Chem., Int. Edit. 2007;46:1094–1096. doi: 10.1002/anie.200603844. [DOI] [PubMed] [Google Scholar]
- 4.Liu VA, Bhatia SN. Biomed. Microdevices. 2002;4:257–266. [Google Scholar]
- 5.Rosenthal A, Macdonald A, Voldman J. Biomaterials. 2007;28:3208–3216. doi: 10.1016/j.biomaterials.2007.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proc. Natl. Acad. Sci. U.S.A. 2006;103:2480–2487. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts C, Chen CS, Mrksich M, Martichonok V, Ingber DE, Whitesides GM. J. Am. Chem. Soc. 1998;120:6548–6555. [Google Scholar]
- 8.Du XP, Plow EF, Frelinger AL, Otoole TE, Loftus JC, Ginsberg MH. Cell. 1991;65:409–416. doi: 10.1016/0092-8674(91)90458-b. [DOI] [PubMed] [Google Scholar]
- 9.Xiong JP, Stehle T, Zhang RG, Joachimiak A, Frech M, Goodman SL, Aranout MA. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 10.Chandra RA, Douglas ES, Mathies RA, Bertozzi CR, Francis MB. Angew. Chem., Int. Edit. 2006;45:896–901. doi: 10.1002/anie.200502421. [DOI] [PubMed] [Google Scholar]
- 11.Bailey RC, Kwong GA, Radu CG, Witte ON, Heath JR. J. Am. Chem. Soc. 2007;129:1959–1967. doi: 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borisenko GG, Zaitseva MA, Chuvilin AN, Pozmogova GE. Nucleic Acids Res. 2009;37:e28. doi: 10.1093/nar/gkn1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Douglas ES, Chandra RA, Bertozzi CR, Mathies RA, Francis MB. Lab Chip. 2007;7:1442–1448. doi: 10.1039/b708666k. [DOI] [PubMed] [Google Scholar]
- 14.Toriello NM, Douglas ES, Thaitrong N, Hsiao SC, Francis MB, Bertozzi CR, Mathies RA. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20173–20178. doi: 10.1073/pnas.0806355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsiao SC, Crow AK, Lam WA, Bertozzi CR, Fletcher DA, Francis MB. Angew. Chem., Int. Edit. 2008;47:8473–8477. doi: 10.1002/anie.200802525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxon E, Bertozzi CR. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 17.Jackson BL, Groves JT. Langmuir. 2007;23:2052–2057. doi: 10.1021/la062667q. [DOI] [PubMed] [Google Scholar]
- 18.DeMond AL, Mossman KD, Starr T, Dustin ML, Groves JT. Biophys. J. 2008;94:3286–3292. doi: 10.1529/biophysj.107.119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang NF, Patel S, Thakar RG, Wu J, Hsiao BS, Chu B, Lee RJ, Li S. Nano Lett. 2006;6:537–542. doi: 10.1021/nl060060o. [DOI] [PubMed] [Google Scholar]
- 20.Vogt RF, Cross GD, Henderson LO, Phillips DL. Cytometry. 1989;10:294–302. doi: 10.1002/cyto.990100308. [DOI] [PubMed] [Google Scholar]
- 21.Fonseca AM, Porto G, Uchida K, Arosa FA. Blood. 2001;97:3152–3160. doi: 10.1182/blood.v97.10.3152. [DOI] [PubMed] [Google Scholar]
- 22.Elliott JF, Lin YA, Mizel SB, Bleackley RC, Harnish DG, Paetkau V. Science. 1984;226:1439–1441. doi: 10.1126/science.6334364. [DOI] [PubMed] [Google Scholar]
- 23.Konoteya Fi. Arch. Int. Med. 1974;133:611–619. [Google Scholar]
- 24.Shinozuka T, Takei S, Yanagida J, Watanabe H, Ohkuma S. Blut. 1988;57:117–123. doi: 10.1007/BF00320150. [DOI] [PubMed] [Google Scholar]
- 25.Barten MJ, Tarnok A, Garbade J, Bittner HB, Dhein S, Mohr FW, Gummert JF. Cell Proliferation. 2007;40:50–63. doi: 10.1111/j.1365-2184.2007.00413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minami Y, Kono T, Miyazaki T, Taniguchi T. Annu. Rev. Immunol. 1993;11:245–268. doi: 10.1146/annurev.iy.11.040193.001333. [DOI] [PubMed] [Google Scholar]
- 27.Roosnek EE, Brouwer MC, Aarden LA. Eur. J. Immunol. 1985;15:652–656. doi: 10.1002/eji.1830150703. [DOI] [PubMed] [Google Scholar]
- 28.Stohl W, Hofman FM, Gray JD. J. Immunol. 1990;145:1078–1087. [PubMed] [Google Scholar]
- 29.Jankowski RJ, Deasy BM, Huard J. Gene Ther. 2002;9:642–647. doi: 10.1038/sj.gt.3301719. [DOI] [PubMed] [Google Scholar]
- 30.Partridge TA. Muscle Nerve. 1991;14:197–212. doi: 10.1002/mus.880140302. [DOI] [PubMed] [Google Scholar]
- 31.Wollert KC, Drexler H. Circ. Res. 2005;96:151–163. doi: 10.1161/01.RES.0000155333.69009.63. [DOI] [PubMed] [Google Scholar]
- 32.McCaig CD, Dover PJ. Biol. Bull. 1989;176:140–144. doi: 10.2307/1541664. [DOI] [PubMed] [Google Scholar]
- 33.Huber A, Pickett A, Shakesheff KM. Eur. Cells Mater. 2007;14:56–63. doi: 10.22203/ecm.v014a06. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto DL, Csikasz RI, Li Y, Sharma G, Hjort K, Karlsson R, Bengtsson T. J. Histochem. Cytochem. 2008;56:881–892. doi: 10.1369/jhc.2008.951228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark P, Coles D, Peckham M. Exp. Cell Res. 1997;230:275–283. doi: 10.1006/excr.1996.3429. [DOI] [PubMed] [Google Scholar]
- 36.Phillippi JA, Miller E, Weiss L, Huard J, Waggoner A, Campbell P. Stem Cells. 2008;26:127–134. doi: 10.1634/stemcells.2007-0520. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.