Abstract
In primitive erythroid cells of human β-globin locus transgenic mice (TgM), the locus control region (LCR)-proximal ε- and γ-globin genes are transcribed, whereas the distal δ- and β-globin genes are silent. It is generally accepted that the β-globin gene is competitively suppressed by γ-globin gene expression at this developmental stage. Previously, however, we observed that ε-globin gene expression was severely attenuated when its distance from the LCR was extended, implying that β-globin gene might also be silenced because of its great distance from the LCR. Here, to clarify the β-globin gene silencing mechanism, we established TgM lines carrying either γ- or ε- plus γ-globin promoter deletions, without significantly altering the distance between the β-globin gene and the LCR. Precocious expression of δ- and β-globin genes was observed in primitive erythroid cells of mutant, but not wild-type TgM, which was most evident when both the ε and γ promoters were deleted. Thus, we clearly demonstrated that the repression of the δ- and β-globin genes in primitive erythroid cells is dominated by competitive silencing by the ε- and γ-globin gene promoters, and that ε- and the other β-like globin genes might be activated by two distinct mechanisms by the LCR.—Okamura, E., Matsuzaki, H., Campbell, A. D., Engel, J. D., Fukamizu, A., Tanimoto, K. All of the human β-type globin genes compete for LCR enhancer activity in embryonic erythroid cells of yeast artificial chromosome transgenic mice.
Keywords: hemoglobin, gene switching, chromosome conformation capture, 3C
The human β-globin gene locus is composed of 5 β-like globin genes (5′-ε-Gγ-Aγ-δ-β-3′, Fig. 1A), which are regulated in a tissue- and developmental-stage-specific fashion. In primitive erythroid cells within the embryonic yolk sac, the ε-globin gene is expressed, whereas the other β-like globin genes are silent. During definitive erythropoiesis in the fetal liver, the 2 γ-globin genes are activated, and the ε-globin gene is concomitantly silenced. At around the time of birth, the site of hematopoiesis changes to the bone marrow and spleen, where the adult δ- and β-globin genes are transcribed, and the γ-globin genes become reciprocally silenced (1). Previous studies have shown that high-level expression of all the β-like globin genes requires a distal cis-regulatory element, the locus control region (LCR), located between 6 and 22 kb 5′ of the ε-globin gene. The LCR is composed of multiple DNase I hypersensitive (HS) sites and acts as a powerful enhancer (2, 3), but its role in the developmental regulation of individual genes within the locus is yet to be fully elucidated.
Figure 1.
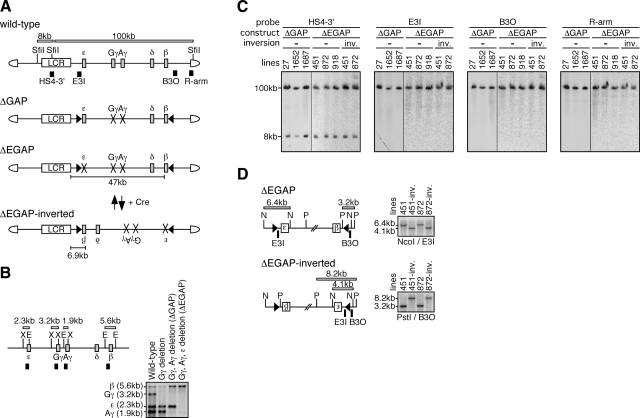
Experimental strategy. A) Schematic representation of the 150-kb wild-type and mutant human β-globin YACs. Positions of the β-like globin genes (gray rectangles) are shown relative to the LCR (open rectangle). SfiI restriction enzyme sites are located 5′ to HS5, between HS4 and HS3 of the LCR, and in the right arm of the YAC. Probes (solid rectangles) used for long-range fragment analysis shown in panel C and expected restriction enzyme fragments with their sizes are shown. In ΔGAP YAC, the Gγ (from −435 to +29 relative to transcriptional start site) and Aγ (from −435 to +29) promoters are missing (cross marks). In ΔEGAP YAC, the ε (from −438 to + 25), Gγ, and Aγ promoters are missing. In both mutant loci, loxP sites (arrowheads) were introduced in inverted orientation. The ΔEGAP-inverted locus was created by Cre-loxP recombination from the ΔEGAP locus in vivo. Distances between the LCR and β-globin gene in each locus are depicted. B) Introduction of the promoter mutations by homologous recombination in yeast. Partial restriction enzyme map of the wild-type human β-globin locus. X, XbaI; E, EcoRI. Probes and expected restriction enzyme fragments with their sizes are shown as solid and open rectangles, respectively. DNA from yeast clones bearing the wild-type or mutant human β-globin YACs was digested with XbaI and EcoRI and separated on agarose gels. After Southern blot transfer to nylon membrane, the DNA was hybridized to the mixed probes. Gγ-, Aγ-, and ε-globin gene promoters were sequentially deleted in this order. C) Long-range structural analysis of the human β-globin YAC in TgM. The whole β-globin locus is contained within 2 SfiI fragments (8 and 100 kb, shown in panel A). DNA from thymus cells of transgenic mice was digested with SfiI in agarose plugs, separated by pulsed-field gel electrophoresis, and Southern blots were hybridized separately to the probes. −, noninverted; inv., inverted. D) Cre-loxP-mediated in vivo inversion of the locus. Detailed structures around the loxP sites in the ΔEGAP and ΔEGAP-inverted transgenes are shown. Positions of the loxP sites (arrowheads), genes (labeled boxes), restriction enzyme sites (N, NcoI; P, PstI), and probes (solid rectangles) are shown proportionally. Expected restriction enzyme fragments with their sizes are shown. Tail DNAs from each mutant TgM line were digested with NcoI or PstI, separated on agarose gels, and Southern blots were hybridized to probes E3I or B3O.
Regulation of human β-like globin gene expression has been intensively studied in transgenic mice (TgM) carrying portions of the human β-globin locus. These studies suggested that developmental expression of the β-like globin genes is regulated through two distinct mechanisms: autonomous silencing and gene competition (4). When directly linked to the LCR, the human embryonic ε-globin gene was expressed only during the primitive stage and became autonomously silenced at later stages in TgM (5). This finding suggested that the sequence information required for turning off ε gene transcription during the fetal stage of erythropoiesis is contained in sequences close to or within the gene. On the other hand, an LCR-linked human γ-globin gene was expressed not only in embryonic but also in adult erythrocytes, albeit at much lower levels. In addition, the LCR-linked adult β-globin gene was expressed at all developmental stages. Therefore, silencing of the human γ- and β-globin genes is not solely ensured by a gene autonomous mechanism. When the LCR, γ-, and β-globin genes were linked in their wild-type order in cis, developmentally controlled gene switching was restored (6, 7). These results were interpreted to mean that the LCR-proximal γ-globin gene competitively suppressed the distal β-globin gene expression during embryonic erythropoiesis, whereas the distal β-globin gene suppressed the proximal γ-globin gene expression in the adult, at least in this small artificial construct. Subsequently, we and others have examined this notion by employing TgM carrying the whole human β-globin locus in cosmids (8) or yeast artificial chromosomes (YACs) (9). We consistently observed that the β-globin gene, when it was artificially located proximal to the LCR within the mutant loci, was precociously expressed in primitive erythroid cells. Based on these and other observations, it is generally believed that the adult β-globin gene is silenced because of a gene competition mechanism in primitive erythroid cells.
In the mouse β-globin locus, the LCR-proximal εy- and βh1-globin genes are abundantly transcribed in the embryonic erythroid cells. Because deletion of both or either of the εy- and βh1-globin gene promoters in the endogenous mouse locus led to accumulation of LCR-distal βmajor- and βminor-globin mRNA by 2- to 3-fold during primitive erythropoiesis (10), it was demonstrated that 2 embryonic genes, εy and βh1, were competitively suppressing the adult βmajor and βminor gene expression at this stage of erythropoiesis. However, the mouse βmajor- and βminor-globin genes differ from the human adult β-globin genes, in that mouse adult genes are intrinsically expressed in primitive erythroid cells, although at a low level. Therefore, it is not certain if the mechanism predicted from observations in the endogenous mouse locus can be extended to explain similar phenomena in the human locus.
Recently, we observed that the level of embryonic ε-globin gene expression was diminished by ∼90% in comparison to that in the wild-type control when its distance from the LCR was simply increased by 2.3 kb in the YAC (11). It is therefore conceivable that LCR enhancer activity may be sensitive to its distance from target genes and may simply be incapable of activating the β-globin gene in embryonic erythroid cells. In this sense, the proximally located β-globin gene in the mutant TgM mentioned above (8, 9) may be activated because of its reduced distance from the LCR, rather than because of the acquisition of a competitive advantage over the other genes within the locus.
We therefore decided to reexamine the silencing mechanism of the human β-globin gene by once again employing YAC TgM. To discriminate whether adult β-globin gene transcription is silenced by gene competition or because of its relatively great distance from the LCR in primitive erythroid cells, we established multiple lines of YAC TgM carrying mutated human β-globin loci. In the TgM bearing deletions of both the Gγ- and Aγ-globin gene promoters in the YAC, the adult δ- and β-globin genes were expressed in primitive erythroid cells, despite its enormous distance from the LCR. Furthermore, when the ε-globin gene promoter was additionally deleted from the YAC, the level of adult δ- and β-globin gene expression increased even further. Any role for the ε-globin gene in competitive regulation of β-globin locus transcription has been incompletely addressed, and thus this is the first evidence demonstrating that the ε-globin gene, in addition to the γ-globin genes, competitively suppresses adult-type β-like globin gene expression in primitive erythroid cells.
When we tested the effect of reducing the distance between the LCR and the β-globin gene on its level of expression in primitive erythroid cells, we observed only moderate induction. These results demonstrated that the adult β-globin genes are silenced principally by gene competition at this developmental stage. To gain insight into the molecular entities participating in the gene competition phenomenon, we compared the spatial organization of the wild-type and mutant human β-globin locus by chromatin conformation capture (3C) (12). The cross-linking frequency between the LCR and β-globin gene regions was clearly higher in the mutant locus, whereas that between the LCR and γ-globin gene sequences was higher in the wild-type locus. These results imply that the physical interaction between the LCR and γ-globin genes interferes with those between the LCR and the β-globin gene in primitive erythroid cells bearing a wild-type human β-globin locus.
MATERIALS AND METHODS
YAC mutagenesis
The targeting vector for deleting the human Gγ-globin promoter was constructed as follows. The 5′ upstream region [nucleotides (nt) 33757-34042; HUMHBB locus, GenBank] of the promoter was PCR amplified with the primers hGg5-del-5S and hg5-del-3A, then digested with XhoI and BamHI. The 3′ downstream region (nt 34508-34808) was amplified with the primers hg3-del-5S and hg3-del-3A, then digested with BamHI and XbaI. These two DNA fragments and XhoI/XbaI-digested pRS306 vector were ligated. The yeast targeting plasmid DNA was linearized with AflII (at nt 33902) and used for mutagenizing the human β-globin YAC (A201F4.3) (13) to make A201F4.3/ΔGγ construct.
The targeting vector for deleting the human Aγ-globin promoter was constructed as follows. The 5′ upstream region (nt 38695-38978) of the promoter was amplified with the primers hAg5-del-5S and hg5-del-3A, then digested with XhoI and BamHI. The 3′ downstream region (nt 39444-39744) was amplified with the primers hg3-del-5S and hg3-del-3A, then digested with BamHI and XbaI. These two DNA fragments and XhoI/XbaI-digested pRS306 vector were ligated. The yeast targeting plasmid DNA was linearized with BstEII (at nt 39568) and used for A201F4.3/ΔGγ mutagenesis, generating ΔGAP (deleted Gγ- and Aγ-globin promoters) construct.
The targeting vector for deleting the human ε-globin gene promoter was constructed as follows. The 5′ upstream region (nt 18777-19047) of the promoter was amplified with the primers he5-del-5S and he5-del-3A, then digested with XhoI and EcoRI. The 3′ downstream region (nt 19515-19794) was amplified with the primers he3-del-5S and he3-del-3A, then digested with EcoRI and HindIII. These two DNA fragments and XhoI/HindIII-digested pRS306 vector were ligated. The yeast targeting plasmid DNA was digested with EcoNI (at nt 19623) and used for ΔGAP mutagenesis, generating ΔEGAP (deleted ε-, Gγ-, and Aγ-globin promoters) construct.
All the vector sequences were verified by DNA sequencing, and successful homologous recombination in yeast was confirmed by Southern blot analyses. Oligo sequences are listed in Supplemental Table 1.
TgM
Generation and structural analysis of human β-globin YAC TgM has been described elsewhere (14). ΔEGAP-inverted TgM was generated by mating ΔEGAP TgM with a TgM ubiquitously expressing Cre recombinase (15). Successful inversion of the locus was confirmed by Southern blot analysis. Animal experiments were carried out in a humane manner and were approved by the Institutional Animal Experiment Committee of the University of Tsukuba. Experiments were conducted in accordance with the Regulation of Animal Experiments of the University of Tsukuba and the Fundamental Guidelines for Proper Conduct of Animal Experiments and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Semiquantitative RT-PCR analysis
Total RNA was extracted from yolk sacs [9.5 d post coitum (dpc)] or phenylhydrazine-induced anemic adult spleens (1 to 2 mo old) by using Isogen (Nippon Gene, Tokyo, Japan). First-strand cDNA was synthesized from 2.5 μg of RNA with ReverTra Ace (Toyobo, Osaka, Japan). One-twentieth of the reaction mixture was subjected to PCR amplification using the following parameters: 94°C for 30 s, 58°C for 1 min, and 72°C for 1 min. Cycle numbers used for PCR analysis were as follows: 16 for δ and 12 for β/α (adult spleen), 18 for ε, 12 for γ/α, 24 for δ, and 18 for β (yolk sac). An aliquot of each PCR product was electrophoresed on 8% polyacrylamide gels, dried, and subjected to X-ray autoradiography and phosphoimaging for quantitative analysis. The PCR primers used for human δ-globin gene amplification were DT-5S2 and DT-3A1 for yolk sac samples, or DT-5S1 and DT-3A2 for adult spleen samples (Supplemental Table 1). The sequences of the other primers used have been described elsewhere (13).
3C assay
The 3C assay was performed as described previously (16, 17), with minor modifications. Here 5 × 106 embryonic blood cells (from 10.5 dpc embryos) were cross-linked in 0.5 ml of 1× PBS(−) with 1% formaldehyde at room temperature for 10 min, followed by the addition of glycine to 0.125 M to quench the reaction. Nuclei were harvested from lysed cells, portioned into 5 aliquots in microtubes, and precipitated by centrifugation. Nuclei pellets were frozen in liquid nitrogen and stored at −80°C. Frozen nuclei (1 tube) were resuspended in restriction enzyme buffer containing 0.3% SDS and incubated for 12 h at 37°C. Triton X-100 was added to 1.8%, and the nuclei were further incubated for 1 h at 37°C to sequester the SDS. The cross-linked DNA (3 μg) was digested with restriction enzyme (EcoRI) for 12 h at 37°C. The restriction enzyme was inactivated by addition of 1.6% SDS and incubation at 65°C for 30 min. The reaction was diluted (to 3.8 ng/μl of DNA) with ligase buffer (17), and Triton X-100 was added to 1% and incubated for 1 h at 37°C. Next, 667 U of T4 DNA ligase (NEB, Ipswich, MA, USA) was added and incubated for 5.5 h at 16°C. Cross-links were reversed by overnight incubation at 65°C in the presence of Proteinase K (37 μg/ml). The DNA was purified by 3 rounds of phenol-chloroform extraction and isopropanol precipitation. Quantification of ligated products was done by quantitative real-time PCR.
We collected blood samples from 2 independent sets of animals for each experiment (Exp. 1 and 2). In each 3C experiment, enzyme digestion and religation of the nuclei was performed at least twice to confirm reproducibility of the results. Quantitative real-time PCR was carried out several times for each DNA template, and the averages ± sd from all the reactions in each experiment were statistically calculated.
Quantitative real-time PCR analysis
Real-time qPCR was performed with the Thermal Cycler Dice Real Time System (Takara Bio, Otsu, Japan) using SYBR Premix EX TaqII (Takara Bio). For gene expression analysis, 1/100 of the first-strand cDNA reaction mixture was used for real-time PCR amplification using the following parameters: 95°C for 10 s, followed by 40 cycles of 95°C for 5 s, and 60°C for 30 s. The PCR primers used for human β-globin gene amplification were same as those for semiquantitative RT-PCR analysis described above. For 3C analysis, 1/30 of the purified DNA was used for real-time PCR amplification using the same parameters as the gene expression analysis described above. To correct for differences in quality and quantity of template, the results were normalized to a control interaction in the unrelated mouse Ercc3 gene (18). The PCR primers used in this study are as follows; 3C-HS2/3/4, 3C-γ, 3C-β, 3C-Ercc3-a, and 3C-Ercc3-b (Supplemental Table 1).
RESULTS
Generation and structual analysis of YAC TgM
We sequentially introduced 3 promoter mutations into a wild-type 150-kb human β-globin YAC, A201F4.3, by homologous recombination in yeast (Fig. 1A). The YAC bearing ∼500-bp deletions at both Gγ- and Aγ-globin promoter regions was termed ΔGAP. ΔEGAP was generated by subsequently deleting the ε-globin promoter in the ΔGAP YAC (Fig. 1A). At each mutagenesis step, successful recombination was confirmed by Southern blot analyses (Fig. 1B). The YAC DNA (ΔGAP and ΔEGAP) was purified by standard methods (19) and injected into fertilized oocytes to generate TgM. Tail DNA from offspring (F0) was screened by PCR using human β-globin gene-specific primers. Several independent TgM lines for ΔGAP (lines 27, 1652, and 1687) and ΔEGAP (lines 451, 872, and 918) were generated.
To verify the integrity of the transgene, thymus cells were embedded in agarose plugs, and high-molecular-weight DNA was prepared. The DNA was digested with SfiI restriction enzyme, fractionated by pulsed-field gel electrophoresis, and Southern blotted. Hybridization was performed on DNA fragment probes from the LCR (HS4–3′), the human ε-globin gene (E3I), the human β-globin gene (B3O), and the YAC right vector arm (R-arm) regions (9). All of the probes detected bands of the expected sizes (8 and 100 kb; Fig. 1A, C), indicating that each TgM line carried intact, unfragmented copies of the transgenes. The end-fragment and copy-number analyses (with a fragment from the endogenous angiotensinogen locus as an internal control; ref. 20) revealed that all of the lines carried single-copy YAC transgenes (data not shown).
Following these structural analyses, 2 lines of ΔEGAP TgM (451 and 872) were subjected to Cre-mediated gene locus inversion to shorten the distance between the LCR and the human δ- and β-globin genes (Fig. 1D). The ΔEGAP animals were mated with TgM that ubiquitously express Cre recombinase, and offspring were analyzed for proper recombination by Southern blot analysis (data not shown). Those animals that had undergone Cre-mediated recombination (Cre-F0) were again mated with wild-type animals to remove the Cre recombinase transgene. Fixed genotypes of Cre-F1 animals (ΔEGAP-inverted) were confirmed by Southern blot analysis (Fig. 1D) and long-range structural analysis (Fig. 1C).
Expression of the human β-like globin genes in the ΔGAP TgM
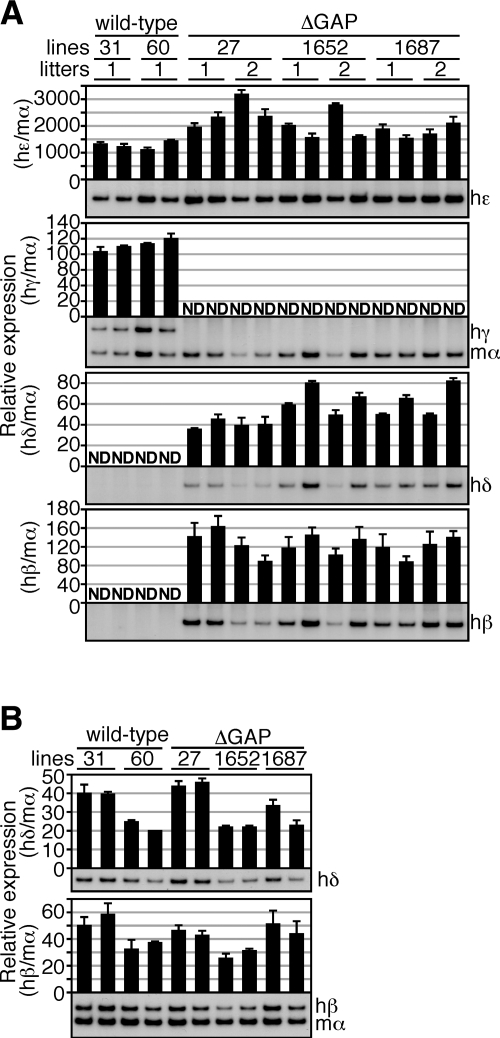
In primitive erythroid cells of yolk sacs (9.5 dpc) of human β-globin YAC TgM, the ε- and both of the γ-globin genes are expressed, whereas the δ- and β-globin genes are silent (wild-type in Fig. 2A). To test the effects of deleting both γ-globin gene promoters, we analyzed human β-like globin genes expression in ΔGAP TgM. Semiquantitative RT-PCR analysis of yolk sac RNA revealed that the human Gγ- and Aγ-globin genes were not transcribed, as expected, whereas the human δ- and β-globin genes were precociously expressed even at the LCR-distal position within the locus (Fig. 2A). These results demonstrated that the human γ-globin genes competitively repress δ- and β-globin gene expression in primitive erythroid cells. It should be noted that the expression level of the human ε-globin gene in the ΔGAP TgM seemed to be slightly increased when compared to that in wild-type TgM (Fig. 2A), implying that ε-globin gene may be also slightly competitively repressed by human γ-globin expression in the yolk sac.
Figure 2.
Expression of the human β-like globin genes in ΔGAP TgM. A) Total RNA was prepared from the yolk sacs of >2 embryos (9.5 dpc) derived from the intercross of male TgM and female wild-type animals. Samples were collected from 2 independent litters of each mutant line. Expression of human ε (hε)-, γ (hγ)-, δ (hδ)-, and β (hβ)-globin compared to endogenous mouse α (mα)-globin genes was separately analyzed by semiquantitative RT-PCR. Signals for hε-globin at 18 cycles, hγ/mα-globin at 12 cycles, hδ-globin at 24 cycles, and hβ-globin at 18 cycles were quantified by PhosphoImager, and ratios of hε/mα, hγ/mα, hδ/mα, and hβ/mα were calculated (mα signal at 12 cycles was set at 100%). B) Total RNA was prepared from the spleens of 1- to 2-mo-old anemic mice. Samples were collected from 2 individuals from each line of TgM. Expression of hδ- and hβ-globin compared to endogenous mα-globin genes was separately analyzed by semiquantitative RT-PCR. Signals for hδ-globin at 16 cycles and hβ/mα-globin at 12 cycles were quantified by PhosphoImager, and ratios of hδ/mα and hβ/mα were calculated (mα signal at 12 cycles was set at 100%). Averages ± sd from ≥3 independent experiments were calculated and are graphically depicted. Representative results are shown at bottom of each panel.
We then analyzed human δ- and β-globin gene expression in definitive erythroid cells. Two animals from each line were made anemic by phenylhydrazine treatment, and RNA samples from spleen were analyzed by semiquantitative RT-PCR (Fig. 2B). Although expression levels of the human δ- and β-globin genes were variable among lines, no significant differences were observed between wild-type and ΔGAP TgM.
Expression of the human β-like globin genes in the ΔEGAP TgM
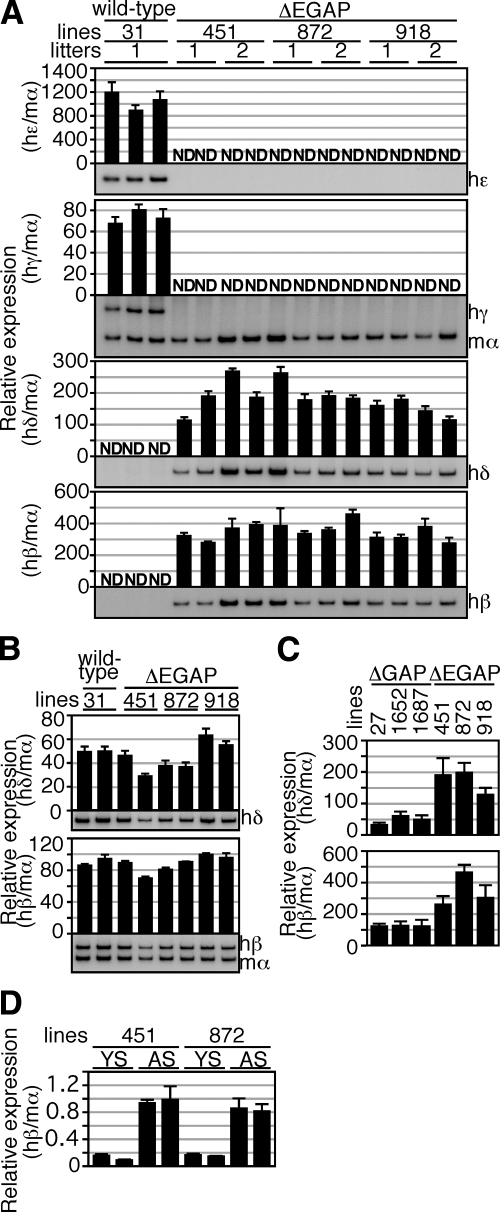
To explore a possible role for the human ε-globin gene in the competitive regulation of human β-like globin genes in primitive erythroid cells, we generated a ΔEGAP TgM, which carried a deletion in the ε-globin promoter region in addition to those in the ΔGAP TgM (Fig. 1A). RT-PCR analysis of RNA from the primitive erythroid cells revealed that human δ- and β-globin genes were precociously expressed, when human ε-, Gγ-, and Aγ-globin genes expression was silenced in the ΔEGAP TgM (Fig. 3A). In the adult spleen, no significant differences in the expression of the human δ- and β-globin genes were observed between the wild-type and ΔEGAP TgM (Fig. 3B). We then compared the level of human δ- and β-globin gene expression in primitive erythroid cells between ΔEGAP and ΔGAP TgM. As shown in Fig. 3C, expression of both genes in ΔEGAP TgM was significantly higher (∼2- to 3-fold) than that in ΔGAP TgM. These results demonstrated that the human ε-globin gene is also involved in competitive repression of the δ- and β-globin genes transcription in primitive erythroid cells of wild-type TgM.
Figure 3.
Expression of the human β-like globin genes in ΔEGAP TgM. A) Total RNA was prepared from the yolk sacs of >2 embryos (9.5 dpc) from 2 independent litters of each mutant line. Expression of hε-, hγ-, hδ-, and hβ-globin compared to endogenous mα-globin genes was separately analyzed by semiquantitative RT-PCR, as described in Fig. 2A. B) Total RNA was prepared from the spleens of 1- to 2-mo-old anemic mice. Samples were collected from 2 individuals from each line of TgM. Expression of hδ- and hβ-globin compared to endogenous mα-globin genes was separately analyzed by semiquantitative RT-PCR, as described in Fig. 2B. C) Comparison of human β-like globin genes expression in the yolk sacs of ΔGAP and ΔEGAP TgM. RNA samples were analyzed by semiquantitative RT-PCR. Averages ± sd of ratios of hδ/mα (top) and hβ/mα (bottom) from 4 individuals of each mutant line were calculated and are graphically depicted. D) Comparison of hβ-globin gene expression in between the yolk sac (YS) and adult spleen (AS) of ΔEGAP TgM. RNA samples were analyzed by real-time PCR. Averages ± sd of the ratio of hβ/mα from ≥3 independent experiments were calculated and are graphically depicted.
We then evaluated the level of human β-globin gene expression in primitive erythroid cells of ΔEGAP TgM by comparing it with that in definitive erythroid cells (Fig. 3D). The real-time PCR analysis showed that, when normalized by the endogenous mouse α-globin gene expression, human β-globin gene expression in primitive erythroid cells of the yolk sac was significantly lower (∼80%) than in definitive erythroid cells of the adult spleen. It should be noted that the difference between the two could be even larger, because the mouse ζ-globin gene, in addition to α-globin gene, is expressed to constitute mouse α-like globin proteins in primitive erythroid cells, and the level of α-globin mRNA at this stage is roughly half of total α-like globin mRNA abundance (21).
Expression of the human β-like globin genes in the ΔEGAP-inverted TgM
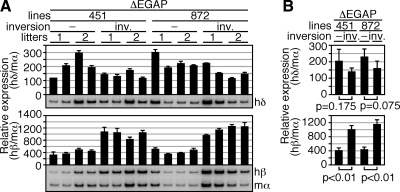
We have previously shown that human ε-globin gene expression in primitive erythroid cells of the TgM is sensitive to its distance from the LCR (11). To test possible distance sensitivity of the human δ- and β-globin genes from the LCR in primitive erythroid cells, we generated an inverted locus by mating ΔEGAP TgM with Cre recombinase TgM (ΔEGAP-inverted TgM; Fig. 1A). In the ΔEGAP locus, distances from the LCR to the δ- and β-globin genes were 40 and 47 kb, respectively, and these were shortened to 14 and 6.9 kb, respectively, in the ΔEGAP-inverted locus (Fig. 1A and not shown). RT-PCR analysis of the YS samples revealed that human δ-globin gene expression was slightly (but not significantly) diminished in primitive erythroid cells (Fig. 4A, B). This result probably represented a combined effect of altering its distance from the LCR and a competitive disadvantage compared to the (closer to the LCR) β-globin gene. On the other hand, the abundance of β-globin gene expression was significantly elevated in the ΔEGAP-inverted locus (2- to 3-fold). The extent of distance-sensitivity of the β-globin gene, however, appeared to be much less than that of the ε-globin gene, whose expression was dramatically diminished (>90% reduction) by extending the distance from the LCR by only 2.3 kb (from 5.8 to 8.1 kb) (11). It must be noted that the inversion of the locus not only alters the distance between the gene and the LCR but also induces other changes, such as the orientation of β-globin gene promoter relative to the LCR, which could also affect transcriptional activity.
Figure 4.
Expression of the human β-like globin genes in ΔEGAP-inverted TgM. A) Total RNA was prepared from the yolk sacs of >2 embryos (9.5 dpc) from 2 independent litters of each mutant line. Expression of hδ- and hβ-globin compared to endogenous mα-globin genes was separately analyzed by semiquantitative RT-PCR as described in Fig. 2A. −, noninverted; inv., inverted. B) Comparison of human β-like globin gene expression between the yolk sacs of ΔEGAP and ΔEGAP-inverted TgM. RNA samples were analyzed by semiquantitative RT-PCR. Averages ± sd of the ratios of hδ/mα (top) and hβ/mα (bottom) from 4 individuals of each mutant line were calculated and are graphically depicted.
Comparison of the spatial organization between ΔEGAP and wild-type human β-globin loci
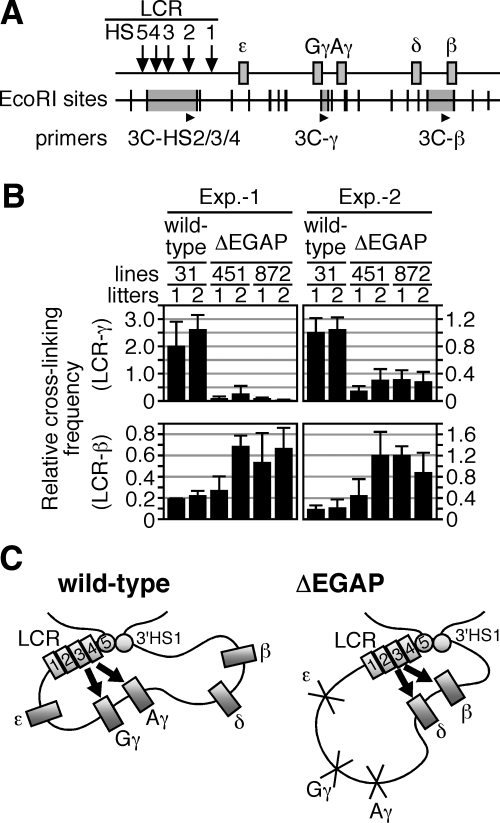
It has been reported that the LCR frequently interacts with the human γ-globin genes in primitive erythroid cells, whereas it contacts the β-globin gene in definitive red cells (18). To determine if precocious β-globin gene expression in the ΔEGAP TgM was accompanied by changes in the locus structure, we compared the spatial organization of ΔEGAP locus with that of the wild-type locus in primitive erythroid cells of TgM. The 3C analysis of the loci (Fig. 5A) revealed that the cross-linking frequency between the LCR and γ-globin gene region was dramatically reduced in ΔEGAP TgM, when compared with that in the wild-type TgM (Fig. 5B, top). In contrast, interactions between the LCR and the β-globin gene region were significantly more frequent in ΔEGAP than in wild-type TgM (Fig. 5B, bottom). These results implied that the promoter regions, which were deleted in the ΔEGAP locus, are mandatory for mediating the interaction between the LCR and the genes, and their physical proximity to the LCR is essential for the genes to be activated. We conclude that the physical interaction between the LCR and the γ-globin gene competitively interferes with that between the LCR and the β-globin gene, and thereby normally acts to suppress β-globin expression in the wild-type locus in primitive erythroid cells.
Figure 5.
Spatial organization of the transgenic human β-globin locus. A) Strategy of the 3C assay in the human β-globin locus. Arrows and gray boxes represent the HS sites and the human β-like globin genes, respectively. EcoRI restriction enzyme sites (vertical lines) and position of the primers used for real-time qPCR (arrowheads) are shown. The 3C-HS2/3/4 primer was designed in the EcoRI fragment (gray highlight) containing HS2–4 of the LCR; 3C-γ and 3C-β primers in the regions around Gγ/Aγ-globin genes and β-globin gene, respectively. B) Comparison of the spatial organization between wild-type and ΔEGAP human β-globin loci in primitive erythroid cells. Chromatin from embryonic blood cells (10.5 dpc) was cross-linked with formaldehyde, digested with EcoRI, and religated. After reversal of cross-linking, purified DNA was subjected to real-time qPCR to determine cross-linking efficiencies between the LCR and γ- (top) or β-globin (bottom) gene regions. To control for digestion and religation efficiencies, as well as the amount of template DNA, between separately processed samples, values were normalized to those obtained with unrelated mouse Ercc3 gene primers (19). Blood samples were collected from 2 independent sets of animals (Exp. 1 and 2). Averages ± sd from ≥4 independent PCR reactions were calculated and are graphically depicted. C) Model of competitive silencing of the β-like globin genes at primitive stage of erythropoiesis. Throughout development, the β-like globin genes, together with the LCR and 3′HS1 (numbered rectangles and circles) are thought to form an active chromatin hub structure (18). In primitive erythroid cells, the spatial organization of individual human β-like globin genes (gray rectangles) relative to the LCR is different between wild-type and ΔEGAP loci. In wild-type TgM (left panel), active γ-globin genes are proximal to the LCR, while δ- and β- globin genes are looped out and silenced. In contrast, in ΔEGAP TgM (right panel), because the interaction with γ-globin gene promoters is disrupted, the LCR can now contact δ- and β-globin genes and potentiate transcription of these genes. Arrows indicate activation by LCR.
DISCUSSION
Although extensive efforts have been made to clarify the mechanisms underlying selective gene activation within the human β-globin locus during definitive erythropoiesis (22), the transcriptional mechanisms employed in primitive erythroid cells has been less intensively studied. Although several studies using TgM have suggested the gene competition model to explain developmental stage-specific gene activation mechanism in the human β-globin locus (6,7,8,9, 23), the molecular details of gene competition are not fully understood. One of the leading hypotheses for globin gene activation by the LCR is a chromatin “looping” model, in which LCR-bound and promoter-bound proteins physically associate to form a complex with the intervening DNA looped out (24). Recently developed techniques, 3C (12) and RNA TRAP (25), revealed a spatial organization of the β-globin locus in erythroid cells that is consistent with the looping hypothesis. In primitive erythroid cells of human β-globin locus TgM, the LCR physically interacted with the γ-globin genes, suggesting that γ-globin genes are activated by a looping mechanism (18). In the current study, we showed that the adult β-globin gene could physically interact with the LCR and was activated even in primitive erythroid cells when interaction between the LCR and γ-globin gene promoters was disrupted (Figs. 2A and 5B). These results implied that, although the β-globin gene has potential to be activated via loop formation during primitive erythropoiesis, some physical constraints between the LCR and the γ-globin genes interfered with a productive interaction between the LCR and the β-globin gene in the wild-type locus (Fig. 5C).
It has not been made clear from the relevant literature whether the ε-globin gene competes for LCR enhancer activity against the other genes within the locus, probably because its transcriptional activity in YAC-TgM is quite low compared with that of the other genes (26), and thus any effects of further decreasing its expression level on other genes were difficult to quantify. In fact, in our previous work, we reported that increased γ-globin gene expression in primitive erythroid cells of TgM carrying mutant human β-globin loci was minuscule, even when the level of ε-globin gene expression was severely (∼10-fold) attenuated (11, 27). Therefore, it was not certain from those results whether the ε-globin gene was or was not under competitive control at this stage of development or whether the slight increase in the level of γ-globin gene expression was simply not detectable because of their intrinsically high expression level in the YAC TgM. In this current study, we explicitly tested the effect of removing ε-globin gene expression on expression of the δ- and β-globin genes in primitive erythroid cells of mutant TgM, levels of which are all reasonably low (Fig. 3D) so that any subtle change could be readily detected. By comparing ΔGAP and ΔEGAP TgM, we revealed that the human ε-globin gene participates in competitive repression of the human δ- and β-globin genes during primitive erythropoiesis (Fig. 3C). Thus, in primitive erythroid cells of human β-globin locus TgM, both the ε- and γ-globin genes contribute to repression of the adult β-like globin genes. It must be noted, however, that ε-globin is exclusively expressed in humans only in embryonic erythroid cells, whereas the other β-like globin genes, including the γ-globin genes, are silent. It is therefore conceivable that the ε-globin gene in the endogenous locus may be silencing the fetal γ-, as well as the adult δ/β-globin genes at this stage of erythropoiesis.
When genes are activated by the LCR over a long distance, they appear to physically interact with the LCR (16, 18, 25). In definitive erythroid cells of wild-type and mutant human β-locus TgM, in which distance between the LCR and β-globin gene was 48 and 6.9 kb, respectively, the level of β-globin gene expression was not significantly different (9). We showed, in this current work, that the β-globin gene could be precociously expressed in primitive erythroid cells of the ΔEGAP TgM (Fig. 3A), and that its level was not extremely elevated (up to 2- to 3-fold) even after the distance from the LCR was drastically reduced (ΔEGAP-inverted in Fig. 4). In contrast, we have shown in previous work that ε-globin, but not γ-globin, gene transcription in primitive erythroid cells was severely decreased (by 90%), when its distance to the LCR was increased by only 2.3 kb in YAC TgM (11). According to these results, while expression of the γ- and β-globin genes, which are almost certainly activated by a looping mechanism, seems to be insensitive to the distance from the LCR, the ε-globin gene may be activated by a distinct, distance-sensitive mechanism (24, 28,29,30,31,32). In this current work, however, we showed that the ε-globin gene promoter could interfere with β-globin gene activation in primitive erythroid cells (Fig. 3C). Assuming that the looping mechanism is the basis of gene competition, by this model the ε-globin gene should also physically interact with the LCR. How can this seemingly contradictory situation be resolved? First, we may need to reevaluate the distance-sensitive feature of ε-globin gene expression, which we demonstrated in our previous work (11). We used a 2.3-kb λ DNA fragment to artificially extend the distance between the LCR and genes. Although we showed no apparent repressor activity in the λ DNA fragment in cell transfection assays, the presence of such an activity in the TgM environment cannot be completely dismissed. Second, 2 features in the ε-globin gene expression, distance sensitivity from the LCR and participation in the gene competition, can also be explained by a “facilitated tracking” mechanism (33), in which the LCR (and its binding protein complexes) tracks along chromatin until it reaches the target gene promoters. Although some literature results are consistent with this model (34), a mechanism for ε-globin gene activation by the LCR in primitive erythroid cells still remains to be elucidated.
In summary, by once again employing the powerful strategy of examining the consequences of mutations in human β-globin locus TgM, we clearly showed that expression of δ- and β-globin genes is competitively silenced by ε- and γ-globin genes in primitive erythroid cells. In addition, it was suggested that ε- and the other β-like globin genes might be activated by the LCR by two distinct mechanisms, an interesting prospect from the point of view of β-globin locus evolution (35).
Supplementary Material
Acknowledgments
We thank Y. Tanimoto for technical assistance in generating TgM. This work was partially supported by research grants from the U.S. National Institutes of Health (HL24415 to J.D.E.) and the Ministry of Education, Culture, Sports, Science and Technology of Japan, Grant-in-Aid for Scientific Research (A) and for Young Scientists (S) (to K.T.).
References
- Stamatoyannopoulos G. Philadelphia/London: W. B. Saunders; The Molecular Basis of Blood Diseases. 1994 [Google Scholar]
- Tuan D, Solomon W, Li Q, London I M. The “beta-like-globin” gene domain in human erythroid cells. Proc Natl Acad Sci U S A. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosveld F, van Assendelft G B, Greaves D R, Kollias G. Position-independent, high-level expression of the human beta-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp Hematol. 2005;33:259–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raich N, Enver T, Nakamoto B, Josephson B, Papayannopoulou T, Stamatoyannopoulos G. Autonomous developmental control of human embryonic globin gene switching in transgenic mice. Science. 1990;250:1147–1149. doi: 10.1126/science.2251502. [DOI] [PubMed] [Google Scholar]
- Behringer R R, Ryan T M, Palmiter R D, Brinster R L, Townes T M. Human gamma- to beta-globin gene switching in transgenic mice. Genes Dev. 1990;4:380–389. doi: 10.1101/gad.4.3.380. [DOI] [PubMed] [Google Scholar]
- Enver T, Raich N, Ebens A J, Papayannopoulou T, Costantini F, Stamatoyannopoulos G. Developmental regulation of human fetal-to-adult globin gene switching in transgenic mice. Nature. 1990;344:309–313. doi: 10.1038/344309a0. [DOI] [PubMed] [Google Scholar]
- Dillon N, Trimborn T, Strouboulis J, Fraser P, Grosveld F. The effect of distance on long-range chromatin interactions. Mol Cell. 1997;1:131–139. doi: 10.1016/s1097-2765(00)80014-3. [DOI] [PubMed] [Google Scholar]
- Tanimoto K, Liu Q, Bungert J, Engel J D. Effects of altered gene order or orientation of the locus control region on human beta-globin gene expression in mice. Nature. 1999;398:344–348. doi: 10.1038/18698. [DOI] [PubMed] [Google Scholar]
- Hu X, Eszterhas S, Pallazzi N, Bouhassira E E, Fields J, Tanabe O, Gerber S A, Bulger M, Engel J D, Groudine M, Fiering S. Transcriptional interference among the murine beta-like globin genes. Blood. 2007;109:2210–2216. doi: 10.1182/blood-2006-06-029868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotsuma M, Matsuzaki H, Tanabe O, Campbell A D, Engel J D, Fukamizu A, Tanimoto K. Linear distance from the locus control region determines epsilon-globin transcriptional activity. Mol Cell Biol. 2007;27:5664–5672. doi: 10.1128/MCB.00602-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Tanimoto K, Liu Q, Bungert J, Engel J D. The polyoma virus enhancer cannot substitute for DNase I core hypersensitive sites 2–4 in the human beta-globin LCR. Nucleic Acids Res. 1999;27:3130–3137. doi: 10.1093/nar/27.15.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto K, Liu Q, Grosveld F, Bungert J, Engel J D. Context-dependent EKLF responsiveness defines the developmental specificity of the human epsilon-globin gene in erythroid cells of YAC transgenic mice. Genes Dev. 2000;14:2778–2794. doi: 10.1101/gad.822500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R N, Ito M, Saunders T L, Camper S A, Jameson J L. Role of Ahch in gonadal development and gametogenesis. Nat Genet. 1998;20:353–357. doi: 10.1038/3822. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra R J, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Vakoc C R, Letting D L, Gheldof N, Sawado T, Bender M A, Groudine M, Weiss M J, Dekker J, Blobel G A. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Palstra R J, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- Bungert J, Dave U, Lim K C, Lieuw K H, Shavit J A, Liu Q, Engel J D. Synergistic regulation of human beta-globin gene switching by locus control region elements HS3 and HS4. Genes Dev. 1995;9:3083–3096. doi: 10.1101/gad.9.24.3083. [DOI] [PubMed] [Google Scholar]
- Clouston W M, Evans B A, Haralambidis J, Richards R I. Molecular cloning of the mouse angiotensinogen gene. Genomics. 1988;2:240–248. doi: 10.1016/0888-7543(88)90008-0. [DOI] [PubMed] [Google Scholar]
- Kingsley P D, Malik J, Emerson R L, Bushnell T P, McGrath K E, Bloedorn L A, Bulger M, Palis J. “Maturational” globin switching in primary primitive erythroid cells. Blood. 2006;107:1665–1672. doi: 10.1182/blood-2005-08-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Han H, Xiang P, Li Q, Stamatoyannopoulos G. Autonomous silencing as well as competition controls gamma-globin gene expression during development. Mol Cell Biol. 2006;26:4775–4781. doi: 10.1128/MCB.00406-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanscombe O, Whyatt D, Fraser P, Yannoutsos N, Greaves D, Dillon N, Grosveld F. Importance of globin gene order for correct developmental expression. Genes Dev. 1991;5:1387–1394. doi: 10.1101/gad.5.8.1387. [DOI] [PubMed] [Google Scholar]
- Li Q, Peterson K R, Fang X, Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D, Chakalova L, Osborne C S, Dai Y F, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- Gaensler K M, Kitamura M, Kan Y W. Germ-line transmission and developmental regulation of a 150-kb yeast artificial chromosome containing the human beta-globin locus in transgenic mice. Proc Natl Acad Sci U S A. 1993;90:11381–11385. doi: 10.1073/pnas.90.23.11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto K, Sugiura A, Omori A, Felsenfeld G, Engel J D, Fukamizu A. Human beta-globin locus control region HS5 contains CTCF- and developmental stage-dependent enhancer-blocking activity in erythroid cells. Mol Cell Biol. 2003;23:8946–8952. doi: 10.1128/MCB.23.24.8946-8952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan D, Kong S, Hu K. Transcription of the hypersensitive site HS2 enhancer in erythroid cells. Proc Natl Acad Sci U S A. 1992;89:11219–11223. doi: 10.1073/pnas.89.23.11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe H L, Monks J, Wijgerde M, Fraser P, Proudfoot N J. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant K E, Routledge S J, Proudfoot N J. Intergenic transcription in the human beta-globin gene cluster. Mol Cell Biol. 2001;21:6507–6514. doi: 10.1128/MCB.21.19.6507-6514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- Engel J D, Tanimoto K. Looping, linking, and chromatin activity: new insights into beta-globin locus regulation. Cell. 2000;100:499–502. doi: 10.1016/s0092-8674(00)80686-8. [DOI] [PubMed] [Google Scholar]
- Blackwood E M, Kadonaga J T. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- Zhu X, Ling J, Zhang L, Pi W, Wu M, Tuan D. A facilitated tracking and transcription mechanism of long-range enhancer function. Nucleic Acids Res. 2007;35:5532–5544. doi: 10.1093/nar/gkm595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison R. Hemoglobins from bacteria to man: evolution of different patterns of gene expression. J Exp Biol. 1998;201:1099–1117. doi: 10.1242/jeb.201.8.1099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.