Abstract
Cyclin B1, an important cell cycle regulator, was up-regulated in lymphocytes of human immunodeficiency virus (HIV)-infected patients. However, the mechanism of cyclin B1 up-regulation and the effects of the up-regulation on the host cells remain unclear. Here, we show that HIV-encoded Tat protein regulates cyclin B1 levels in two different ways: first, Tat stimulates the transcription of cyclin B1, which increases cyclin B1 levels and promotes the cells apoptosis; and second, Tat stimulates polyubiquitination-mediated degradation of cyclin B1 through binding to the N-terminal of cyclin B1 (aa 61–129) that is just downstream of the D box, which prevents excessive levels of cyclin B1 in the cells. These results suggest that Tat-regulating cyclin B1 affects the status of HIV: Tat stimulates cyclin B1 expression to slow down the host cell cycle progress and to promote the host cell apoptosis, which might facilitate HIV release; Tat stimulates cyclin B1 degradation to prevent overaccumulation of cyclin B1, which might facilitate HIV replication. Taken together, our results reveal for the first time how HIV-Tat regulates cyclin B1 and keeps its balance in the cells.—Zhang, S.-M., Sun, Y., Fan, R., Xu, Q.-Z., Liu, X.-D., Zhang, X., Wang, Y., Zhou, P.-K. HIV-1 Tat regulates cyclin B1 by promoting both expression and degradation.
Keywords: apoptosis, ubiquitination
Cyclin B1 is an important cell cycle regulator, which associates with CDK1 (previously named CDC2) to control mitotic entry (1). Cyclin B1 levels rise in the S phase and peak at the G2/M boundary (2). Overexpression of cyclin B1 was found to be involved in sensitizing the cells to apoptosis induction (3,4,5). It was previously reported that unscheduled cyclin B1 expression and CDK1 activation were detected in T lymphocytes from human immunodeficiency virus (HIV)-infected patients (6, 7). In addition, high levels of cyclin B1 returned to normal after effective antiretroviral therapy, suggesting a correlation between viral replication and cyclin B1 levels (7). However, the mechanism of the up-regulated level of cyclin B1 in HIV-infected cells and the effects of the regulation of cyclin B1 in the infected cells remain unclear.
Tat (transactivator of transcription) is an 86- to 101-residue regulatory protein encoded by HIV, which plays an essential role in efficient transcription of viral genes and in viral replication through multifunctions (8). Tat protein can also be released from the infected host cells into the extracellular space through a noncanonic, Golgi-independent pathway of secretion and affects the bystander noninfected normal cells or tissues, resulting in direct or indirect damage on numerous cell types and different organs in host systems. Previously, we have shown that cyclin B1 is consistently overexpressed in TE671 cells transfected with HIV-1 Tat (9). In this study, we further explored the mechanism by which Tat affects the protein level of cyclin B1. We found that HIV-encoded Tat protein regulates cyclin B1 in two ways: by stimulating the transcription of cyclin B1 to increase the cyclin B1 levels, and by stimulating polyubiquitination of cyclin B1 through binding to the N-terminal of cyclin B1 to promote cyclin B1 degradation. Because high levels of cyclin B1 could result in apoptosis, these results indicate that Tat induces apoptosis at least partially by up-regulating cyclin B1. Our results provide the evidence that Tat regulates cyclin B1, which might be important for HIV function in host cells.
MATERIALS AND METHODS
Cell lines and cell synchronization
Human rhabdomyosarcoma TE671 cells were kindly provided by Dr. Lung-Ji Chang (University of Florida, Gainesville, FL, USA). TE671-Tat and TE671-pCI were established in our laboratory as described previously (9). HEK293T cells were purchased from American Type Culture Collection (Manassas, VA, USA). All cells were grown in Dulbecco’s modified Eagle’s medium (Life Technologies, Inc., Carlsbad, CA, USA) supplemented with 10% fetal bovine serum, 100 U/ml of penicillin, and 100 μg/ml of streptomycin. TE671-Tat and TE671-pCI were grown in the presence of 400 μg/ml G418. Cells were synchronized by double thymidine. The cells were treated with 2 mM thymidine for 16 h. After being released to normal growing medium for 8 h, the cells were treated with 2 mM thymidine for an additional 16 h. After being released with normal growing medium, the cells were treated with 100 ng/ml nocodazole for 17 h and the cells were arrested in mitosis.
Plasmids and siRNA
pCI-Tat was made as described in our previous study (9). The vectors encoding wild-type or mutant cyclin B1 were constructed based on the sequence of human cyclin B1 cDNA. PCR products were inserted into pCMV-HA, pGEX-5T, pEGFP-N1, pDsRed-N1, and pET22b plasmid, respectively. The vector pCycB(-287)-LUC containing 287 bp of upstream sequence of cyclin B1 and firefly luciferase gene was kindly provided by Dr. Karen S. Katula (University of North Carolina, Chapel Hill, NC, USA; ref. 10). The vectors encoding wild-type and mutant Tat were constructed based on the sequence of HIV Tat cDNA. PCR products were inserted into pGEX-5T. The plasmid encoding His-ubiquitin was kindly provided by Dr. Junjie Qian (Beijing Insitute of Radiation Medicine, Beijing, China). The point mutations of ubiquitin, His-UbK11R, His-UbK48R, and His-UbK63R, were generated by PCR based on the sequence of His-ubiquitin. The primer sequences are shown in Supplemental Table S1. siRNA (5′-AAGAAAUGUACCCUCCAGAAA-3′) targeting 776–796 of cyclin B1 (si-CB1) and negative control RNA (si-NC) (5′-UUCUCCGAACGUGUCACGUTT-3′) were synthesized by GenePharma (Shanghai, China).
Expression and purification of Tat protein
Escherichia coli Rosetta-gami B (DE3) were transformed with plasmid pET22b-Tat, pGEX5T-Tat, or pGEX5T-1 (control one). When the culture reached the midlog phase (OD600≈0.6), isopropyl-β-d-thiogalactopyranoside was added at the final concentration of 0.5 mM for 3 h to induce the expression of target protein. The cells were collected and lysed by an ultrasonic liquid processor (Sonxi, Shanghai, China) at 4°C for 30 min. The supernatant was collected by centrifugation at 12,000 g for 20 min. The supernatant was filtered with 0.45 μm membrane and loaded onto an Ni2+ affinity column preequilibrated with buffer B (50 mM sodium phosphate; 0.3 M NaCl, pH 8.0; and 10 mM imidazole). After being washed with buffer B, His-Tat was eluted with buffer C (50 mM sodium phosphate; 0.3 M NaCl, pH 8.0; and 250 mM imidazole).
GST pulldown
The cDNA of cyclin B1 or Tat inserted into the pGEX5T vector was expressed as GST fusion protein in E. coli BL21 (DE3). The glutathione (GST)-Sepharose beads were prewashed with TEN100 buffer (20 mM Tris, pH 7.4; 0.1 mM EDTA; and 100 mM NaCl) 4 times and equilibrated in TEN100 buffer. The GST or GST-fusion proteins were rotated with GST-Sepharose beads (Pharmacia, Piscataway, NJ, USA) at 4°C for 1 h (for immobilization). Whole-cell extract or purified protein with the immobilized GST or GST-fusion protein was rotated at 4°C for 1 h. The beads were washed 4 times with NETN buffer (0.5% Nonidet P-40; 0.1 mM EDTA; 20 mM Tris, pH 7.4; and 300 mM NaCl) for whole-cell extract, or with TEN buffer (20 mM Tris, pH 7.4; 0.1 mM EDTA; and 100 mM NaCl) for purified proteins. The protein-bound beads were boiled in Laemmli sample buffer, and the proteins were verified by Western blot.
Luciferase reporter assay
Plasmids pRL-TK and pMIR-reporter were kindly provided by Dr. Hui Zhang (Thomas Jefferson University, Philadelphia, PA, USA). Different deleted mutants of cyclin B1 promoter region (CB1P-381, CB1P-287, CB1P-180, or CB1P-90) were cloned into the upstream of Renilla luciferase in the plasmid pRL-TK. TE671 cells were cotransfected with pMIR-reporter (firefly luciferase) and pRL-TK with or without pCI-Tat. The luciferase reporter assay was carried out by using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA).
Confocal microscopy of expression plasmids
293T cells were grown on slide covers in tissue culture dishes. The cells were transfected with the plasmid by using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). At 48 h after transfection, the cells were rinsed with PBS and subjected to fixation using 4% (v/v) paraformaldehyde for 30 min. The cells were stained with DAPI. Images were viewed with a confocal fluorescent microscope connected to a Bio-Rad Radiance2100 laser scanner (Bio-Rad, Richmond, CA, USA).
Coimmunoprecipitation and antibodies used in Western blot
293T cells were transfected with pCMV-cyclin B1 in the presence or absence of pCI-Tat. The cells were collected at 48 h after transfection and lysed with the buffer (50 mM Tris-HCl; 150 mM NaCl; 1% Nonidet P-40; 0.1% SDS; 1 mM DTT, pH 7.5); and protease inhibitor cocktail (Roche, Indianapolis, IN, USA) at 4°C for 1 h. After centrifugation, the lysates with anti-HA antibody (Clontech Laboratories, Palo Alto, CA, USA) were rotated at 4°C for 1 h. Protein G/A-Sepharose beads (Amersham Biosciences, Piscataway, NJ, USA) were then added to the mixture and were rotated at 4°C for 1 h. The samples were washed and boiled in Laemmli buffer and analyzed by Western blots. Anti-Tip60 polyclonal antibody, anti-cyclin B1 monoclonal antibody, anti-CDK1 monoclonal antibody, anti-β-actin polyclonal antibody, and luminal analysis reagents were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-p-Cdk1-Tyr15 monoclonal antibody was purchased from Cell Signaling Technology (Danvers, MA, USA). Anti-His tag monoclonal antibody was purchased from Novagen (San Diego, CA, USA). Anti-HA polyclonal antibody was purchased from Clontech.
Polyubiquitination assay
293T cells were plated in 10-cm dishes and transfected with His-ubiquitin in the presence or absence of pCI-Tat or pCMV-cyclin B1. Cells were collected at 24 h after transfection and lysed in 1 ml of buffer A (6 M guanidium-HCl; 100 mM Na2HPO4/NaH2PO4, pH 8.0; and 10 mM imidazole). The lysates were briefly sonicated to reduce the viscosity. After centrifugation, extracts were placed in 50 ml of nickel-NTA-agarose beads (Qiagen, Valencia, CA, USA) at room temperature for 3 h. The beads were then washed twice in buffer A and twice in buffer B (50 mM Tris, pH 6.8, and 20 mM imidazole). The protein-bound beads were boiled with 2× Laemmli sample buffer supplemented with 20 mM imidazole, and the supernatants were analyzed by Western blotting.
Cell cycle analysis
293T or TE671 cells were transfected with HA-cyclin B1 or pCI-Tat, or treated with recombinant Tat protein (rTat). At different times, the cells were collected and fixed with 70% ethanol. The cells were washed 3 times with PBS and treated with RNase A (62 μg/ml) at 37°C for 30 min. The cells were stained with propidium iodide solution (40 μg/ml propidium iodide and 0.1% Triton X-100 in PBS buffer) and analyzed by flow cytometry.
RESULTS
Tat up-regulates cyclin B1 level
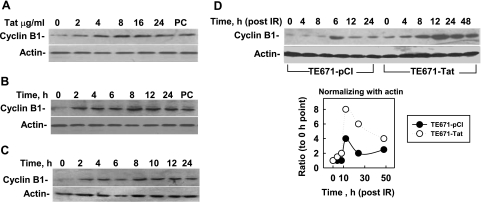
Cyclin B1 was found to consistently overexpress in peripheral CD4+ and CD8+ cells from HIV-infected patients, which is the main feature of HIV-associated cell-cycle deregulation (6, 7, 11, 12). We previously reported an increased level of cyclin B1 in stably Tat-expressing TE671-Tat cells (9). These results suggest that Tat up-regulates cyclin B1. To further study how Tat affects cyclin B1, we examined the effects of the rTat on cyclin B1 levels in cultured cells. The results showed that expression of cyclin B1 was stimulated by rTat even at a low concentration (2 μg/ml; Fig. 1A) and that the stimulating effect lasted at least 24 h (Fig. 1B). These results suggest that exogenous Tat could up-regulate cyclin B1 in a bystander manner. We then examined the effects of the medium containing secreted Tat from Tat-expressing TE671-Tat cells on cyclin B1 levels in non-Tat-expressing TE671-pCI cells. The results showed that after 24 h of incubation, the medium from TE671-Tat cell cultures stimulated cyclin B1 expression as strongly as rTat (Fig. 1C). Ionizing radiation (IR)-induced DNA damage could stimulate cyclin B1 accumulation. To study the effects of Tat on IR-induced cyclin B1 accumulation, we compared cyclin B1 levels between TE671-pCI cells and TE671-Tat cells at different time points after IR (4 Gy). The results showed that expression of cyclin B1 in TE671-pCI cells was elevated at 12 h after IR and then declined (Fig. 1D). However, expression of cyclin B1 in irradiated TE671-Tat cells was not only elevated earlier but was also increased at a higher level and maintained longer than in irradiated TE671-pCI cells (Fig. 1D). These results confirm that Tat up-regulates levels of cyclin B1. To investigate whether Tat-stimulating cyclin B1 was specific, we compared the levels of CDK1, WEE1, CDC25C, and the cyclin-B1-associated proteins between TE671-pCI and TE671-Tat cells after IR. The results indicate that there were no apparent differences of WEE1, CDC25C, and CDK1 between the 2 cell lines (Supplemental Fig. S1), except that IR-induced phosphorylation of CDK1 in TE671-Tat cells occurred earlier than in TE671-pCI. These results support that Tat specifically stimulates the high expression of cyclin B1.
Figure 1.
Tat protein stimulates cyclin B1 levels. Cyclin B1 expression levels were analyzed by Western blot. Actin was used as the internal loading control. A) rTat was added to the culture of TE671-pCI cells at different concentrations, and cells were collected 24 h later. Tat stably expressed cell line (TE671-Tat) was used as positive control (PC). B) rTat was added to the medium of TE671-pCI at a final concentration of 8 μg/ml, and cells were collected at different times. C) TE671-pCI cell cultures were changed with the medium in which Tat-expressing TE671-Tat cells were growing for 24 h (without G418), and cells were collected at different time points. D) Influence of irradiation and Tat expression on expression of cyclin B1. TE671-pCI and TE671-Tat cells were plated 12 h before exposure to 4 Gy irradiation. After irradiation, cells were collected at different times, and the expression level of cyclin B1 was detected.
Tat stimulates cyclin B1 expression at transcriptional level
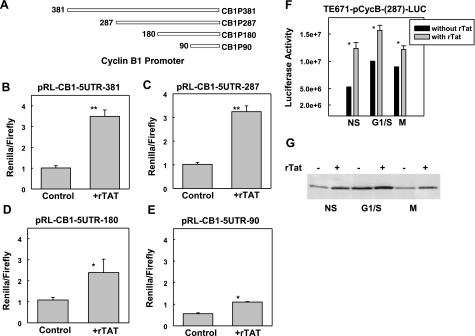
To study whether Tat stimulated high levels of cyclin B1 through transcription regulation, we examined the effects of Tat on the promoter of cyclin B1. We made different truncation mutants of cyclin B1 promoter that were cloned into the upstream of Renilla luciferase (Fig. 2A, CB1P-RL constructs). We cotransfected 293T cells with the luciferase reporter and pMIR-reporter with different CB1P-RL constructs, and then we treated the cells with rTat at 6 h after transfection for 24 h. We measured the activities of firefly and Renilla luciferase and calculated the ratio of Renilla/firefly luciferase activity. The results showed that rTat protein strongly simulated the expression of Renilla luciferase under the driving of cyclin B1 promoter with 381 or 287 bp (Fig. 2A–C). The activity stimulated by rTat could maintain until truncated to 90 bp (Fig. 2D, E). However, when cyclin B1 promoter was further shortened, the stimulated effects diminished (data not shown), suggesting that the 90 bp of cyclin B1 promoter is minimally required for Tat targeting. To exclude the possibility that Tat induced up-regulation of cyclin B1 as a consequence of Tat-induced G2/M-phase arrest, we transfected TE671 cells with pCycB- (287)-LUC, and then the cells were synchronized at the G1/S boundary by the double-thymidine blocking method or at M phase by nocodazole. We examined the luciferase activity and cyclin B1 levels with or without rTat. The results showed that rTat protein could stimulate the expression of cyclin B1, while the cells were blocked in G1/S boundary or M phase (Fig. 2F, G), excluding the possibility that Tat-induced up-regulation of cyclin B1 is an indirect effect. These results clearly indicate for the first time that Tat stimulates cyclin B1 expression through transcription regulation.
Figure 2.
Tat up-regulates the expression of cyclin B1 at transcriptional level. A) Schematic representation of deletion mutants of cyclin B1 promoter. Different deletion mutants of cyclin B1 were cloned into the upstream of Renilla luciferase. B) TE671 cells were transfected with pMIR-reporter (firefly luciferase) and CB1P-381 with rTat or without rTat (control). C) TE671 cells were transfected with pMIR-reporter and CB1P-287 with or without rTat. D) TE671 cells were transfected with pMIR-reporter and CB1P-180 with or without rTat. E) TE671 cells were transfected with pMIR-reporter and CB1P-90 with or without rTat. rTat was added to the cell culture 6 h after transfection. At 48 h post-transfection, dual luciferase activity was measured, and the ratio of 2 luciferase activities was determined. Data are averages ± sd from 4 samples in 2 independent experiments. F) TE671 cells were transfected with pCycB-(287)-LUC and synchronized at the G1/S boundary by the double-thymidine approach or synchronized in mitosis by incubation with 100 ng/ml nocodazole for 17 h, as described in Materials and Methods. rTat was added to the culture, and firefly luciferase activity was assayed 12 h later. NS, nonsynchronized cells; G1/S, G1/S boundary cells; M, M-phase cells. Data are averages ± sd from 3 replicas. *P < 0.05, **P < 0.01 vs. control. G) Cyclin B1 levels were measured by Western blot; same cells as described in F.
Tat-stimulated cyclin B1 involves apoptosis
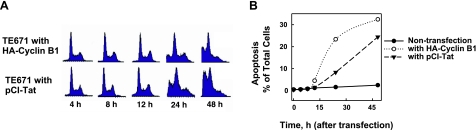
Either Tat or cyclin B1 alone could stimulate apoptosis. To investigate whether Tat-induced cyclin B1 expression was involved in apoptosis, we analyzed cell cycle distribution and apoptosis in the cells with or without exogenous cyclin B1 or Tat. TE671 cells were transfected with HA-cyclin B1 or pCI-Tat, and cell cycle distribution was analyzed at different times after transfection. The results showed that either cyclin B1 or Tat alone could induce apoptosis (Fig. 3, sub-G1 peak). Cotransfection of pCI-Tat and HA-cyclin B1 into TE671 cells showed a similar result as pCI-Tat transfection alone (data not shown), supporting the hypothesis that Tat-stimulated cyclin B1 expression contributes to Tat-induced apoptosis of host cells.
Figure 3.
Tat stimulates apoptosis. A) Cell cycle distribution of TE671 cells at different times after transfection with HA-cyclin B1 or pCI-Tat, measured by flow cytometric analysis (FACS). Sub-G1 peak represents apoptosis. B) Apoptosis induced by overexpression of Tat or cyclin B1; plot based on data shown in A.
Tat also stimulates cyclin B1 degradation through the polyubiquintination pathway
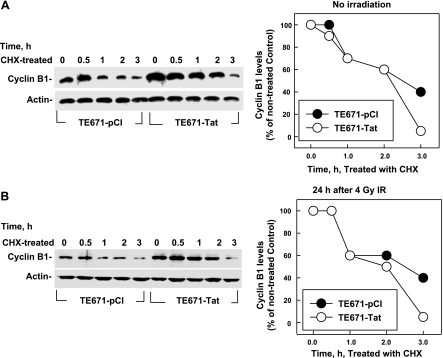
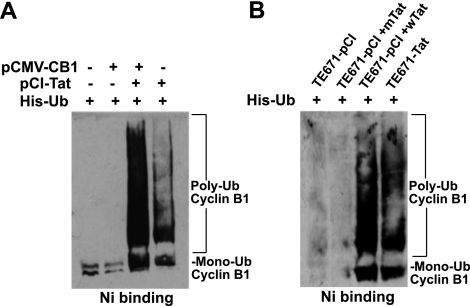
Although Tat stimulates transcription of cyclin B1, Tat-induced high levels of cyclin B1 remained only for a short time and then gradually decreased (Fig. 1), even when additional Tat was added to the culture (data not shown), suggesting that Tat might also stimulate cyclin B1 degradation, keeping the balanced level of cyclin B1 for HIV requirements. To test this hypothesis, we measured the half-life of cyclin B1 protein in TE671-pCI and TE671-Tat cells by blocking protein synthesis with cycloheximide (CHX). At the 0 h of CHX treatment, the cyclin B1 level in TE671-Tat cells was much higher than in TE671-pCI cells, and it decreased slowly in TE671-Tat cells for the first 2 h after CHX treatment (Fig. 4A). However, the cyclin B1 level in TE671-Tat cells at 3 h after CHX treatment dramatically decreased, resulting in even lower levels of cyclin B1 in TE671-Tat cells than in TE671-pCI cells (Fig. 4A). These results suggest that Tat stimulates cyclin B1 degradation. To confirm this hypothesis, we compared cyclin B1 stability in the irradiated cells. Both TE671-Tat and TE671-pCI cells were irradiated (4 Gy) and treated with CHX at 24 h after irradiation. The results (Fig. 4B) were similar to those shown in Fig. 4A. These data indicate that Tat stimulates cyclin B1 degradation when cyclin B1 reaches a high level. It is known that cyclin B1 is degraded through the ubiquitin/proteasome pathway. To test whether Tat stimulates cyclin B1 degradation through the same pathway, we examined the effects of Tat on cyclin B1 polyubiquitination. Cyclin B1 and 6 histidine-tagged ubiquitin (His-Ub) were expressed in the cells with or without the presence of Tat, and cyclin B1 ubiquitination was detected by capturing His-Ub from cell extracts with nickel beads under denaturing conditions. The results showed that the signals of polyubiquintinated cyclin B1 in the cells with Tat were much stronger than in the cells without Tat (Fig. 5). These data strongly support the hypothesis that Tat not only stimulates cyclin B1 expression but also stimulates cyclin-B1-polyubiquitinated degradation.
Figure 4.
Effects of Tat on the half-life of cyclin B1. Levels of cyclin B1 were measured by Western blot. Actin was used as the internal loading control. A) CHX was added to the culture of TE671-pCI and TE671-Tat cells. B) TE671-pCI and TE671-Tat cells were exposed to 4 Gy. Cells were treated with 200 ng/ml of CHX and were collected at different times.
Figure 5.
Tat stimulates the polyubiquitination of cyclin B1. A) 293T cells were cotransfected with vectors of His-Ub and HA-cyclin B1, myc-Tat, or both. Ubiquitinated cyclin B1 was retained on nickel beads and analyzed by immunoblotting with an anti-cyclin B1 antibody. B) TE671-pCI and TE671-Tat cells were transfected with vector of His-Ub and TE671-pCI treated with wild Tat or mutant Tat. The level of cyclin B1 ubiquitination was measured as described in A.
Tat stimulates polyubiquitination of cyclin B1 through interaction with cyclin B1
It was reported that APCCdc20 generates a degradation signal on cyclin B1 that consists of a heterogeneous mixture of ubiquitin-ubiquitin linkages through Lys 11 and Lys 63 in addition to Lys 48 (13). To investigate the polyubiquitination pattern of cyclin B1 induced by Tat, we constructed the expression vectors of ubiquitin with different mutations (K11R, K48R, and K63R; Supplemental Fig. S2A). 293T cells were transfected with expression vectors for wild-type Ub or its 3 different mutants with or without pCI-Tat cotransfection. Ubiquitinated cyclin B1 was retained on nickel beads and analyzed by Western blot. The results showed that polyubiquitination of cyclin B1 was not affected by any of the mutant ubiquitins in the presence of Tat (Supplemental Fig. S2B), indicating that Tat uses a different mechanism from APC to stimulate cyclin B1 polyubiquitinated degradation.
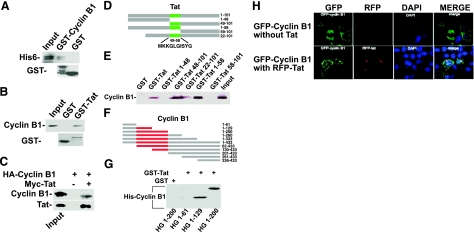
Tat was reported to interact directly with p300/CBP to induce the polyubiquitination and degradation of Tip60 (14). We were interested in studying whether Tat-stimulating cyclin-B1-polyubiquitinated degradation was through interacting with cyclin B1. For this purpose, we performed GST pulldown and coimmunoprecipitation assays. GST fusion proteins were expressed in E. coli and immobilized on GST-Sepharose beads. The results showed that E. coli expressed His-tagged Tat bound to the immobilized GST-cyclin B1 but not to the immobilized GST (Fig. 6A). In addition, the results showed that the cyclin B1 expressed in E. coli or TE671 cells bound to immobilized GST-Tat but not to GST (Fig. 6B) and that cyclin B1 was efficiently precipitated by anti-myc-tag antibody and protein A/G resin in cotransfected cells but not in cells that were transfected with cyclin B1 alone (Fig. 6C). To identify the interaction domains between these 2 proteins, a series of truncation mutants of Tat fused with GST and mutants of cyclin B1 were constructed and expressed in E. coli (Fig. 6D, F). GST pulldown revealed that aa 49–58 of Tat are indispensable for interaction with cyclin B1 (Fig. 6E) and aa 61–129 of cyclin B1 have at least one main domain for interacting with Tat (Fig. 6G). Most important, the mutant Tat that lost the interaction with cyclin B1 could not stimulate the polyubiquitination of cyclin B1 (Fig. 5B), indicating that the mechanism of Tat-stimulated cyclin-B1-polyubiquitinated degradation might also be through interacting with this protein.
Figure 6.
Tat directly interacts with cyclin B1. A) Recombinant GST and GST-cyclin B1 bound to glutathione-Sepharose beads were incubated with recombinant His6-tagged Tat protein. Bound fractions were analyzed by immunoblotting with anti-His or anti-GST antibody. B) Recombinant GST and GST-Tat bound to glutathione-Sepharose beads were incubated with recombinant His-tagged cyclin B1 protein. Bound fractions were analyzed by immunoblotting with anti-cyclin B1 or anti-GST antibody. C) 293T cells were transfected with HA-cyclin B1 with or without myc-Tat. At 48 h after transfection, cells were collected. Tat was immunoprecipitated with anti-myc antibody, and cyclin B1 was detected by immunoblotting analysis with anti-HA antibody. D) Schematic instruction of deletion mutants of Tat protein. Design of these mutants was based on different function domains of Tat. E) Recombinant GST and GST-Tat and its mutants bound to glutathione-Sepharose beads were incubated with total TE671 cell lysates. Bound fractions were analyzed by immunoblotting with anti-cyclin B1 antibody. F) Schematic representation of deletion mutants of cyclin B1 protein. G) Recombinant GST and GST-Tat bound to glutathione-Sepharose beads were incubated with recombinant His-deletion mutant of cyclin B1 proteins (HG). Bound fractions were analyzed by immunoblotting with anti-His antibody. H) Colocalization of Tat and cyclin B1 fused with GFP or RFP. 293T cells were transfected with GFP-cyclin B1 (row 1) or cotransfected with GFP-cyclin B1 and RFP-Tat (row 2). Cells were collected at 48 h post-transfection, fixed, and prepared for observation with a fluorescence microscopy.
Tat stimulates the nucleus localization of cyclin B1
To further study the relationship of Tat and cyclin B1, we inserted Tat or Cyclin B1 the full length of cDNA into the expression vector to allow production of RFP or GFP fusion proteins, respectively. We observed the cellular localization of different proteins by using fluorescence confocal microscopy. 293T cells were transfected with GFP-cyclin B1 or cotransfected with GFP-cyclin B1 and RFP-Tat. At 48 h after transfection, cells were fixed and then stained with DAPI for visualization of nuclear DNA. The results showed that GFP-cyclin B1 mainly localized in the cytoplasm when the cells were transfected with this vector alone (Fig. 6H, row 1). Interestingly, when the cells were cotransfected with GFP-cyclin B1 and RFP-Tat vectors together, RFP-Tat mainly localized in a special region of the nucleus, probably in the nucleolus or heterchromatin region, and some GFP-cyclin B1 merged to the nuclear region, where Tat mainly localized (Fig. 6H, row 2). The dynamic localization of cyclin B1 was demonstrated to be tightly regulated by the cell cycle (15); therefore, the unscheduled location of cyclin B1 (Fig. 6H) is believed to be due to the association of cyclin B1 with Tat.
DISCUSSION
In this study, our results demonstrate that Tat (both endogenous and exogenous) stimulates cyclin B1 transcription and results in a relatively high level of cyclin B1. These data provide a mechanism accounting, at least in part, for a consistent overexpression of cyclin B1 detected in peripheral CD4+ and CD8+ cells from HIV-infected patients (6, 7, 11, 12). MicroRNA (miRNA) plays a very important role in gene post-transcriptional regulation through binding to the sequences of the 3′-untranslated region (UTR) of the target gene. To study whether Tat up-regulation of cyclin B1 was also involved in miRNA, we inserted the 3′-UTR sequences of cyclin B1 into the 3′-UTR of firefly luciferase in vector pMIR-reporter (pMIR-CB1–3′UTR). The pRL-TK (Renilla luciferase) and pMIR-CB1–3′UTR were cotransfected into 293T cells. We did not observe any influence of Tat on the expression of reporter genes (data not shown), excluding the possibility that Tat up-regulation of cyclin B1 expression is also through miRNA. Tat is able to stimulate the expression of TNF, IL-2, IL-6, IL-8, IL-10, TGF-α, and TGF-β (16,17,18,19,20) and inhibits the expression of IL-2, IL-2 receptor, p53 and major histocompatibility complex class I (19, 21, 22). The possible mechanisms by which Tat regulates gene expression at the transcriptional level are through the following pathways: indirectly binding to transcriptional factor SP1 (23), directly binding to TATA box binding protein (TBP; ref. 24), associating with TBP-associated factor TAFII55 (25), and binding to the RNA leader sequence or interacting with CAAT enhancer-binding protein-β (20). Interestingly, the promoter of cyclin B1 includes the SP1 binding site (−144 to −139) and CAAT box (−81 to −77 and −49 to −45), which was essential for S-phase induction (10). Our results indicate that the level of reporter gene expression is much higher when both the SP1 binding site and CAAT box are present (Fig. 2), indicating that both SP1 and CAAT box are required for Tat to stimulate cyclin B1.
It was reported that the increased expression of cyclin B1 is caused by defective degradation in the presence of a normal rate of synthesis (7). We show a different result in this study, namely, that Tat stimulates polyubiquitination-mediated degradation of cyclin B1. These different results might be due to the fact that Tat stimulation of cyclin B1 degradation requires a high level of cyclin B1 accumulation. With a lower level of cyclin B1, Tat could not actively stimulate the polyubiquitination-mediated degradation of cyclin B1. These different results might also be due to comparing in vitro with in vivo results, because in addition to Tat, several other pathways contribute to cyclin B1 ubiqitin-mediated degradation in vivo. Tat interacts with proteasomal α- and β-subunits (26), which increases the activity of proteasomes (27,28,29). It was also reported that Tat uses p300/CBP-associated E4-type ubiquitin-ligase activity to promote Tip60 polyubiquitination through direct interaction with p300/CBP (14). Our results indicate that Tat promotes the polyubiquitination of cyclin B1, which might also depend on its interaction with cyclin B1 (Fig. 5). The binding site of cyclin B1 is very close to the D box (RTALGDIGN), which is required for degradation in metaphase (30) by anaphase-promoting complex/cyclosome (APC/C) (31). Degradation of cyclin B1 is reported to be fulfilled by the ubiquitination-mediated APCCdc20 proteasomal degradation pathway (13). However, mutation at the polyubiquitination sites of cyclin B1 by APC does not affect the polyubiquitination of cyclin B1 by Tat (Supplemental Fig. S2), indicating that the mechanism of Tat-induced polyubiquitination of cyclin B1 is different from that by which APC induces polyubiquitination of cyclin B1. The detailed mechanism needs future study.
Apoptosis is one of the most severe effects in HIV-1-infected individuals, resulting in the depletion of CD4+ T cells and tissue damage. Tat is one of the major factors for HIV to induce apoptosis, which is involved in several signal transduction pathways, such as up-regulating the Fas/Fas-ligand-dependent pathway, caspase-8, and TRAIL (32,33,34); down-regulating BCL-2 (35, 36); destroying the mitochondria membrane potential (37); targeting microtubules and LIS1 (38, 39); and inhibiting superoxide dismutase (32). In addition, apoptosis occurs predominantly in bystander cells and not in productively infected cells in HIV- and SIV-infected lymphocytes. This phenomenon might be mainly due to Tat, because Tat is the only protein encoded by HIV that can transfer into or out of cells freely. Cyclin B1 was reported to be involved in the induction of apoptosis or mitotic catastrophe (40). The persistent expression of cyclin B1 acts as an apoptotic signal to cells after radiation (41) or chemotherapy (5, 40). After combining these results with our data, we believe that Tat up-regulated cyclin B1 is one of the major factors for Tat to induce apoptosis. Our findings explain why high cyclin B1 expression and CDK1 activation was detected in T lymphocytes from HIV-infected patients (6) and why cyclin B1 expression returned to normal after effective antiretroviral therapy (7).
Nuclear localization of cyclin B1 was reported to regulate DNA-damage-induced apoptosis (41, 42). Interestingly, we observed an unscheduled localization in the nucleus of cyclin B1 induced by Tat. This result will, at least partially, explain the apoptosis involved in Tat-promoted cyclin B1. In addition, our results also suggest that the interaction of Tat with cyclin B1 might also promote cyclin B1 degradation through polyubiqitine-mediated degradation. We believe that the dual effects of Tat on cyclin B1, stimulating cyclin B1 expression and cyclin B1 degradation, affect HIV stature: Tat-stimulating cyclin B1 degradation benefits HIV to replicate; and Tat-stimulating cyclin B1 expression (resulting in the host cells apoptosis) benefits HIV to release.
Taken together, our results demonstrate that Tat regulates cyclin B1 by stimulating transcriptional expression on the one hand and by stimulating polyubiquitination-mediated degradation on the other hand, which affects cells apoptosis.
Acknowledgments
The authors thank Dr. Lung-Ji Chang (University of Florida, Gainesville, FL, USA), Junjie Qian (Beijing Insitute of Radiation Medicine, Beijing, China), Hui Zhang (Thomas Jefferson University, Philadelphia, PA, USA) and Karen S. Katula (University of North Carolina, Chapel Hill, NC, USA) for providing reagents. This work was supported by grants from China [National BioResource Project (NBRP), Ministry of Science and Technology (MOST), 973 Program 2007CB914603; China National Natural Science Foundation (CNNSF), grants 30500267 and 30400120; Outstanding Youth Science Foundation, Natural Science Foundation of China (NFSC), grant 30825011; International Atomic Energy Agency Coordinated Research Project E3.30.22, grant 12510/R4) and from the United States (National Institutes of Health, grant G-M080771).
References
- Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- Pines J, Hunter T. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashal R D, Lester S, Corless C, Richie J P, Chandra R, Propert K J, Dutta A. Expression of cell cycle-regulated proteins in prostate cancer. Cancer Res. 1996;56:4159–4163. [PubMed] [Google Scholar]
- Kawamoto H, Koizumi H, Uchikoshi T. Expression of the G2-M checkpoint regulators cyclin B1 and cdc2 in nonmalignant and malignant human breast lesions: immunocytochemical and quantitative image analyses. Am J Pathol. 1997;150:15–23. [PMC free article] [PubMed] [Google Scholar]
- Gomez L A, de las Pozas A, Reiner T, Burnstein K, Perez-Stable C. Increased expression of cyclin B1 sensitizes prostate cancer cells to apoptosis induced by chemotherapy. Mol Cancer Ther. 2007;6:1534–1543. doi: 10.1158/1535-7163.MCT-06-0727. [DOI] [PubMed] [Google Scholar]
- Piedimonte G, Corsi D, Paiardini M, Cannavò G, Ientile R, Picerno I, Montroni M, Silvestri G, Magnani M. Unscheduled cyclin B expression and p34 cdc2 activation in T lymphocytes from HIV-infected patients. AIDS. 1999;13:1159–1165. doi: 10.1097/00002030-199907090-00003. [DOI] [PubMed] [Google Scholar]
- Cannavo' G, Paiardini M, Galati D, Cervasi B, Montroni M, De Vico G, Guetard D, Bocchino M L, Picerno I, Magnani M, Silvestri G, Piedimonte G. Abnormal intracellular kinetics of cell-cycle-dependent proteins in lymphocytes from patients infected with human immunodeficiency virus: a novel biologic link between immune activation, accelerated T-cell turnover, and high levels of apoptosis. Blood. 2001;97:1756–1764. doi: 10.1182/blood.v97.6.1756. [DOI] [PubMed] [Google Scholar]
- Dayton A, Sodroski J, Rosen C, Goh W, Haseltine W. The trans-activator gene of the human T cell lymphotropic virus type III is required for replication. Cell. 1986;44:941–947. doi: 10.1016/0092-8674(86)90017-6. [DOI] [PubMed] [Google Scholar]
- Sun Y, Huang Y-C, Xu Q-Z, Wang H-P, Bai B, Sui J-L, Zhou P-K. HIV-1 Tat depresses DNA-PKCS expression and DNA repair, and sensitizes cells to ionizing radiation. Int J Radiat Oncol Biol Phys. 2006;65:842–850. doi: 10.1016/j.ijrobp.2006.02.040. [DOI] [PubMed] [Google Scholar]
- Katula K S, Wright K L, Paul H, Surman D R, Nuckolls F J, Smith J W, Ting J P-Y, Yates J, Cogswell J P. Cyclin-dependent kinase activation and S-phase induction of the cyclin B1 gene are linked through the CCAAT elements. Cell Growth Differ. 1997;8:811–820. [PubMed] [Google Scholar]
- Paiardini M, Cervasi B, Galati D, Dominici S, Albrecht H, Sfacteria A, Magnani M, Silvestri G, Piedimonte G. Early correction of cell cycle perturbations predicts the immunological response to therapy in HIV-infected patients. AIDS. 2004;18:393–402. doi: 10.1097/00002030-200402200-00004. [DOI] [PubMed] [Google Scholar]
- Galati D, Paiardini M, Cervasi B, Albrecht H, Bocchino M, Costantini A, Montroni M, Magnani M, Piedimonte G, Silvestri G. Specific changes in the posttranslational regulation of nucleolin in lymphocytes from patients infected with human immunodeficiency virus. J Infect Dis. 2003;188:1483–1491. doi: 10.1086/379249. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick D S, Hathaway N A, Hanna J, Elsasser S, Rush J, Finley D, King R W, Gygi S P. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Col E, Caron C, Chable-Bessia C, Legube G, Gazzeri S, Komatsu Y, Yoshida M, Benkirane M, Trouche D, Khochbin S. HIV-1 Tat targets Tip60 to impair the apoptotic cell response to genotoxic stresses. EMBO J. 2005;24:2634–2645. doi: 10.1038/sj.emboj.7600734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A, Zon W, Rosenthal C, Wolthuis R. Cyclin B1-Cdk1 activation continues after centrosome separation to control mitotic progression. PLoS Biol. 2007;5:e123. doi: 10.1371/journal.pbio.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry K, Reddy H, Pandita R, Totpal K, Aggarwal B. HIV-1 tat gene induces tumor necrosis factor-beta (lymphotoxin) in a human B-lymphoblastoid cell line. J Biol Chem. 1990;265:20091–20093. [PubMed] [Google Scholar]
- Ambrosino C, Ruocco M R, Chen X, Mallardo M, Baudi F, Trematerra S, Quinto I, Venuta S, Scala G. HIV-1 Tat induces the expression of the interleukin-6 (IL6) gene by binding to the IL6 leader RNA and by interacting with CAAT enhancer-binding protein beta (NF-IL6) transcription factors. J Biol Chem. 1997;272:14883–14892. doi: 10.1074/jbc.272.23.14883. [DOI] [PubMed] [Google Scholar]
- Badou A, Bennasser Y, Moreau M, Leclerc C, Benkirane M, Bahraoui E. Tat protein of human immunodeficiency virus type 1 induces interleukin-10 in human peripheral blood monocytes: implication of protein kinase c-dependent pathway. J Virol. 2000;74:10551–10562. doi: 10.1128/jvi.74.22.10551-10562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehret A, Li-Weber M, Frank R, Krammer P H. The effect of HIV-1 regulatory proteins on cellular genes: derepression of the IL-2 promoter by Tat. Eur J Immunol. 2001;31:1790–1799. doi: 10.1002/1521-4141(200106)31:6<1790::aid-immu1790>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Mahieux R, Lambert P F, Agbottah E, Halanski M A, Deng L, Kashanchi F, Brady J N. Cell cycle regulation of human interleukin-8 gene expression by the human immunodeficiency virus type 1 tat protein. J Virol. 2001;75:1736–1743. doi: 10.1128/JVI.75.4.1736-1743.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howcroft T, Strebel K, Martin M, Singer D. Repression of MHC class I gene promoter activity by two-exon Tat of HIV. Science. 1993;260:1320–1322. doi: 10.1126/science.8493575. [DOI] [PubMed] [Google Scholar]
- Li C J, Wang C, Friedman D J, Pardee A B. Reciprocal modulations between p53 and Tat of human immunodeficiency virus type 1. Proc National Acad Sci U S A. 1995;92:5461–5464. doi: 10.1073/pnas.92.12.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loregian A, Bortolozzo K, Boso S, Caputo A, Palu G. Interaction of Sp1 transcription factor with HIV-1 Tat protein: looking for cellular partners. FEBS Lett. 2003;543:61–65. doi: 10.1016/s0014-5793(03)00399-5. [DOI] [PubMed] [Google Scholar]
- Kashanchi F, Piras G, Radonovich M F, Duvall J F, Fattaey A, Chiang C-M, Roeder R G, Brady J N. Direct interaction of human TFIID with the HIV-1 transactivator. Tat Nature. 1994;367:295–299. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]
- Chiang C, Roeder R. Cloning of an intrinsic human TFIID subunit that interacts with multiple transcriptional activators. Science. 1995;267:531–536. doi: 10.1126/science.7824954. [DOI] [PubMed] [Google Scholar]
- Apcher G S, Heink S, Zantopf D, Kloetzel P-M, Schmid H-P, Mayer R J, Krüger E. Human immunodeficiency virus-1 Tat protein interacts with distinct proteasomal α and β subunits. FEBS Lett. 2003;553:200–204. doi: 10.1016/s0014-5793(03)01025-1. [DOI] [PubMed] [Google Scholar]
- Seeger M, Ferrell K, Frank R, Dubiel W. HIV-1 Tat Inhibits the 20S proteasome and its 11S regulator-mediated activation. J Biol Chem. 1997;272:8145–8148. doi: 10.1074/jbc.272.13.8145. [DOI] [PubMed] [Google Scholar]
- Huang X, Seifert U, Salzmann U, Henklein P, Preissner R, Henke W, Sijts A J, Kloetzel P-M, Dubiel W. The RTP site shared by the HIV-1 Tat protein and the 11 S regulator subunit α is crucial for their effects on proteasome function including antigen processing. J Mol Biol. 2002;323:771–782. doi: 10.1016/s0022-2836(02)00998-1. [DOI] [PubMed] [Google Scholar]
- Gavioli R, Gallerani E, Fortini C, Fabris M, Bottoni A, Canella A, Bonaccorsi A, Marastoni M, Micheletti F, Cafaro A, Rimessi P, Caputo A, Ensoli B. HIV-1 Tat protein modulates the generation of cytotoxic T cell epitopes by modifying proteasome composition and enzymatic activity. J Immunol. 2004;173:3838–3843. doi: 10.4049/jimmunol.173.6.3838. [DOI] [PubMed] [Google Scholar]
- Clute P, Pines J. Temporal and spatial regulation of cyclin B1 destruction in metaphase. Nat Cell Biol. 1999;1:82–87. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- Herbert M, Levasseur M, Homer H, Yallop K, Murdoch A, McDougall A. Homologue disjunction in mouse oocytes requires proteolysis of securin and cyclin B1. Nat Cell Biol. 2003;5:1023–1025. doi: 10.1038/ncb1062. [DOI] [PubMed] [Google Scholar]
- Westendorp M O, Frank R, Ochsenbauer C, Stricker K, Dhein J, Walczak H, Debating K-M, Krammer P H. Sensitization of T cells to CD95-mediated apoptosis by HIV-1 Tat and gp120. Nature. 1995;375:497–500. doi: 10.1038/375497a0. [DOI] [PubMed] [Google Scholar]
- Bartz S R, Emerman M. Human immunodeficiency virus type 1 tat induces apoptosis and increases sensitivity to apoptotic signals by up-regulating FLICE/caspase-8. J Virol. 1999;73:1956–1963. doi: 10.1128/jvi.73.3.1956-1963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Li X, Pang X, Ding L, Wood O, Clouse K, Hewlett I, Dayton A I. Identification of a potential HIV-induced source of bystander-mediated apoptosis in T cells: upregulation of trail in primary human macrophages by HIV-1 tat. J Biomed Sci. 2001;8:290–296. doi: 10.1007/BF02256603. [DOI] [PubMed] [Google Scholar]
- Sastry K J, Marin M C, Nehete P N, McConnell K, el-Naggar A K, McDonnell T J. Expression of human immunodeficiency virus type I tat results in down-regulation of bcl-2 and induction of apoptosis in hematopoietic cells. Oncogene. 1996;13:487–493. [PubMed] [Google Scholar]
- Gougeon M-L. Apoptosis. as. an. HIV. strategy. to. escape. immune. attack. Nat Rev Immunol. 2003;3:392–404. doi: 10.1038/nri1087. [DOI] [PubMed] [Google Scholar]
- Macho A, Calzado M, Jiménez-Reina L, Ceballos E, León J, Muñoz E. Susceptibility of HIV-1-TAT transfected cells to undergo apoptosis. Biochemical mechanisms. Oncogene. 1999;18:7543–7551. doi: 10.1038/sj.onc.1203095. [DOI] [PubMed] [Google Scholar]
- Chen D, Wang M, Zhou S, Zhou Q. HIV-1 Tat targets microtubules to induce apoptosis, a process promoted by the pro-apoptotic Bcl-2 relative Bim. EMBO J. 2002;21:6801–6810. doi: 10.1093/emboj/cdf683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epie N, Ammosova T, Sapir T, Voloshin Y, Lane W S, Turner W, Reiner O, Nekhai S. HIV-1 Tat interacts with LIS1 protein. Retrovirology. 2005;2:6. doi: 10.1186/1742-4690-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgne A, Versteege I, Mahe M, Studeny A, Leonce S, Naime I, Rodriguez M, Hickman J A, Meijer L, Golsteyn R M. Analysis of cyclin B1 and CDK activity during apoptosis induced by camptothecin treatment. Oncogene. 2006;25:7361–7372. doi: 10.1038/sj.onc.1209718. [DOI] [PubMed] [Google Scholar]
- Porter L A, Singh G, Lee J M. Abundance of cyclin B1 regulates gamma -radiation-induced apoptosis. Blood. 2000;95:2645–2650. [PubMed] [Google Scholar]
- Porter L A, Cukier I H, Lee J M. Nuclear localization of cyclin B1 regulates DNA damage-induced apoptosis. Blood. 2003;101:1928–1933. doi: 10.1182/blood-2002-04-1103. [DOI] [PubMed] [Google Scholar]