Abstract
Our recent study established the NMR structure of the recombinant bAα406-483 fragment corresponding to the NH2-terminal half of the bovine fibrinogen αC-domain and revealed that at increasing concentrations this fragment forms oligomers (self-associates). The major goals of the present study were to determine the structure and self-association of the full-length human fibrinogen αC-domains. To accomplish these goals, we prepared a recombinant human fragment, hAα425-503, homologous to bovine bAα406-483, and demonstrated using NMR, CD, and size-exclusion chromatography that its overall fold and ability to form oligomers are similar to those of bAα406-483. We also prepared recombinant hAα392-610 and bAα374-568 fragments corresponding to the full-length human and bovine αC-domain, respectively, and tested their structure, stability, and ability to self-associate. Size-exclusion chromatography revealed that both fragments form reversible oligomers in a concentration-dependent manner. Their oligomerization was confirmed in sedimentation equilibrium experiments, which also established the self-association affinities of these fragments and revealed that the addition of each monomer to assembling αC-oligomers substantially increases the stabilizing free energy. In agreement, unfolding experiments monitored by CD established that self-association of both fragments results in a significant increase of their thermal stability. Analysis of CD spectra of both fragments revealed that αC self-association results in the increase of regular structures implying that the COOH-terminal half of the αC-domain adopts ordered conformation in αC-oligomers and that this domain contains two independently folded sub-domains. Altogether, these data further clarify the structure of the human and bovine αC-domains and the molecular mechanism of their self-association into αC-polymers in fibrin.
Fibrinogen is a polyfunctional plasma protein that plays a prominent role in haemostasis and participates in wound healing, inflammation, angiogenesis, atherosclerosis, thrombosis, and other physiological and pathological processes. Fibrinogen is a chemical dimer consisting of two identical subunits, each composed of three non-identical polypeptide chains, Aα, Bβ and γ (1). The chains assemble to form a number of structural and functional domains that interact with various proteins and cell types thereby enabling fibrinogen participation in the above mentioned processes. The COOH-terminal portion of each fibrinogen Aα chain forms a compact αC-domain attached to the bulk of the molecule with a flexible αC-connector (2-4). According to the current view, in fibrinogen, two αC-domains interact intramolecularly with each other and with the central region of the molecule, while in fibrin they switch to an intermolecular interaction to form αC-polymers, which are reinforced by covalent cross-linking with factor XIIIa (4, 5). Besides their contribution to the fibrin assembly process (5, 6), the αC-domains are involved in the initiation of fibrinolysis through their tPA- and plasminogen-binding sites (7, 8) and promote cell adhesion and migration through their RGD sequences (9, 10).
Although X-ray studies of fibrinogen crystals established the three-dimensional structure of more than two-thirds of the fibrinogen molecule, they failed to define any αC-domain structure and resulted in a conclusion that the αC-domains are disordered (11-13). Conversely, comparative studies of fibrinogen and its proteolytically modified variant, fragment X, through differential scanning calorimetry and electron microscopy revealed the presence of a compact structure in the αC-domains (2, 3, 14, 15). To directly test the structure of these domains, we expressed an Aα392-610 fragment corresponding to the human αC-domain in Escherichia coli (16). However, we were able to find conditions for refolding of this fragment only after we expressed and studied its bovine analogue containing residues bAα374-568 and a truncated variant of this analogue, bAα374-538 (17). After such conditions had been identified, we refolded all three recombinant fragments and demonstrated that they all contain compact structures (17). Since bovine bAα374-538 was the smallest fragment with a compact structure, it was selected for further structural studies by NMR. Analysis of the NMR data revealed a β-hairpin formed by the Cys423-Cys453 linked loop and suggested that the region next to this hairpin is also structured (18). Subsequent NMR study of a shorter bovine αC-fragment, bAα406-483, revealed a second loose β-hairpin in this region yielding a mixed parallel/antiparallel β-sheet structural motif (19). Since our ultimate goal is to establish the structure of the human αC-domain, whose sequence differs from that of the bovine αC-domain (17, 20), one of the goals of the present study was to characterize the structure of the corresponding human αC-domain fragments.
Our study with bovine bAα406-483 established that this fragment forms ordered oligomers in a concentration-dependent and reversible manner (19). The study also revealed that the structure of this fragment in oligomers is stabilized and intermolecular interactions causing bAα406-483 oligomerization are thermodynamically driven (18, 19). Based on these and other findings, we hypothesized that the interaction between monomeric units in bAα406-483 oligomers could be utilized for formation of αC-polymers in fibrin and these oligomers may mimic fibrin αC-polymers (19). However, bAα406-483 represents only about a half of the bovine αC-domain. Thus, to test these hypotheses it is necessary to demonstrate that the full-length bovine αC-domain, bAα374-568, as well as its human counterpart, hAα392-610, can also form ordered oligomers in a similar manner. This was another goal of the present study.
EXPERIMENTAL PROCEDURES
Preparation of the Recombinant Fibrinogen αC-Domains and Their Truncated Variants
Recombinant bAα374-568 and hAα392-610 fragments corresponding to the full-length bovine and human αC-domains, respectively, were expressed in Escherichia coli and subsequently purified and refolded by the procedures described previously (17). A truncated variant of the human αC-domain, hAα425-503 fragment, homologous to the previously characterized bovine bAα406-483 fragment (19), was expressed in Escherichia coli using the pET-20b expression vector (Novagen Inc.). The cDNA encoding this fragment was amplified by polymerase chain reaction using a plasmid carrying the full-length human αC region sequence (16, 17). The following oligonucleotides were used as primers: 5′-AGAGACATATGACTGGTAAAGAGAAGGTC-3′ and 5′-AGAGAAAGCTTTTACCAAGTGTCGAAGAAGGCAGC-3′. The forward primer incorporated the NdeI restriction site immediately before the coding region; the final three bases of the NdeI site, ATG, code for the fMet residue that initiates translation. The reverse primer included a TAA stop codon immediately after the coding segment, followed by a HindIII site. The amplified cDNA fragment was purified by electrophoresis in agarose gel, digested with NdeI and HindIII restriction enzymes, and ligated into the pET-20b expression vector. The resulting plasmid was used for transformation of DH5α and then B834(DE3) pLysS E. coli host cells. The cDNA fragment was sequenced in both directions to confirm the integrity of the coding sequence. The expressed hAα425-503 fragment was found in inclusion bodies, from which it was purified by the procedure described earlier (7). The purified fragment was refolded by slow dialysis from urea at 4 °C using the protocol described in (17), and the unfolded material was removed by size-exclusion chromatography performed at 4 °C on a Superdex 75 column equilibrated with TBS1 (20 mM Tris buffer, pH 7.4, with 150 mM NaCl) and 0.2 mM PMSF. The refolded fragments were concentrated to 1-2 mg/mL with a Centriprep 10 concentrator (Millipore), filtered through 0.2 μm filter unit, and stored at 4 °C.
Human 15N-labeled hAα392-610 and hAα425-503 fragments were expressed in Escherichia coli in minimal media supplemented with 15NH4Cl. The 15N-hAα425-503 fragment was subsequently purified and refolded from inclusion bodies as described above; the 15N-hAα392-610 fragment was purified and refolded as described earlier (17). Bovine 15N-labeled bAα374-538 and bAα406-483 fragments were prepared and refolded by the procedures described previously (18, 19). All refolded fragments were concentrated to ~3 mg/mL and dialyzed against 10 mM KPO4 buffer, pH 6.5, containing 150 mM NaCl and 10% D2O.
Protein Concentration Determination
Concentration of the recombinant hAα425-503 fragment was determined spectrophotometrically using extinction coefficient E280, 1% = 6.32 calculated from the amino acid composition with the equation: E280, 1% = (5,690W + 1,280Y + 120S-S)/(0.1 M), where W, Y and S-S represent the number of Trp and Tyr residues and disulfide bonds, respectively, and M represents the molecular mass (21, 22). Molecular mass of this fragment equal to 8,704 Da was calculated based on its amino acid composition. Note that this value takes into account the NH2-terminal fMet residue (see above) while the numbering of this fragment does not. The molecular masses and E280, 1% for the recombinant hAα392-610 and bAα374-568 fragments were determined previously (17).
NMR Data Collection and Structure Elucidation
NMR data were recorded using the 15N-labeled hAα392-610, hAα425-503, bAα374-538 and bAα406-483 fragments in 20 mM KPO4, pH 6.5, with 150 mM NaCl and 10% D2O. The NMR experiments were performed at fragment concentrations of ~3 mg/mL. All NMR spectra were recorded at 282 K on a Bruker DRX-600 MHz spectrometer equipped with a triple-resonance cryo-probe and Z-axis gradient as described previously (18, 19). Assignments were made by standard methods utilizing combined data obtained from the following experiments: 15N HSQC, sensitivity enhanced HNCO, C(CO)NH, H(CO)NH, HCCH-TOCSY, HN(CO)CACB, HNCACB, and HBHA(CO)NH (for description, see ref 23). Due to the large size of the disordered regions of the fragments and aggregation at greater than 3 mg/mL protein concentration, not all peaks could be assigned. The human 15N-hAα392-610 fragment aggregated more rapidly and gave broader, weaker signals than the corresponding bovine fragment, 15N-bAα374-538. Backbone 15N relaxation measurements of the 15N-hAα425-503 fragment were performed at 282 K as described previously (18).
Circular Dichroism Study
Circular dichroism (CD) measurements were made with a Jasco-810 spectropolarimeter. CD spectra of all recombinant αC-fragments in the indicated conditions were recorded using a 0.01-cm path-length quartz cuvette at 4 °C. Analysis of the CD spectra was performed using the secondary structure prediction program supplied with the spectropolarimeter, which is based on the previously published method (24). Thermally induced unfolding curves were obtained by monitoring the ellipticity at 225 nm while increasing the temperature at a rate of 1 °C/min with a Peltier type PFD-425S attachment. Unfolding experiments were performed in TBS using a 0.1-cm path-length quartz cuvette. All CD data were expressed as the mean residue ellipticity, [θ], in units of degrees square centimeter per decimole.
Size-Exclusion Chromatography
Analytical size-exclusion chromatography was used to analyze the aggregation state of the prepared recombinant αC-domain fragments. The experiments were performed with a fast protein liquid chromatography system (FPLC, Pharmacia) on a Superdex 75 column at flow rate of 0.5 mL/min and 4 °C. Typically, 50 μl of the fragment at different concentrations were loaded onto the column equilibrated with TBS or other buffer and followed by elution with the same buffer. Protein elution was monitored by measuring absorbance at 280 nm.
Analytical Ultracentrifugation
Samples for analytical ultracentrifugation were prepared by overnight dialysis of the hAα392-610, hAα406-503, and bAα374-568 fragments at the three indicated concentrations versus TBS. Sedimentation equilibrium experiments were performed in a Beckman Optima XL-A analytical ultracentrifuge (Beckman Instruments, Palo Alto, CA) equipped with absorbance optics and an An60 Ti rotor, as previously described (25, 26). Due to their increased absorbance, samples of all three fragments at the concentrations higher that 2 mg/mL were monitored in double-sector cells with 3 mm centerpieces; standard 12 mm centerpiece cells were used for samples at the concentrations of 2 mg/mL or lower. Data for each fragment concentration were collected at 6,000 rpm and 8,000 rpm, as 3 sequential scans at 3 hour intervals following a 24 hour equilibration period at 4 °C, then at 2 hour intervals following an 18 hour period at 4 °C at the higher rotor speed. Sedimentation equilibrium data were analyzed with HeteroAnalysis (version 1.1.28, J. W. Cole and J. W. Lary, Analytical Ultracentrifugation Facility, Biotechnology/Bioservices Center, University of Connecticut, Storrs, CT) to obtain weight-average molecular weights (Mw) and to characterize the self-association of each fragment with an isodesmic model.
Molecular Modeling
The homology modeling of the 3D structure of the human fibrinogen hAα425-503 fragment was carried out using the structure of the bovine bAα406-483 fragment (PBD entry 2JOR) as a template. Prior to the modeling, the sequences of the bovine and human fibrinogen αC-domain were aligned to evaluate the homology of the regions corresponding to the bAα406-483 and hAα425-503 fragments. The alignment was computed with the T-Coffee program package (27) running on the Tcoffee@igs server provided by Hewlett Packard computers and the Centre National de la Recherche Scientifique (28). The initial raw model of hAα425-503 was built manually on the computer graphics. After the substitution and preliminary adjustment of non-matching side chains, which was done using rotamer library approach with PROPAK (29), the model was subjected to the refinement by two steps molecular dynamics at a constant temperature with CNS (30). At the first step of dynamics the harmonic potential restraints were applied to all atoms except for substituting side chains and the areas around deletions within two residues margins. The second step was performed with all the restraints removed and followed by the final refinement with conjugate gradient energy minimization.
RESULTS
NMR Study of the Human αC-Domain Fragments
In our previous NMR study (18), we identified a disulfide-linked β-hairpin and an ordered region within the recombinant bAα374-538 fragment corresponding to the bovine fibrinogen αC-domain. However, the resonance ambiguity arising from the disordered portions of this fragment precluded complete structural definition of the ordered region. We overcame this problem by recombinantly removing the disordered portions and establishing the complete NMR structure of the resultant bAα406-483 fragment (19). In the present study, we used the same approach to investigate the structure of the human αC-domain. Specifically, we prepared the 15N-labeled hAα392-610 fragment corresponding to the full-length human αC-domain and a truncated variant of this fragment, hAα425-503, homologous to its bovine counterpart, bAα406-483, and examined their structure by NMR techniques.
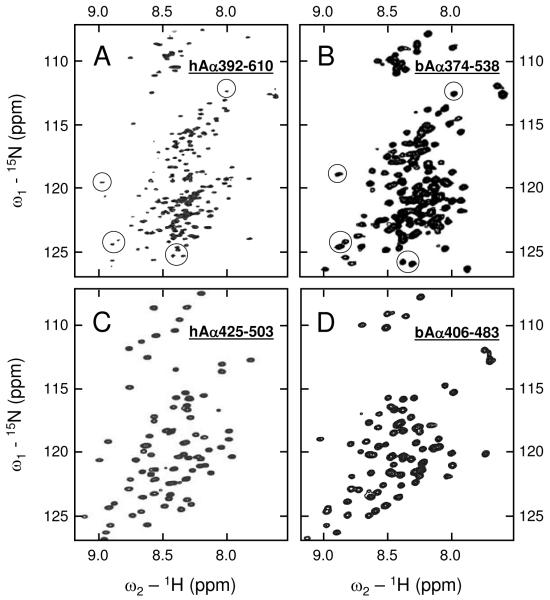
Analysis of 15N-HSQC spectrum of the human 15N-hAα392-610 fragment revealed a large number of sharp intense signals between 7.9 and 8.5 1H ppm characteristic of a random coil structure, as well as numerous broader signals outside the random coil region indicating the presence of a well ordered conformation (Figure 1A). This spectrum resembles that of the bovine 15N-bAα374-538 fragment (Figure 1B) whose structure was previously characterized (18). Particularly, several isolated peaks of 15N-hAα392-610 occur at similar positions as those assigned in 15N-bAα374-538 to its first disulfide-linked β-hairpin (circled in Figure 1A and B). This observation alone does not prove that the two fragments adopt similar β-hairpin conformations because HSQC resonance position is sensitive to local geometry as well as nearest neighbor amino acid residues. However, since the amino acid sequences of these two fragments are highly conserved around the β-hairpin stabilizing disulfide (18), comparison of these spectra suggests that the bovine and human αC-domains may have similar structure.
Figure 1.
1H-15N HSQC NMR spectra of the hAα392-610 (panel A), bAα374-538 (panel B), hAα425-503 (panel C), and bAα406-483 (panel D) fragments. Spectrum A was taken with slightly smaller spectral width in the 15N dimension in an attempt to enhance peak resolution. Several peaks assigned to the first disulfide-linked β-hairpin in bAα374-538 are circled in panel B; those occurring at similar positions in hAα392-610 are circled in panel A.
The 15N-HSQC spectrum of the truncated human fragment, 15N-hAα425-503, had the same general quality as that of the bovine 15N-bAα406-483 fragment (Figure 1, panels C and D), whose NMR solution structure was established earlier (19). Again, as in the case with the larger fragments, the peak dispersion of the two spectra are similar suggesting that this human fragment has a compact structure that may be similar to that of the bovine fragment. To identify this structure, we prepared and analyzed the 15N- and 13C-labeled hAα425-503 fragment. However, due to the large disordered regions of the fragment (signal overlap) and the low protein concentrations (weak signal intensity) required to avoid aggregation, we were unable to directly correlate many of the NMR spectral data to the respective amino acid residues. The majority of unassigned resonances are associated with side-chain atoms; this lack of inter-residue spatial data precluded the three-dimensional structure determination of the fragment. At the same time, closer inspection of the spectra in Figure 1C and D shows that human hAα425-503 has at least similar resonance dispersion in the downfield 1H region, compared to the bovine counterpart. This general feature suggests that structured regions are present in this fragment.
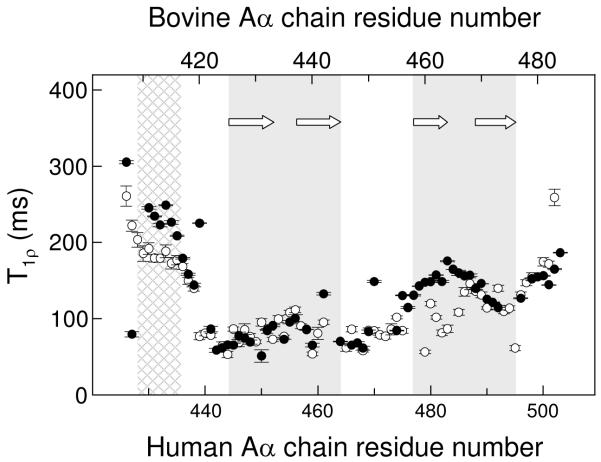
To further characterize human hAα425-503, the dynamical characteristics were then evaluated. 15N NMR backbone T1ρ relaxation experiments conducted on 15N-hAα425-503 revealed that most of its residues corresponding to the first β-hairpin in bovine bAα406-483 have T1ρ values of about 50-100 ms (filled circles in Figure 2) indicating slow concerted motion. The human fragment residues corresponding to the second β-hairpin in the bovine fragment have higher T1p values, about 110-160 ms; at the same time, these values are significantly lower than those for the residues at the termini. These T1ρ values and thereby motion among the residues of the hAα425-503 fragment can be correlated to the distribution of T1p values, according to the two β-hairpin locations, in the previously studied bovine 15N-bAα406-483 fragment (empty circles in Figure 2). This finding further reinforces the above suggestion that the bovine and human fragments may have similar structure. It should be noted that the average T1p value for residues 477-495 of human hAα425-503 is higher than that for the corresponding residues of bovine bAα406-483 forming the second β-hairpin. This implies that residues corresponding to the second β-hairpin in the human fragment are more mobile and thus form less stable conformation than that in the bovine counterpart.
Figure 2.
15N backbone relaxation data demonstrating similarity in relaxation and dynamics of the human hAα425-503 and bovine bAα406-483 fragments. T1p relaxation data for hAα425-503 and bAα406-483 are shown by filled and empty circles, respectively; vertical bars represent experimental errors. The regions corresponding to the previously identified first and second β-hairpins in the bovine bAα406-483 fragment (19) are shaded in gray, β-strands forming these β-hairpins are shown by arrows; the NH2-terminal region of slower motion in this fragment (19) is shown by cross-hatched pattern.
Altogether, the experiments described above suggest that the overall fold of the human hAα425-503 and bovine bA406-483 fragments is similar. This means that hAα425-503, like its bovine counterpart bAα406-483 (19), should contain two β-hairpins forming a mixed parallel/antiparallel β-sheet. However, the stability of the second β-hairpin in hAα425-503 may be lower than that in bAα406-483 precluding its structural determination in this study. To further test these speculations, we studied the structure and stability of the hAα425-503 fragment by circular dichroism (CD).
CD Study of the Structure and Stability of the Human hA α425-503 Fragment
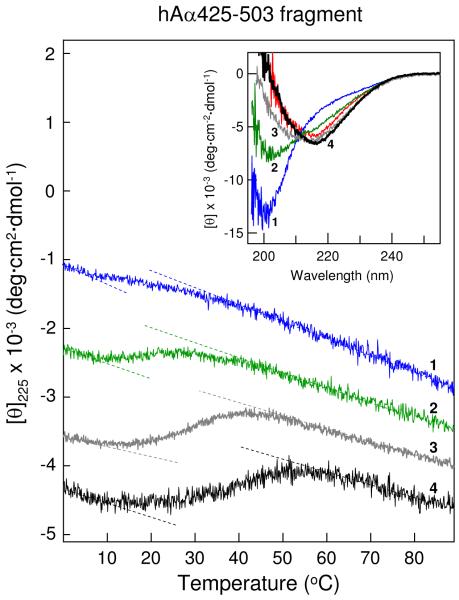
In our previous study, the bovine bAα406-483 fragment at a concentration of 3 mg/mL, at which the NMR experiments were performed, exhibited CD spectra with a negative band at 213-217 nm, characteristic of β-sheet structures (19). In contrast, CD spectra of the human hAα425-503 fragment at a similar concentration, 3.2 mg/mL, contained a negative maximum at about 200 nm characteristic for random coil and a slight shoulder at 210-220 nm (Figure 3, inset, blue spectrum 1). Analysis of this spectrum using the secondary structure prediction program supplied with the CD instrument revealed 58% regular structures (44% β-sheets and 14% turns) and 42% random coil. For comparison, the analysis of the CD spectrum of bAα406-483 (Figure 3, inset, red spectrum) revealed only 25% random coil. Thus, the shoulder may reflect the presence of the first disulfide-linked β-hairpin while the negative maximum may be connected with the destabilized and partially disordered second β-hairpin. When the hAα425-503 fragment at 3.2 mg/mL was heated in the spectropolarimeter while the ellipticity at 225 nm was monitored, it exhibited a weak sigmoidal transition with a midpoint (Tm) at 21.3 °C (Figure 3, blue curve 1). This is again in contrast to bAα406-483, which in the previous study (19) exhibited a well pronounced transition with Tm = 26.8 °C. At the same time, addition to the buffer of NaCl, which was previously found to stabilize the αC-domain fragments (17), resulted in a dramatic transformation of CD spectrum of hAα425-503 (Figure 3, inset). Namely, in 1 M NaCl the intensity of its negative band occurring at 200 nm decreased and that of the negative shoulder at 210-220 nm increased (green spectrum 2); in 2 M NaCl (grey and black spectra 3 and 4), the negative band disappeared, the shoulder was transformed into a negative band with a maximum at 217 nm, and the spectrum became very similar to that of bAα406-483. In agreement, the secondary structure prediction analysis of spectrum 4 revealed that the content of random structures decreased and became similar to that determined for bAα406-483. Further, when hAα425-503 at 3.4 mg/mL was heated in 2 M NaCl, it exhibited well pronounced sigmoidal transitions with Tm = 33.9 °C; at lower concentration, 1.6 mg/mL, Tm was shifted to 29.6 °C (Figure 3, curves 3 and 4, and Table 1). Altogether, these results indicate that the human hAα425-503 fragment is less stable than its bovine counterpart, bAα406-483, in agreement with the above speculation, and that increasing concentrations of NaCl stabilize its structure.
Figure 3.
CD-detected thermal unfolding of the hAα425-503 fragment. The unfolding experiments were performed in 20 mM Tris buffer, pH 7.4, containing 0.15 M NaCl at 3.2 mg/mL hAα425-503 (blue curve 1) and in the same buffer containing either 1 M NaCl at 3.2 mg/mL hAα425-503 (green curve 2) or 2 M NaCl at 1.6 mg/mL and 3.4 mg/mL hAα425-503 (gray curve 3 and black curve 4, respectively). The unfolding curves have been arbitrary shifted along the vertical axis to improve visibility; the dashed straight lines represent linear extrapolations of the CD values before and after transitions to highlight their sigmoidal character. Inset shows CD spectra of the hAα425-503 fragment obtained in the above mentioned conditions; the numbering and color coding of these spectra correspond to those of the unfolding curves. The CD spectrum of the bovine bAα406-483 fragment at 3.0 mg/ml in Tris-buffer, pH 7.4, containing 0.15 M NaCl is shown in red for comparison. All spectra were obtained at 4 °C.
Table 1.
CD-Detected Thermal Stability of the Recombinant αC-Domain Fragments and their Aggregation State Determined by Size-Exclusion Chromatography
| Conditions |
||||
|---|---|---|---|---|
| Fragmenta | NaCl (M) | Fragment conc. (mg/ml) |
Tm (°C) b | Oligomers (%)b |
| hAα425-503 | 0.15 | 3.2 | 21.3 ± 1.2 | 1.7 ± 0.6 |
| 0.15 | 6.3 | 24.4 ± 1.0 | 6.6 ± 0.4 | |
| 1.0 | 3.2 | 28. 4 ± 1.1 | 26.2 ± 0.1 | |
| 2.0 | 1.6 | 29.6 ± 1.3 | 43.6 ± 1.0 | |
| 2.0 | 3.4 | 33.9 ± 1.8 | 60.0 ± 0.5 | |
| hAα392-610 | 0.15 | 1.9 | 39.6 ± 0.8 | 15.4 ± 0.8 |
| 0.15 | 3.8 | 42.7 ± 0.8 | 32.0 ± 0.1 | |
| 2.0 | 4.0 | 46.4 ± 0.4 | 58.5 ± 2.3 | |
| bAα374-568 | 0.15 | 1.8 | 30.4 ± 0.9 | 9.5 ± 0.8 |
| 0.15 | 3.7 | 33.2 ± 0.1 | 27.2 ± 0.5 | |
| 2.0 | 3.8 | 40.3 ± 0.3 | 54.4 ± 0.6 | |
All fragments were in 20 mM Tris buffer, pH 7.4, containing the indicated concentrations of NaCl.
Values are means ± the standard deviation of at least two independent experiments.
Oligomerization of the Human hA α425-503 Fragment
Since the bovine bAα406-483 fragment exhibited concentration-dependent and reversible oligomerization, we also tested the ability of human hAα425-503 to form oligomers. Size-exclusion chromatography experiments revealed that in TBS the hAα425-503 fragment at the concentrations of 3.2 and 6.3 mg/mL contained about 2% and 7% oligomers, respectively (Figure 4 A-B and Table 1). The detected fractions of oligomers were lower than those observed earlier for its bovine counterpart, bAα406-483, which exhibited at similar concentrations 8 and 13% oligomers, respectively (19). Oligomers formed by hAα425-503 eluted with similar elution volume (~ 8 mL) as those formed by bAα406-483 (19) suggesting the same number of monomers, five to six. In 2 M NaCl, in which the structure of hAα425-503 was stabilized, as mentioned above, this fragment at 1.6 and 3.4 mg/mL exhibited higher contents of oligomers, about 44 and 60%, respectively (Figure 4 C-D and Table 1). Thus, the higher stability of hAα425-503 in 2 M NaCl may be explained by the increase of its oligomeric fraction. This suggests that the stabilizing effect of NaCl is connected with its ability to promote formation of hAα425-503 oligomers. Further, when hAα425-503 at 3.4 mg/mL in 2 M NaCl was dialyzed overnight versus TBS and then analyzed by size-exclusion chromatography, the fraction of oligomers substantially decreased (not shown). Altogether, the above results indicate that, similar to the bovine bAα406-483 fragment, human hAα425-503 forms oligomers in a concentration-dependent and reversible manner.
Figure 4.
Size-exclusion chromatography of the hAα425-503 fragment performed in various conditions. Panels A and B show elution profiles of hAα425-503 in Tris buffer, pH 7.4, containing 0.15 M NaCl, at 3.2 and 6.3 mg/mL, respectively. Panels C and D show elution profiles of hAα425-503 in the same buffer containing 2 M NaCl at 1.6 and 3.4 mg/mL, respectively. All experiments were performed at 4 °C using the Superdex 75 column; arrows indicate the free volume of the column.
Structure, Stability, and Oligomerization of the Recombinant Full-Length Bovine and Human αC-Domains
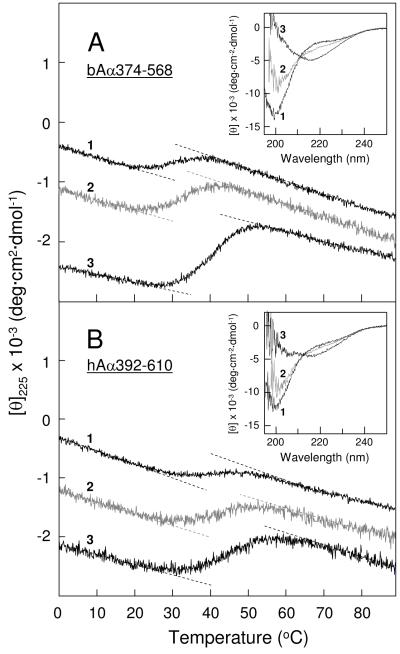
To test the structure and stability of the full-length bovine and human αC-domains and their ability to form oligomers, we studied the recombinant bAα374-568 and hAα392-610 fragments corresponding to these domains by CD and size-exclusion chromatography. The thermally-induced unfolding experiments revealed that the stability of both fragments increased with increasing concentrations or upon addition of 2 M NaCl. Specifically, bAα374-568 in TBS at 1.8 and 3.7 mg/mL exhibited unfolding transitions with Tm of 30.4 and 33.2 °C, respectively; in 2 M NaCl at 3.8 mg/mL Tm was shifted to 40.3 °C (Figure 5A, and Table 1). Human hAα392-610 exhibited similar unfolding transitions, although its thermal stability was higher than that of its bovine counterpart (Tm was 39.6 and 42.7 °C in TBS at 1.9 and 3.8 mg/mL, respectively, and 46.4 °C at 4 mg/mL in 2 M NaCl) (Figure 5B and Table 1). Size-exclusion chromatography experiments (not shown) revealed that in TBS bovine bAα374-568 contained about 10 and 27% oligomers at 1.8 and 3.7 mg/mL, respectively; the amount of oligomers increased to ~54% in 2 M NaCl (Table 1). When bAα374-568 in 2 M NaCl was diluted to 1 mg/mL and dialyzed overnight versus TBS, the amount of oligomers substantially decreased (to 14%) suggesting that its oligomerization was reversible. The human hAα392-610 fragment exhibited similar tendency for the concentration- and NaCl-induced oligomerization (Table 1) and this oligomerization was also reversible. Thus, as in the case with the smaller bAα406-483 and hAα425-503 fragments, the observed thermal stabilization of both bAα374-568 and hAα392-610 correlated well with the increase of their oligomeric fractions. This further reinforces the above suggestion that the stabilizing effect of NaCl is connected with its ability to promote oligomer formation.
Figure 5.
CD-detected thermal unfolding of the full-length bovine and human αC-domain fragments. The unfolding of the bovine bAα374-568 fragment (panel A) was performed in 20 mM Tris-buffer, pH 7.4, containing 0.15 M NaCl, at the fragment concentrations of 1.8 and 3.7 mg/mL (curve 1 and 2, respectively), and in the same buffer containing 2 M NaCl at the fragment concentration of 3.8 mg/mL (curve 3). The unfolding of the human hAα392-610 fragment (panel B) was performed in Tris-buffer, pH 7.4, containing 0.15 M NaCl, at the fragment concentrations of 1.9 and 3.8 mg/mL (curve 1 and 2, respectively), and in the same buffer containing 2 M NaCl at the fragment concentration of 4.0 mg/mL (curve 3). The unfolding curves have been arbitrary shifted along the vertical axis to improve visibility; the dashed straight lines represent linear extrapolations of the CD values before and after transitions to highlight their sigmoidal character. Insets in both panels shows CD spectra of the corresponding fragments obtained in the above mentioned conditions at 4 °C; the numbering of these spectra correspond to that of the unfolding curves.
The bovine bAα374-568 fragment in TBS at 1.8 mg/ml exhibited a CD spectrum with a negative maximum at about 200 nm and a slight shoulder at 210-225 nm; at higher concentration, 3.7 mg/mL, the intensity of the negative maximum decreased while that of the shoulder increased (Figure 5A, inset). The spectrum further changed in 2 M NaCl, in which the negative maximum disappeared and the shoulder at 210-225 nm was transformed into a negative maximum at 217 nm characteristic for β-sheet structures. A similar transformation of a CD spectrum was observed with the human hAα392-610 fragment (Figure 5B, inset). This transformation is reminiscent of that observed with the human hAα425-503 fragment (Figure 3, inset), however, the interpretation of these changes may be different. Namely, since in the bovine bAα374-568 and bAα406-483 fragments both β-hairpins are folded (18, 19), the negative maximum at 200 nm may be connected with the unordered COOH-terminal half of bAα374-568. The disappearance of this maximum and appearance of that at 217 nm may reflect formation of regular structures (most probably β-sheets) by this half in concentration- and NaCl-induced bAα374-568 oligomers. In agreement, analysis of the CD spectra of bAα374-568 obtained in TBS and 2 M NaCl using the secondary structure prediction program revealed that their transformation is accompanied by the increase of regular structures from 53 to 66%. Similarly, the analysis of hAα392-610 CD spectra obtained in TBS and 2 M NaCl revealed the increase of regular structures from 47 to 61%. Altogether the above results suggest a significant increase of regular structures in the bAα374-568 and hAα392-610 fragments upon their oligomerization.
Sedimentation Equilibrium Study of the αC-Domain Fragments
To further characterize oligomerization of the full-length bovine and human αC-domain fragments we performed analytical ultracentrifugation. The results of sedimentation equilibrium experiments with the human hAα392-610 fragment are presented in Figure 6 A-C, which depicts the concentration gradients obtained during sedimentation equilibrium at three fragment concentrations, each at two rotor speeds. Molecular weight determinations with HeteroAnalysis using these data yielded a value of 266,910. This value indicates that hAα392-610 forms oligomers consisting, on average, of 11 monomers (Table 2). These data were also used to determine the equilibrium association constant (Ka) for self-association of hAα392-610 using the isodesmic association model Mn−1 + M = Mn, where Ka is the same for all species if n > 2 (31, 32). The resultant Ka of 8.32 × 104 M−1 was utilized to calculate the equilibrium dissociation constant (Kd) and the free energy of self-association (ΔG) using the equations Kd = 1/Ka and ΔG = −RT ln Ka, respectively. The calculated values (Table 2) indicate that the hAα392-610 fragment self-associates with the Kd = 12 μM and that addition of each monomer to an assembling hAα392-610 oligomer may add as much as 6.7 kcal/mol of the stabilizing free energy. The bovine bAα374-568 fragment exhibited similar behavior during sedimentation equilibrium (Figure 6 D-F) although its oligomerization parameters were found to be slightly different (Table 2).
Figure 6.
Analysis of oligomerization of the human hAα392-610 (panels A-C) and bovine bAα374-568 (panels D-F) fragments by analytical ultracentrifugation at 6,000 rpm (open circles) and 8,000 rpm (grey triangles). The experiments were performed in TBS at three concentrations of the fragments, 2.0, 4.1 and 8.1 mg/mL (panels A, B and C, respectively), and 1.8, 3.8 and 7.2 mg/mL (panels D, E and F, respectively). The data presented in panels A and D were obtained in a 12 mm path length cell, whereas those presented in panels B, C, E and F were obtained at higher concentrations in 3 mm path length cells. Sedimentation equilibrium data for both fragments obtained at all three concentrations demonstrate oligomerization. HeteroAnalysis of the data obtained for the hAα392-610 (A-C) and bAα374-568 (D-F) fragments yielded Mw = 266,910 and 179,029, respectively; the solid lines describe an isodesmic self-association model with Ka = 8.32 × 104 M−1 and 2.61 × 104 M−1 for hAα392-610 and bAα374-568, respectively. This model accounts for both rotor speed and concentration-dependence as evidenced by the correspondence between data and fitted lines in all panels.
Table 2.
Oligomerization Parameters of the Recombinant αC-Domain Fragments Determined by Analytical Ultracentrifugationa
| Fragment | Mo calc.b | Mw obs.c,d | Mo/Mw | Ka (M−1) |
Kd (μM) |
ΔGd (kcal/mol) |
|---|---|---|---|---|---|---|
| hAα392-610 | 23,582 | 266,910 ± 1,504 | 11.3 | 8.32 × 104 | 12.0 | −6.69 ± 0.01 |
| bAα374-568 | 21,334 | 179,029 ± 1,029 | 8.4 | 2.61 × 104 | 38.3 | −6.00 ± 0.01 |
| hAα425-503 | 8,704 | 30,612 ± 2,455 | 3.5 | 0.88 × 103 | 1,136 | −4.00 ± 0.18 |
| bAα406-483e | 8,960 | 52,290 ± 990 | 5.8 | 5.28 × 103 | 188.7 | −4.98 ± 0.04 |
All experiments were performed in TBS at 4 °C.
Molecular weight of monomeric fragments calculated from their amino acid sequence that includes NH2-terminal Met.
Experimentally observed molecular weight of oligomeric fractions.
Values are means ± the standard deviation based on a global fit of data obtained at two rotor speeds and three fragment concentrations.
Data for the bAα406-483 at 4 °C were obtained earlier (19).
Similar experiments were also performed with the smaller human αC-fragment, hAα425-503. Analysis of the sedimentation equilibrium data obtained at three hAα425-503 concentrations, 1.5, 3.0 and 6.3 mg/mL, each at two rotor speeds, 6000 and 8000 rpm (not shown) resulted in Ka = 0.88 × 103 M−1 and ΔG = −4.0 kcal/mol (Table 2). The much lower resultant values of Ka and ΔG for this fragment compared to those determined for larger hAα392-610 correlate well with its lower stability and tendency to self-associate (Table 1). It should be noted that the previously determined ΔG and Ka values for its bovine counterpart, the bAα406-483 fragment (19), were also lower than those for bAα374-568 (Table 2). Altogether, the above results indicate that both human and bovine full-length αC-domain fragments, hAα392-610 and bAα374-568, have higher self-association affinity and tendency for oligomerization than their truncated variants, hAα425-503 and bAα406-483.
DISCUSSION
In our previous studies, we expressed various fragments of the human and bovine fibrinogen αC-domains, tested their folding status, determined the NMR solution structure of one of these fragments, bAα406-483, and characterized its self-association (oligomerization) (17-19). These studies confirmed the presence of ordered structures in the NH2-terminal half of the αC-domains and provided some clue about the mechanism of their self-association; however, the structure and interaction of the full-length αC-domain remained unclear. The major goals of the present study were to clarify the structure of the full-length human and bovine αC-domains and the molecular mechanism of their polymerization in fibrin.
Although human and bovine fibrinogens have high sequence homology and their overall fold determined by X-ray analysis is similar (13, 33), the sequence homology of their αC-domains, whose three-dimensional (3D) structure have not been identified by X-ray, is lower and their sizes are different due to a number of deletions in the bovine species (17, 20) (Figure 7A). From this arises a question of how the observed difference in the amino acid sequence of the human and bovine αC-domains affects their 3D structure. To address this question, we first expressed the human hAα425-503 fragment and compared its structure and properties with those of the previously studied bovine bAα406-483 fragment (19). Although we were unable to identify the NMR structure of hAα425-503 due to the reasons described in the previous section, the similarity of some of the resonances in its 15N-HSQC spectrum and T1ρ values to those of bovine bAα406-483 (19) strongly suggests that their overall fold is similar. In agreement, the hAα425-503 fragment formed oligomers in a concentration-dependent and reversible manner, i.e. exhibited behavior similar to that of bAα406-483. Furthermore, our CD experiments revealed that although monomeric hAα425-503 is less stable and more disordered than bovine bAα406-483, upon oligomerization its CD spectrum becomes very similar to that of bAα406-483. Altogether, these data strongly suggest that both hAα425-503 and bAα406-483 have similar 3D structures.
Figure 7.
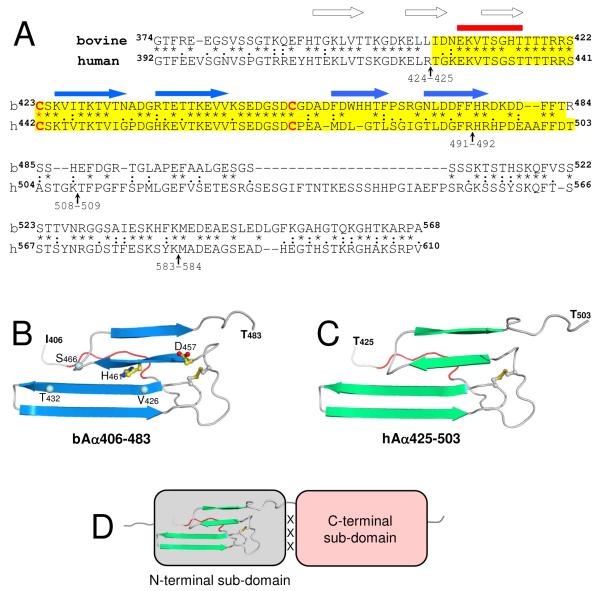
Structural organization of the fibrinogen αC-domain. Panel A, alignment of the human and bovine fibrinogen αC-domain sequences2 performed as in (27). The degree of conservation is represented by asterisks, colons and dots, which denote identical residues, conserved and semi-conserved substitutions, respectively; the sequences corresponding to the bovine bAα406-483 and human hAα425-503 fragments are highlighted in yellow; vertical arrows indicate the identified plasmin cleavage sites (1); locations of the NH2-terminal region of slower motion (see Figure 2 and ref. 19) and β-sheet strands identified in bAα406-483 (19) are shown by red horizontal bar and blue horizontal arrows, respectively. Location of predicted β-strands3 in the bovine Aα374-422 region is shown by empty arrows. Panel B, the ribbon diagram of the bovine bAα406-483 fragment based upon its NMR structure (19); arrows indicate β-strands, the region of slower motion is shown in red. Asn457 and His461 missing in the human sequence are shown by balls and sticks; the location of Val426, Thr432, and Ser461 mentioned in the text are shown by blue balls. Panel C, the homology model of the human hAα425-503 fragment built based on the 3D structure of bovine bAα406-483. Panel D, schematic representation of the αC-domain consisting of the N-terminal sub-domain, which includes the previously identified β-sheet (19), and the C-terminal sub-domain whose structure is not established yet; interaction between these domains (see text) is denoted by ‘XXX’. Figures on panels B and C were prepared using PyMOL (38).
To further test this suggestion, we analyzed the sequences of the human hAα425-503 and bovine bAα406-483 fragments (highlighted in yellow in Figure 7A) and performed homology modeling of the 3D structure of hAα425-503 using the structure of bAα406-483 fragment as a template. The sequences in the area of the first β-hairpin restricted at the base by the disulfide linkage (Cys423-Cys453 and Cys442-Cys472 in the bovine and human species, respectively) display more than 90% homology implying unambiguously a nearly identical fold. The sequences in the area of the second loose β-hairpin are less conserved and the human sequence contains two single residue deletions (Figure 7A). One of these deletions, Asp457 (bovine numbering), is located before the second β-hairpin identified in the bovine fragment (Figure 7B) and may result in shortening of the turn between the disulfide bridge and the first β-strand of this hairpin. The second deletion, corresponding to bovine His461, shortens this strand itself. Nevertheless, both deletions allow modeling the structure of the second β-hairpin in human hAα425-503 without introducing significant distortions to the template. The homology model of the hAα425-503 fragment built as described in Experimental Procedures is presented in Figure 7C. This model confirms the structural similarity between the bovine and human fragments. It is also in agreement with the observed reduced structural stability of the human hAα425-503 fragment, which may be connected with the deletions discussed above. Namely, the deletion of His461 and shortening of the β-strand should affect the previously identified interactions between His461and Val426 and between Ser466 and Thr432 (19) (Figure 7B). Since these interactions are involved in stabilizing the β-sheet in bovine bAα406-483 (19) their weakening or elimination in human hAα425-503 may contribute to the reduced stability of the latter.
Our finding that the bAα406-483 fragment, as well as hAα425-503, is folded into compact β-sheet structure suggests that the corresponding NH2-terminal portion in the bovine and human αC-domain is folded independently, i.e. represents an independently folded structural unit or domain. In contrast, our previous NMR study with the monomeric bovine bAα374-538 fragment suggests that its COOH-terminal portion is disordered (18). The similarity of 15N-HSQC spectrum of the monomeric human hAα392-610 fragment with that of bovine bAα374-538 (Figure 1 A-B), as well as the similarity of their CD spectra (Figure 5, insets), implies that in the monomeric human αC-domain this portion is also disordered. At the same time, CD experiments also revealed that formation of oligomers by both bAα374-568 and hAα392-610 results in a dramatic transformation of their CD spectra indicating decrease of random coil and formation of additional regular structures, most probably β-sheets. Furthermore, analysis of these spectra suggests a substantial increase in regular structures, up to 66%. Such changes cannot be attributed to only the NH2-terminal portion, which represents les than half of the αC-domain, and therefore imply that the COOH-terminal portion also adopts regular conformation upon oligomerization and, similarly to the NH2-terminal portion, may also form an independently folded domain. To reflect the presence of two domains in the αC-domain while preserving the recommended fibrin(ogen) nomenclature (34), we propose to denote them as N- and C-terminal sub-domains (Figure 7D).
The existence of two sub-domains in the αC-domain is in agreement with the results of proteolytic degradation of fibrin(ogen) by plasmin. It was reported that in spite of the presence of more than 20 potential plasmin cleavage sites (Arg-X or Lys-X) in the Aα392-610 region of human fibrinogen only four of them, at Arg424, Arg491, Lys508 and Lys583 (shown by vertical arrows in Figure 7A), are cleaved by this enzyme (1). Furthermore, two potential resulting fragments of plasminolysis, Aα425-491 and Aα509-583, correspond to the major portions of the NH2- and COOH-terminal halves of the αC-domain, respectively, and the former includes practically all regular structure identified in the N-terminal sub-domain (Figure 7A). Since proteolytic cleavage usually occurs between compact protein domains and limited proteolysis is often used for testing domain structure of multidomain proteins, these observations further support the presence of two sub-domains in the αC-domain. It should be noted that although this and the previous study (18) did not identify ordered structures in the C-terminal sub-domain of the monomeric αC-domain fragments, this does not mean that this sub-domain is unfolded in fibrinogen or fibrin. Indeed, in fibrinogen the αC-domains interact intramolecularly to form a dimer while in fibrin they interact intermolecularly to form αC-polymers (2, 4). Such interactions may increase their stability and maintain folded structure of their C-terminal sub-domains.
Structural organization of the Aα chain region corresponding to the αC-domain has long been a matter of dispute. While some have suggested that this region is ordered and folded into a compact structure (2-4, 14, 15), others have argued that the region is mostly disordered and unfolded (11, 12, 20). The data presented in the current study, as well as our previous studies (17-19), clearly indicate that even in the isolated monomeric αC-domain - which, as mentioned above, is less stable due to the loss of interactions present in fibrinogen and fibrin - about two-thirds of its NH2-terminal half are folded into a compact N-terminal sub-domain (Fig. 7). One cannot exclude that the remaining one-third may contribute to the structure of this sub-domain. In agreement, secondary structure prediction using the Jnet algorithm (35) suggests that about half of this third may form two additional β-strands (Fig. 7A). It should be noted that this prediction also suggests that the region of slower motion, whose exact 3D structure has not been determined in our previous study (19), may also adopt β-strand conformation (Fig. 7A). The folding status of the C-terminal sub-domain is less defined. Our CD data discussed above suggest formation of regular structures in this sub-domain upon αC-domain self-assembly. However, these data do not allow us to accurately evaluate the content of these structures because the samples analyzed contained a mixture of monomeric and oligomeric αC-domains. Thus, the question of whether in fibrin(ogen) this sub-domain is ordered and compact or contains ordered and disordered regions remains to be answered. Further investigation of αC-oligomers described in the present study may help to address this question.
Another important finding of the current study is that hAα392-610 and bAα374-568 fragments form oligomers in a concentration-dependent and reversible manner. It should be noted that the oligomerization was observed both in TBS, which mimics physiological conditions, and at high concentrations of NaCl, which was used to increase the fraction of oligomers. Such character of oligomerization suggests that the interaction between αC-domains is specific and most probably mimics the interaction between the αC-domains upon formation of αC-polymers in fibrin. This also implies that the structure of αC-oligomers mimics that of fibrin αC-polymers. In our previous study of the bAα406-483 fragment we hypothesized that the mechanism of its oligomerization, which may include β-hairpin swapping, could be utilized for formation of αC-polymers in fibrin (19). The observed similarity of the oligomerization process of the full-length αC-domain fragments with that of bAα406-483 supports this hypothesis and highlights the role of the N-terminal sub-domain in this process.
The present study also demonstrates significant contribution of the C-terminal sub-domain to the structure and stability of αC-oligomers. Indeed, our results suggest that this sub-domain adopts ordered conformation in αC-oligomers, as mentioned above, and significantly increases the stability of both bovine and human αC-domains upon their oligomerization. Namely, the addition of each monomer to assembling bAα374-568 and hAα392-610 oligomers adds respectively 6.0 and 6.7 kcal/mol of the stabilizing free energy (Table 2). These values are much higher than those determined for the smaller bAα406-483 and hAα425-503 fragments (4.98 and 4.0 kcal/mol, respectively) lacking this sub-domain. The thermal stability of bAα374-568 and hAα392-610 is also much higher than that of the smaller fragments (Table 1). Furthermore, the fact that unfolding of these fragments starts at higher temperatures than that of the smaller fragments (Figure 3 and 5) suggests that the folded C-terminal sub-domain interacts with the N-terminal sub-domain and that this interaction increases the overall stability of the αC-domain.
It should be noted that thermal stability of the full-length human αC-domain is higher than that of its bovine counterpart. For example, heat-induced unfolding of the hAα392-610 and bAα374-568 fragments, both at similar low concentrations, occurs with Tm at 39.6 and 30.4 °C, respectively (Table 1). Such a difference cannot be explained by the higher fraction of oligomers in the human species (~15% in human versus ~10% in bovine) since bAα374-568 at higher concentration, at which ~27% oligomers was detected, still unfolds at lower temperature (Tm = 33.2 °C). Since the stability of the human hAα425-503 fragment was found to be lower than that of bovine bAα406-483, the higher overall stability of the full-length human αC-domain over the bovine one may be explained by a more significant stabilizing effect of its C-terminal sub-domain. In agreement, the bovine C-terminal sub-domain is shorter than its human counterpart due to the present deletions (Figure 7A) and therefore its own stability, as well as its stabilizing effect, may be lower. This finding further highlights the important role of the C-terminal sub-domain in stabilizing the overall structure of the αC-domain.
Our data indicate that the C-terminal sub-domain not only significantly contributes to the overall stability of the αC-domains but also increases their affinity to each other. Indeed, the value of the equilibrium dissociation constants for self-association of the full-length human and bovine αC-domain fragments, hAα392-610 and bAα374-568, were found to be 12 and 38.3 μM, respectively. These Kd values are much lower than those determined for self-association of the smaller hAα425-503 and bAα406-483 fragments (Table 2) and thus confirm contribution of the C-terminal sub-domain to the increased affinity of the full-length αC-domains. It should be noted that the Kd = 12 μM for human hAα392-610 is comparable with physiological concentrations of fibrinogen (6-12 μM). Such an affinity may not be sufficient for two αC-domains to form a stable dimer in the fibrinogen molecule and, therefore, additional interactions with the central region through fibrinopeptides, hypothesized long ago (36) and confirmed recently in direct experiments (37), are required to stabilize their dimerization. This implies that upon fibrin assembly thrombin-mediated removal of fibrinopeptides should result in dissociation of the dimer and destabilization of the monomeric αC-domains. However, the monomeric state of the αC-domains is transient due to rapid polymerization of fibrin monomers and in polymeric fibrin, in which the local concentration of the αC-domains dramatically increases, this affinity should be sufficient for effective self-association of these domains into αC-polymers, in which their structure is stabilized. Thus, the switch of the αC-domains from intra- to intermolecular interactions hypothesized earlier (4, 5) may be driven by their comparatively low affinity to each other and the need to restore their stability.
In summary, the present study established that the overall fold of the hAα425-503 fragment corresponding to the NH2-terminal portion of the human αC-domain is similar to that of the corresponding bovine fragment, whose NMR structure was established earlier (19). The study suggests that the full-length human and bovine αC-domains each consists of two independently folded sub-domains, the N-terminal sub-domain formed by the parallel/antiparallel β-sheet and the less stable C-terminal domain whose structure remains to be determined. The study also revealed that the full-length αC-domains form ordered oligomers in a concentration-dependent and reversible manner, and that both sub-domains contribute to the affinity of αC-domains to each other and their higher stability in oligomers. Such character of the oligomerization implies that this process mimics polymerization of the αC-domains in fibrin and the structure of αC-oligomers may mimic that of fibrin αC-polymers. Finally, the results of this study further clarify the molecular mechanism of the previously proposed intra- to intermolecular switch of the αC-domains upon fibrin assembly.
Footnotes
This work was supported by National Institutes of Health Grant HL-56051 to L.M., American Heart Association, Mid-Atlantic Affiliate Grant-in-Aid 055527U to R.R.H., and by the Intramural Research Program of the NIH, National Heart, Lung, and Blood Institute to N.T.
- TBS
- 20 mM Tris buffer
- pH 7.4
- with 150 mM NaCl
- PMSF
- phenylmethylsulphonyl fluoride
- PBS
- 20 mM potassium phosphate buffer
- pH 6.5
- 150 mM NaCl
- CD
- Circular Dichroism
Note, that numbering of the presented bovine αC-domain sequence is based on the bovine Aα chain sequence deposited by M. Murakawa (UniProtKB/Swiss-Prot, accession number P02672, release 37.0); this numbering has been used in our previous publications (17-19). The updated sequence of this chain deposited later (UniProtKB/Swiss-Prot, accession number P02672, release 39.12) contains extra 13-residues in the connector region. Thus, 13 residues should be added to correlate the presented sequence with the updated one.
Secondary structure prediction was performed on the Jpred 3 prediction server (www.compbio.dundee.ac.uk/www-jpred/) utilizing the Jnet algorithm (35).
REFERENCES
- 1.Henschen A, McDonagh J. Fibrinogen, fibrin and factor XIII. In: Zwaal RFA, Hemker HC, editors. Blood Coagulation. Elsievier Science Publishers; Amsterdam: 1986. pp. 171–241. [Google Scholar]
- 2.Medved LV, Gorkun OV, Privalov PL. Structural organization of C-terminal parts of fibrinogen Aα-chains. FEBS Lett. 1983;160:291–295. doi: 10.1016/0014-5793(83)80985-5. [DOI] [PubMed] [Google Scholar]
- 3.Erickson HP, Fowler WE. Electron microscopy of fibrinogen, its plasmic fragments and small polymers. Ann. N. Y. Acad. Sci. 1983;408:146–163. doi: 10.1111/j.1749-6632.1983.tb23242.x. [DOI] [PubMed] [Google Scholar]
- 4.Weisel JW, Medved L. The structure and function of the αC domains of fibrinogen. Ann. N. Y. Acad. Sci. 2001;936:312–327. doi: 10.1111/j.1749-6632.2001.tb03517.x. [DOI] [PubMed] [Google Scholar]
- 5.Medved LV, Gorkun OV, Manyakov VF, Belitser VA. The role of fibrinogen αC-domains in the fibrin assembly process. FEBS Lett. 1985;181:109–112. doi: 10.1016/0014-5793(85)81123-6. [DOI] [PubMed] [Google Scholar]
- 6.Gorkun OV, Veklich YI, Medved LV, Henschen AH, Weisel JW. Role of the αC domains of fibrin in clot formation. Biochemistry. 1994;33:6986–6997. doi: 10.1021/bi00188a031. [DOI] [PubMed] [Google Scholar]
- 7.Tsurupa G, Medved L. Identification and characterization of novel tPA- and plasminogen-binding sites within fibrin(ogen) αC-domains. Biochemistry. 2001;40:801–808. doi: 10.1021/bi001789t. [DOI] [PubMed] [Google Scholar]
- 8.Medved L, Nieuwenhuizen W. Molecular mechanisms of initiation of fibrinolysis by fibrin. Thromb. Haemost. 2003;89:409–419. [PubMed] [Google Scholar]
- 9.Cheresh DA, Berliner SA, Vicente V, Ruggeri ZM. Recognition of distinct adhesive sites on fibrinogen by related integrins on platelets and endothelial cells. Cell. 1989;58:945–953. doi: 10.1016/0092-8674(89)90946-x. [DOI] [PubMed] [Google Scholar]
- 10.Belkin AM, Tsurupa G, Zemskov E, Veklich Y, Weisel JW, Medved L. Transglutaminase-mediated oligomerization of the fibrin(ogen) αC domains promotes integrin-dependent cell adhesion and signaling. Blood. 2005;105:3561–3568. doi: 10.1182/blood-2004-10-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Z, Mochalkin I, Veerapandian L, Riley M, Doolittle RF. Crystal structure of native chicken fibrinogen at 5.5- Å resolution. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3907–3912. doi: 10.1073/pnas.080065697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z, Kollman JM, Pandi L, Doolittle RF. Crystal structure of native chicken fibrinogen at 2.7 Å resolution. Biochemistry. 2001;40:12515–12523. doi: 10.1021/bi011394p. [DOI] [PubMed] [Google Scholar]
- 13.Kollman JM, Pandi L, Sawaya MR, Riley M, Doolittle RF. Crystal structure of human fibrinogen. Biochemistry. 2009;48:3877–3886. doi: 10.1021/bi802205g. [DOI] [PubMed] [Google Scholar]
- 14.Privalov PL, Medved LV. Domains in the fibrinogen molecule. J. Mol. Biol. 1982;159:665–683. doi: 10.1016/0022-2836(82)90107-3. [DOI] [PubMed] [Google Scholar]
- 15.Veklich YI, Gorkun OV, Medved LV, Nieuwenhuizen W, Weisel JW. Carboxyl-terminal portions of the α chains of fibrinogen and fibrin. Localization by electron microscopy and the effects of isolated αC fragments on polymerization. J. Biol. Chem. 1993;268:13577–13585. [PubMed] [Google Scholar]
- 16.Matsuka YV, Medved LV, Migliorini MM, Ingham KC. Factor XIIIa-catalyzed cross-linking of recombinant αC fragments of human fibrinogen. Biochemistry. 1996;35:5810–5816. doi: 10.1021/bi952294k. [DOI] [PubMed] [Google Scholar]
- 17.Tsurupa G, Tsonev L, Medved L. Structural organization of the fibrin(ogen) αC-domain. Biochemistry. 2002;41:6449–6459. doi: 10.1021/bi025584r. [DOI] [PubMed] [Google Scholar]
- 18.Burton RA, Tsurupa G, Medved L, Tjandra N. Identification of an ordered compact structure within the recombinant bovine fibrinogen αC-domain fragment by NMR. Biochemistry. 2006;45:2257–2266. doi: 10.1021/bi052380c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burton R, Tsurupa G, Hantgan RR, Tjandra N, Medved L. NMR solution structure, stability, and interaction of the recombinant bovine fibrinogen αC-domain fragment. Biochemistry. 2007;46:8550–8560. doi: 10.1021/bi700606v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doolittle RF, Kollman JM. Natively unfolded regions of the vertebrate fibrinogen molecule. Proteins. 2006;63:391–397. doi: 10.1002/prot.20758. [DOI] [PubMed] [Google Scholar]
- 21.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 22.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 23.Cavanagh J, Fairbrother WJ, Palmer AGI, Skelton NJ. Protein NMR Spectroscopy: principles and practice. 1st ed. Academic Press; San Diego, CA: 1996. [Google Scholar]
- 24.Yang JT, Wu CS, Martinez HM. Calculation of protein conformation from circular dichroism. Methods Enzymol. 1986;130:208–269. doi: 10.1016/0076-6879(86)30013-2. [DOI] [PubMed] [Google Scholar]
- 25.Hantgan RR, Paumi C, Rocco M, Weisel JW. Effects of ligand-mimetic peptides Arg-Gly-Asp-X (X = Phe, Trp, Ser) on αIIbβ3 integrin conformation and oligomerization. Biochemistry. 1999;38:14461–14474. doi: 10.1021/bi9907680. [DOI] [PubMed] [Google Scholar]
- 26.Hantgan RR, Rocco M, Nagaswami C, Weisel JW. Binding of a fibrinogen mimetic stabilizes integrin αIIbβ3′s open conformation. Protein Sci. 2001;10:1614–1624. doi: 10.1110/ps.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 28.Poirot O, O’Toole E, Notredame C. Tcoffee@igs: A web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003;31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ponder JW, Richards FM. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J. Mol. Biol. 1987;193:775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 30.Brünger AT, Adams PD, Clore GM, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges N, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system (CNS), a new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Chun PW, Kim SJ, Stanley CA, Ackers GK. Determination of the equilibrium constants of associating protein systems. 3. Evaluation of the weight fraction of monomer from the weight-average partition coefficient (application to bovine liver glutamate dehydrogenase) Biochemistry. 1969;8:1625–1632. doi: 10.1021/bi00832a044. [DOI] [PubMed] [Google Scholar]
- 32.Na GC, Timasheff SN. Stoichiometry of the vinblastine-induced self-association of calf brain tubulin. Biochemistry. 1980;19:1347–1354. doi: 10.1021/bi00548a013. [DOI] [PubMed] [Google Scholar]
- 33.Brown JH, Volkmann N, Jun G, Henschen-Edman AH, Cohen C. The crystal structure of modified bovine fibrinogen. Proc. Natl. Acad. Sci. U. S. A. 2000;97:85–90. doi: 10.1073/pnas.97.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medved L, Weisel JW, Fibrinogen and Factor XIII Subcommittee of Scientific Standardization Committee of International Society on Thrombosis and Haemostasis Recommendations for nomenclature on fibrinogen and fibrin. J. Thromb. Haemost. 2009;7:355–359. doi: 10.1111/j.1538-7836.2008.03242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Veklich YI, Gorkun OV, Medved LV, Nieuwenhuizen W, Weisel JW. Carboxyl-terminal portions of the α chains of fibrinogen and fibrin. Localization by electron microscopy and the effects of isolated αC fragments on polymerization. J. Biol. Chem. 1993;268:13577–13585. [PubMed] [Google Scholar]
- 37.Litvinov RI, Yakovlev SV, Tsurupa G, Gorkun OV, Medved L, Weisel J. Direct evidence for specific interactions of the fibrinogen αC-domains with the central E region and with each other. Biochemistry. 2007;46:9133–9142. doi: 10.1021/bi700944j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeLano WL. The PyMol molecular graphic system. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]