Abstract
Calcium has long been recognized as an important regulator of cell cycle transitions although the mechanisms are largely unknown. A functional genomic screen has identified genes involved in the regulation of early cell cycle progression by calcium. These genes when overexpressed confer the ability to bypass the G1/S arrest induced by Ca2+- channel antagonists in mouse fibroblasts. Overexpression of the cystine-glutamate exchanger, xCT, had the greatest ability to evade calcium antagonist-induced cell cycle arrest. xCT carries out the rate limiting step of glutathione synthesis in many cell types and is responsible for the uptake of cystine in most human cancer cell lines. Functional analysis indicates that the cystine uptake activity of xCT overcomes the G1/S arrest induced by Ca2+- channel antagonists by bypassing the requirement for calcium signaling. Since cells overexpressing xCT were found to have increased levels and activity of the AP-1 transcription factor in G1, redox stimulation of AP-1 activity accounts for the observed growth of these cells in the presence of calcium channel antagonists. These results suggest that reduced calcium signaling impairs AP-1 activation and that xCT expression may directly affect cell proliferation.
Keywords: calcium, glutathione, AP-1, redox, cell cycle, growth factor
INTRODUCTION
Although calcium ion is arguably the most important second messenger in excitable cell signaling, a considerable amount of work has indicated an important role for Ca2+ signaling in the regulation of proliferation in non-excitable cells (for review [1]). Calcium transients induced by growth factors, hormones and cytokines have been shown to be important for the regulation of both normal and pathological cell proliferation (for review see [2–4]. Ca2+ signaling elicited by mitogenic stimulation has been detected in all normal and transformed cell lines and is among the most conserved events induced upon receptor activation. Multiple points in the mammalian cell cycle including entry from G0, the G1/S transition, and mitosis require Ca2+ transients for cell cycle progression [5–9]. In most cases, these are requisite events for cell cycle progression. For example, when human fibroblasts are stimulated by growth factors, depletion of extracellular calcium at any time during the first eight hours of the cell cycle results in a reversible inhibition of DNA synthesis [10].
In mouse fibroblasts, growth factor stimulation elicits a calcium response characterized by rapid mobilization of internal Ca2+ stores followed by influx of Ca2+ from the cell exterior[11]. The calcium influx phase of this response is inhibited both by inorganic calcium channel blockers and therapeutically important dihydropyridines. Although poorly understood, these inhibitors as wells as benzodiazepine and phenylalkamine-type calcium channel antagonists all arrest cell cycle progression in G1 [12–14]. Although the targets of the calcium channel antagonists in non-excitable cells are unknown, their use has been proposed as an approach to the treatment of proliferative diseases such as atherosclerosis and tumorigenesis [15–17].
To identify gene products involved in the regulation of proliferation by growth factor-induced calcium signaling, we have carried out genetic screens for genes that, when overexpressed, allow cells to evade the cell cycle arrest induced by calcium channel antagonists. Several genes related to cell signaling were isolated in these screens. The gene that produced the greatest impact on the cells’ ability to bypass cell cycle arrest is the human cystine-glutamate exchanger, xCT. xCT carries out an essential step in the synthesis of glutathione (GSH). We show that the expression levels of xCT directly affect cellular cystine uptake and in turn GSH levels in mammalian cells. Our results indicate that growth factor-induced calcium signaling contributes to cell proliferation at least, in part, via the establishment of a cellular redox environment conducive to entry into S phase.
EXPERIMENTAL/MATERIALS AND METHODS
Cell culture
HT1080 fibrosarcoma cells and LinXE ecotropic packaging cells were cultured in DMEM 10% Fetal Calf Serum. NIH 3T3 and Swiss 3T3 cells were cultured in DMEM 5% Calf Serum and DMEM 10% Cosmic Calf serum respectively. NIH 3T3 cells that overexpress MDR1 were selected with 20ng/ml vinblastine for 10 days after infection with retrovirus produced by LinX E cells transfected with pLMDR1L6 (a gift of Dr. Igor Roninson). xCT was isolated in a cDNA library screen using the MaRX retrovirus system [18]. A HygroMaRX (HM) II-borne cDNA library prepared from HT1080 fibrosarcoma cells was packaged in an ecotropic virus-packaging cell line LinX E, and used to infect early passage NIH 3T3 cells. A total of approximately 107 cells were infected. The infected cells were selected with 75µg/ml hygromycin and then subjected to 100µM CoCl2 for one week. Integrated proviruses were excised from genomic DNA with Cre recombinase as described. The resulting excised proviral plasmids were amplified in E. coli and tested after reintroduction into NIH 3T3 cells for the ability to confer resistance to Co2+. DNA sequence of both strands of positive cDNA inserts was determined using an Applied Biosystems Inc. fluorescent DNA sequencer.
Plasmid constructions
The CD98 heavy chain, a gift of Dr.Jeffery Leiden, was cloned into the EcoRI site of pBABEpuro. A full length version of E16, a gift of Dr. Lukas Kuhn, was cloned into the EcoRI and HindIII sites of pHM IV. shRNA sequences targeting the mouse and human xCT genes, six different for each, were designed based on the human miRNA, miR30, structure [19]. Synthetic oligonucleotides encoding the designed shRNAs were PCR-amplified by primers harboring the 5' and 3' miR30 sequences, respectively. The amplified shRNAs were cut by XhoI and EcoRI and cloned into a modified pSHAG-MAGIC 2 (pSM2) vector. A non-specific hairpin targeting the firefly (Photinus pyralis) luciferase gene was also built as above for use as a control. At least three different pSM2 clones for each shRNA were sequence verified and used for transfections.
The pHM IV -CGFP plasmid was constructed using a green fluorescent protein ORF missing the initial ATG. Full length xCT was positioned upstream of this sequence by PCR such that the last amino acid of xCT was followed by GFP sequence.
Flow Cytometry
Vector control and cells overexpressing xCT were trypsinized, washed twice with PBS, fixed in 70% ethanol and stored at −20°C or processed immediately for propidium iodide(PI) staining and FACS as follows: After washing 2 times with PBS cell pellets were resuspended in 300–500 µl propidium iodide +RNase solution (BD Biosciences) and incubated 30 minutes in the dark. FACS analysis was performed using a Beckman Coulter Eics altra flow cytometer. Red fluorescence from PI was detected through a 610nM long pass filter. Data from a minimum of 20000 cells were acquired and processed using Expo 32 software.
Measurement of [3H] Thymidine Incorporation
Cells were plated at a density of 1x104 cells/cm2 in 24-well plates, grown for 24h in serum-containing medium. The cells were serum-starved for 48h, and individual wells were treated with serum in the absence or presence of 20µM nifedipine for 12hrs. 0.2µCi of [3H] thymidine (85 Ci/mmol) was added and cells were incubated for another 12hr. Cells were extracted with 5% trichloracetic acid, washed twice and then solubilized in 0.1M NaOH and analyzed for 3H radioactivity by scintillation counting on a 3H channel.
Measurement of amino acid uptake
Adherent NIH 3T3 cells infected with either HM II or HM II-xCT were grown at 37°C in 12 well dishes. Cells were washed twice with modified transport buffer [20] (140 mM NaCl, 5.4 mM KCl, 0.8 mM MgSO4, 1.8 mM CaCl2, 5 mM glucose, 25mM HEPES, pH 7.4) and incubated with radioactive amino acids. The amino acids were L-[14C]-alanine (Amersham), L-[35S]-cystine (Amersham), L-[14C]-glutamate (NEN), L-[3H]-leucine (Amersham), and L-[3H]-serine (Amersham). Cells were incubated at 25°C for two minutes and immediately washed three times with ice-cold transport buffer. Cells were solubilized in 0.2 N NaOH and 0.2% SDS and counted with a scintillation cocktail on the appropriate channel. Uptake activities represent the average of at least three trials of experiments qualitatively reproduced at least twice and are shown as a percentage normalized to vector controls ± SEM.
Determination of cellular glutathione
Glutathione (GSH) levels were determined using a spectrophotometric method (Oxis, Inc.). Briefly, NIH 3T3 cells expressing either vector or retrovirally encoded xCT were washed with PBS and lysed in 5% metaphosphoric acid. Cell lysates were cleared by centrifugation, and cellular GSH conjugated to chromogenic substrate, and quantified by absorbance at 400nm. Normalization of GSH values to either cell number or protein mass gave identical results.
Calcium Measurements
Swiss 3T3 cells were seeded in clear bottom, 96-well dishes (Costar 3603) and serum starved for 24 hours. Serum starved cells were loaded with 5 µM fura-2 acetoxymethyl ester (fura-2 AM) in serum-free DMEM for 1 hour. Fura-2 AM loaded cells were washed 2X with Ca2+-free modified Krebs Ringers HEPES [11] supplemented with 1mM glucose and cystine (KRH) and incubated for 30 minutes to allow for complete cleavage of the acetoxymethyl ester. Fura-2 loaded, serum starved cells were stimulated with 10ng/ml PDGF in KRH +/− 2mM CaCl2 as indicated. Fluorescence was measured using a BioTek Synergy HT plate reader with excitation filters at 340 and 380 and emission filters at 508 nm. At the end of each measurement, maximal and minimal fluorescence was measured by the addition of 5µM ionomycin in KRH, 3mM CaCl2 and 10 mM EGTA respectively. [Ca2+]i was calculated using the Grynkiewicz equation [21] where KD is 224 nM.
Antibody Production and Immunoblotting
Peptides corresponding to the C-terminal 8 amino acids of human xCT were synthesized with an N-terminal cysteine residue (CVVPEEDKL) and coupled to keyhole-Limpet hemocyanin. Antibodies were raised in New Zealand White rabbits by Covance Research Products Inc (CRPinc) Denver, PA using their 118 day standard protocol. Antibodies for p42/44 ERK, AKT, pThr AKT, pSer AKT, p27, cyclin D1 and GAPDH were purchased from Santa Cruz. Immunoblots were performed using standard protocols. Cells were lysed in RIPA buffer containing complete protease inhibitor tablets (Roche), proteins were resolved by SDS-PAGE [22] and transferred to PVDF membranes. Membranes were blocked in Tris buffered saline containing .1% Tween and 5% nonfat powdered milk (T:TBS, 5% milk). Primary antibody incubation (1:1000) was performed in T:TBS, 5% milk overnight at 4 degrees. Proteins were visualized using a goat anti-rabbit secondary antibody conjugated to HRP (Santa Cruz) and a chemiluminescence detection system (Pierce).
Luciferase Reporter Assays
NIH 3T3 vector control and cells overexpressing xCT were transiently transfected with luciferase reporter plasmids for relevant cell cycle related transcription factors including, myc, AP1, Rb, E2F, p53, NFAT, and NF-κB or the control plasmid pTA (Clontech) using lipid transfection reagents (FuGene, Roche). Cells were transfected with reporter construct plus pRL to normalize for transfection efficiency. Firefly (reporter) and Renilla (Renilla reniformis; pRL) luciferase activity was measured by the Dual-Luciferase Reporter Assay kit (Promega) using a BioTek Synergy HT reader after 48 hours according to manufacturer’s protocol (Promega). Each experiment was performed in triplicate and average luciferase units are presented.
RNA Preparation and qPCR
Total RNA was isolated from vector control and xCT-overexpressing NIH3T3 cell lines using Trizol (Invitrogen) according to manufacturers instructions. 2 µg total RNA was subjected to RT using oligodT primers and M-MLV reverse transcriptase (Promega). Real-time quantitative PCR was performed using ITaq SYBR mix (Promega) in an ABI Prism 7900 HT Sequence Detection System (PerkinElmer) with the following thermal conditions: a single cycle of 95.C for 2.5 min, followed by 40 cycles of 95 .C for 15 s and 60oC for 60s. Primer sequences obtained The c-jun, forward primer, 5-AGCAGGGACCCATGGAAGTT-3, and reverse primer, 5-AAAGATGACCTTTGCTTGTGCAT-3 were normalized to GAPDH control. Determination of absolute abundance of mRNAs was performed using Sequence Detection System (SDS 2.1) software
RESULTS
The human cystine transporter, xCT, enables cells to overcome cell cycle arrest induced by calcium channel antagonists
Although the importance of Ca2+ in mitogenic signaling is well established, pharmacological approaches using calcium channel blockers have failed to identify the molecular mechanisms that are responsible [13, 16, 23–26]. Using retrovirus-based cDNA libraries [18], we performed genetic screens to identify the mechanism of calcium-based regulation of proliferation. The prolonged G1 arrest caused by these agents results in cell death which allows for the selection of variants capable of cell cycle progression [6, 7,27]. Genes were sought whose overexpression allowed cells to overcome the growth arrest induced by calcium channel antagonists extensively described in murine 3T3 fibroblasts [5, 11]. For these experiments 107 cells contained on approximately fifty 10 cm plates were infected with a HygroMaRXII library harboring cDNA inserts prepared from HT1080 cells. After infection and selection for the presence of the virus, cells were subjected to selection with calcium antagonists for one week. 165 proviruses were rescued from potential positive clones as described in Methods and were retested. Those clones capable of conferring resitance to channel blockers upon retest are listed in Table I. The activated version of N-Ras (K61) found in HT1080 cells [28] and the c-Jun genes, both of which have known roles in regulating proliferation were also recovered in this screen.
Table 1.
Genes conferring resistance to cell cycle arrest induced by calcium channel blockers.
| Gene | # of clones recovered | growth in nifedipine |
|---|---|---|
| N-ras K61 | 4 | +++ |
| C-jun | 1 | ++ |
| A2b2 purinergic receptor | 1 | ++ |
| Calpactin II LC (p11) | 1 | + |
| Slc7a11 (xCT) | 1 | +++++ |
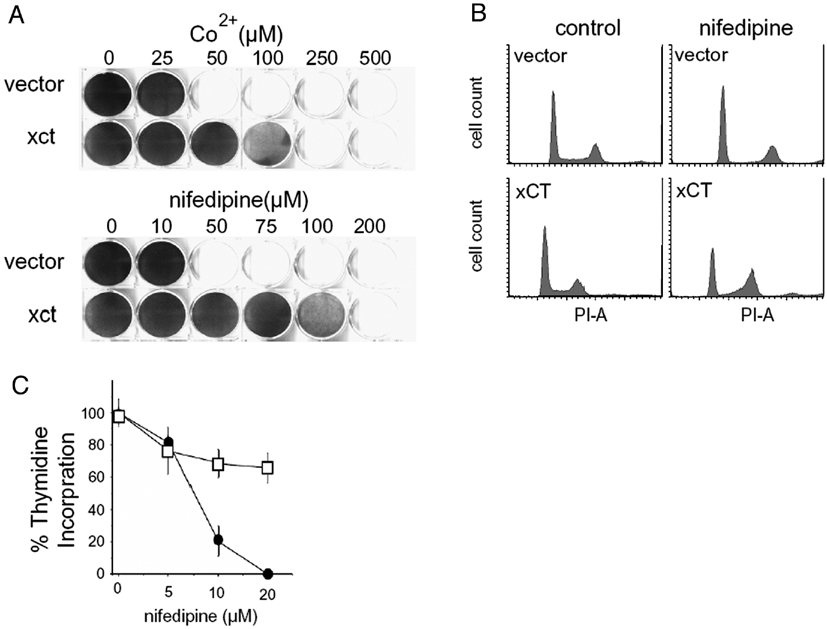
The gene which conferred the greatest ability of cells to bypass the arrest imposed by calcium channel antagonists was the SLC7All gene. Overexpression of SLC7All, which encodes the human cystine transporter, xCT, allowed cells to proliferate in the presence of the inorganic calcium channel blocker, cobalt, and calcium channel blockers of the dihydropyridine family, such as nifedipine (Figure 1A and data not shown). The in vitro cell growth phenotype conferred by this insert was directly related to the drug-induced cell cycle arrest, since overexpression counteracted the pre-S phase cell cycle arrest imposed by CoCl2 and nifedipine (Figure 1 B,C). Cells infected with this virus were resistant to the related dihydropyridines nifedipine, nitrendipine and nimodipine but not to verapamil, diltiazem, or an oxidized metabolite of nifedipine, 2,6-Dimethyl-4-(2’-nitrophenyl)-3,5 pyridinecarboxylic acid dimethylester (data not shown). Overexpression of xCT failed to confer increased resistance to several other drugs that inhibit proliferation such as chlorambucil, cisplatin, doxorubicin, etoposide, malphalan, menadione, methyl methane sulfonate, paraquat or other inorganic ions (supplementary Figure 1). Moreover, expression of the human p-glycoprotein (MDR1), which is responsible for generalized detoxification [29], did not confer resistance to nifedipine (supplementary Figure 2).
Figure 1.
Overexpression of xCT permits bypass of the calcium channel antagonist-imposed G1/S block. A. Growth of NiH 3T3 cells infected with empty HygroMaRX II retrovirus or HygroMaRX II-xCT. Cells at a density of 104 cells/well were treated with concentrations shown. Growth after 5 days was visualized by crystal violet staining. B. Expression of xCT bypasses the G1/S block imposed by the calcium channel blocker nifedipine. Cells grown in 50 µM nifedipine for 12 hours were trypsinized, fixed, and stained with propidium iodide for flow cytometric analysis as described. An increase in the number of xCT cells in S phase in the presence of drug is shown. C. Thymidine incorporation assays were performed on vector and xCT overexpressing cells stimulated with serum in the absence or presence of 20µM nifedipine for 12hrs. Mean incorporation of four independent trials is shown as a percentage of nifedipine-treated to untreated control + SEM.
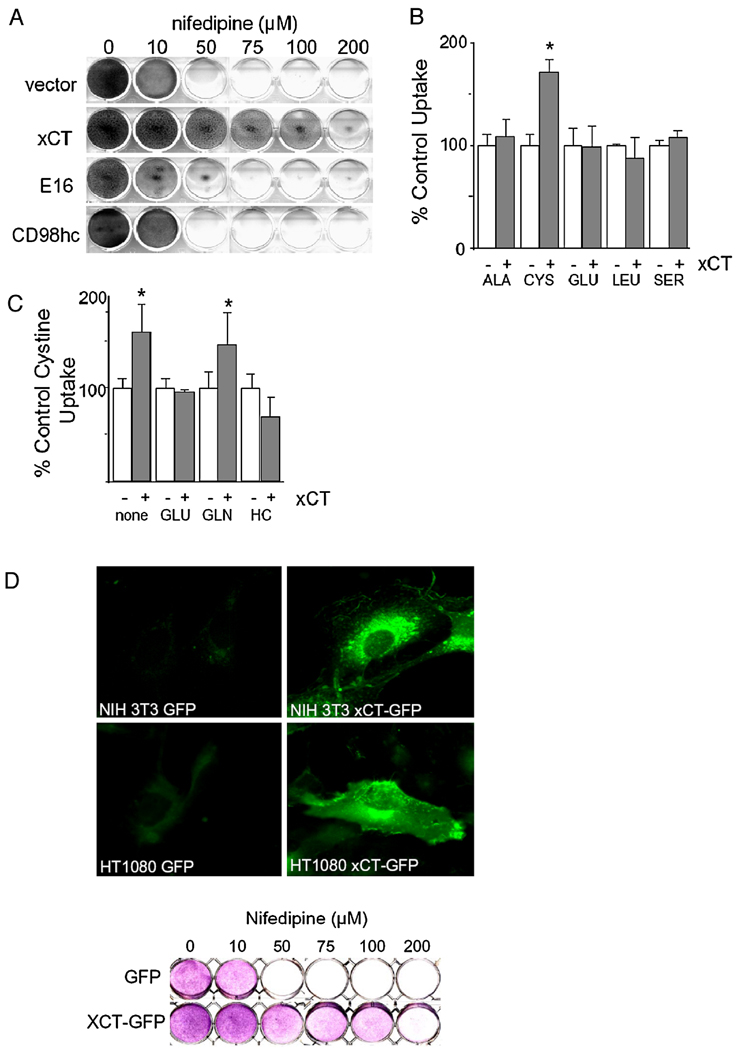
xCT encodes the light chain of a heterodimeric amino acid transport system that exchanges extracellular cystine for intracellular glutamate. The heavy chain of this transport system is the CD98 protein, which is encoded by the SLC3A2 gene and is involved in a number of cellular processes [30]. Most cells are thought to express a number of light chains [30], each of which physically associates with the CD98 heavy chain. To determine whether the increase in drug resistance was a consequence of increased xCT activity or some artifact of xCT overexpression, such as a dominant negative effect, we tested the ability of CD98 heavy chain and the most closely related light chain to confer resistance to calcium channel antagonists. Full-length cDNAs for CD98hc and the most closely related human light chain, E16, were introduced into NIH 3T3 cells by retroviral gene transfer. Expression of either gene alone failed to confer resistance to nifedipine (Figure 2A). Moreover, overexpression of CD98hc and xCT together failed to confer higher levels of nifedipine resistance than overexpression of xCT alone (data not shown). This indicates that the increase in drug resistance is due specifically to increased xCT function. In addition, these results agree with previous studies that indicate that the CD98 light chains are functionally distinct and that the heavy chain is not required for light chain function [31, 32]. That overexpression of xCT has a positive influence on cell cycle progression may be important since both xCT and CD98, are frequently overexpressed in tumors [33, 34].
Figure 2.
The ability to confer calcium channel antagonist resistance is due to increased activity of xCT. A. NIH 3T3 cells infected with retroviruses directing the expression of the closely related E16 light chain gene or CD98 heavy chain are not resistant to nifedipine. B. xCT is functional. Specificity of amino acid transport in NIH 3T3 cells. Mean uptake activities of four independent trials are shown as a percentage normalized to vector controls + SEM. C. Increased cystine uptake in NIH 3T3 cells is inhibited by homocysteine and glutamate but not glutamine. 1mM inhibitor was added to assay described above. Means + SEMs of three independent trials are shown. D. xCT is expressed on the plasma membrane. NIH 3T3 or HT1080 cells infected with xCT- GFP fusion protein exhibit plasma membrane staining (top panel). The gene fusion between xCT and GFP is functional when expressed in NIH 3T3 cells (lower panel). Growth of cells plated at a density of 104 cells/well and treated with concentrations of nifedipine shown was visualized by crystal violet staining.
Several lines of evidence indicate that xCT overexpression leads to increased xCT function in mouse fibroblasts. As shown in Figure 2B, NIH 3T3 fibroblasts that overexpress xCT demonstrate a statistically significant (P = 0.002) increase in the uptake of cystine but not other amino acids. As expected for xCT activity, the observed increase in cystine uptake was largely inhibited by the addition of glutamate or homocysteic acid but not glutamine (Figure 2C). The modest increase in cystine uptake over vector control cells is not surprising considering that NIH 3T3 cells possess endogenous cystine uptake activity [35]. In addition, a gene fusion that places the green fluorescent protein (GFP) in frame at the carboxyl terminus of xCT is functional in its ability to confer nifedipine resistance (Figure 2D). These cells exhibit low level GFP fluorescence at the plasma membrane and in intracellular vesicles. Expression of the xCT-GFP construct in HT1080 cells, from which the cDNA library was prepared and which are known to express CD98hc, resulted in pronounced staining of the plasma membrane with intense signal at membrane ruffles. Overexpression of CD98hc in the NIH 3T3 cells expressing the xCT-GFP was without effect on the GFP fluorescence (data not shown). Taken together our results indicate that the retrovirus-encoded xCT is indeed functional in the NIH 3T3 cells and that increased xCT activity is responsible for the observed bypass of calcium antagonist imposed G1 arrest.
xCT overexpression increases intracellular reduced thiol levels which allows cells to proliferate
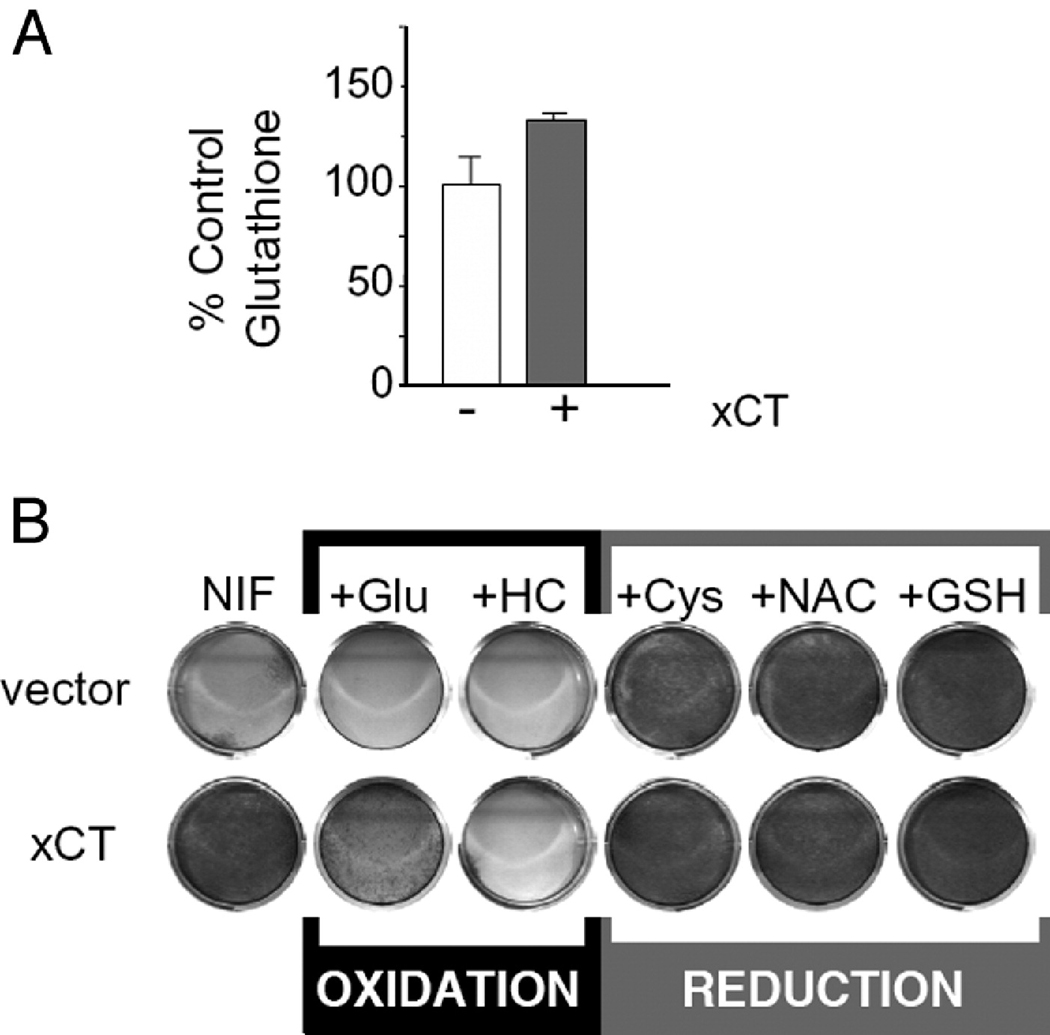
Since xCT is responsible for both cystine uptake and glutamate efflux, both activities were tested for the ability to provide increased resistance to calcium channel blockers. Glutamate efflux caused by increased xCT activity has been shown to generate Ca2+ signaling in neurons [36]. Addition of the glutamatergic antagonists NBQX (1,2,3,4-Tetrahydro-6-nitro-2,3-dioxo-benzo[f]quinoxaline-7-sulfonamide) or DL-AP3 (DL-2-Amino-3-phosphonopropionic acid) at 10 µM had no effect on the level of nifedipine resistance in NIH 3T3 cells overexpressing xCT (data not shown). Moreover, the addition of glutamate to these cells caused a marked decrease in the level of nifedipine resistance rather than an expected increase (Figure 3). This indicated that autocrine glutamate signaling was not involved in the drug resistance conferred by xCT.
Figure 3.
The xCT phenotypes are do due to its effects on intracellular redox. A. xCT expression affects cellular GSH levels. NIH 3T3 cells infected with empty HM II retrovirus (open bars) or HM II expressing xCT (solid bars). Cells overexpressing xCT contained greater levels of GSH than vector controls. B. The addition of anti-oxidants phenocopies the action of xCT in nifedipine (Nif) resistance. The addition of cystine (cys), glutathione (GSH), or N-acetylcysteine (NAC) to growth media increases cellular nifedipine resistance. The addition of the glutamate (glu) or homocysteine (HC), which act to oxidize cells, decreases cellular nifedipine resistance.
Because extracellular glutamate counteracted the drug resistance conferred by xCT overexpression, it was likely that the increased level of drug resistance was due to the cystine uptake function of xCT. Cystine, the oxidized form of cysteine, is the major source of cellular free thiol. Intracellular cystine is rapidly reduced to cysteine and incorporated into thiol containing proteins and GSH. xCT activity correlates with cellular GSH levels in a variety of cell types [37] and 3T3 cells that overexpress xCT were found to contain greater levels GSH than vector control cells (Figure. 3A). This increase in intracellular reduced thiols is responsible for the ability of cells to bypass the cell cycle arrest imposed by calcium channel antagonists. The addition of extracellular cystine phenocopied the drug resistance effect of xCT overexpression in cells containing only vector. The addition of other antioxidants, such as N-acetyl-cysteine and GSH to growth media also increased cellular nifedipine resistance. Extracellular homocysteine, which acts as an oxidant by inhibiting the uptake of cystine through the xc− system [38], exacerbated the drug sensitivity (Figure. 3B). These results indicate that it is the intracellular levels of GSH or other reduced thiols resulting from cystine that is taken up by xCT that gives rise to the drug resistant phenotype. It is somewhat surprising that resistance was not conferred to oxidants such as paraquat or chlorambucil that are normally detoxified by GSH. This may indicate that other enzymes in the GSH pathway are required for the detoxification of these compounds.
xCT overexpression reduces cellular free Ca2+ concentrations
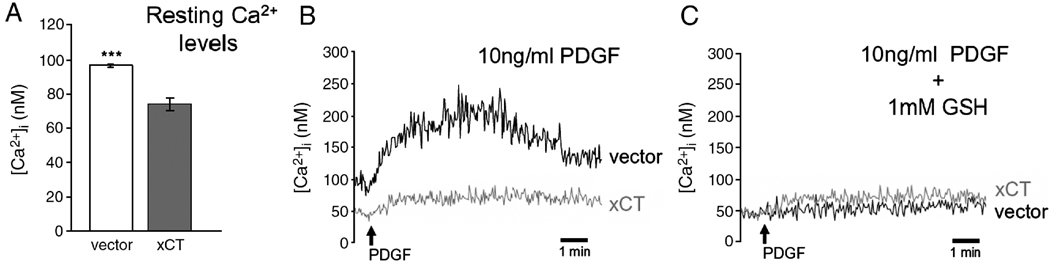
xCT was isolated by virtue of its overexpression allowing cells to evade cell cycle arrest induced by cobalt and dihyrdropyridine-type calcium channel antagonists. These agents have been shown to inhibit the capacitative phase of growth factor-induced Ca2+ signaling required for cell cycle progression [11]. Effects of xCT overexpression on these signaling events were investigated by measuring intracellular resting Ca2+ concentrations and Ca2+ signaling in response to growth factor stimulation in Swiss 3T3 fibroblasts. Swiss 3T3 fibroblasts display the same xCT overexpression phenotypes as NIH 3T3 cells yet exhibit a more readily observed calcium signal in response to mitogenic stimulation than do NIH 3T3 cells (Fig. 4 and data not shown). Surprisingly, we found, that in the absence of any drugs, cells overexpressing xCT have less cytoplasmic free calcium than vector control cells (Figure 4A). The average resting calcium levels determined in unstimulated cell cultures (n=53) was 23% lower in xCT overexpressing cells (73.5±3.63 nM), compared to vector controls (95.8 ± 0.78 nM). Cells in which xCT was overexpressed also exhibited a marked decrease in the magnitude of calcium transients after PDGF addition (Figure 4B). Calcium levels were also lower in xCT overexpressing cells after the addition of the sarco-endoplasmic reticulum ATPase (SERCA) inhibitor, thapsigargin, which causes stores to empty and store operated channels to open (data not shown). Since it is through effects on reduced thiols that xCT confers resistance to the channel blockers (Figure 3B), we tested whether incubation with GSH phenocopies the effect of xCT overexpression on calcium levels. Application and washout of 1 mM GSH prior to PDGF stimulation decreased Ca2+ signals in both xCT overexpressors and controls (Figure 4C). Significantly, GSH application to vector controls reduced Ca2+ levels both resting and after stimulation with PDGF to the same level as xCT overexpressing cells. This was not simply an effect of GSH on the fura-2 AM calcium indicator, because the addition of GSH to the fura-2 sodium salt produced an effect where it appeared that slightly more calcium was bound to fura-2 (data not shown).
Figure 4.
A. Swiss 3T3 fibroblasts overexpressing xCT contain significantly lower [Ca2+]i than vector control cells (p<.001). Fura-2 loaded Swiss 3T3 fibroblasts expressing either the HM IV control vector or the HM IV-xCT were subjected to fluorescent calcium indicator analysis as outlined in Methods. Resting levels are shown in A. Changes in [Ca2+]i recorded upon addition of 10ng/ml PDGF are shown in B. C. The addition of 1mM GSH to control cells phenocopies the effects of xCT overexpression on [Ca2+]i. Experiments were performed multiple times and representative traces are shown.
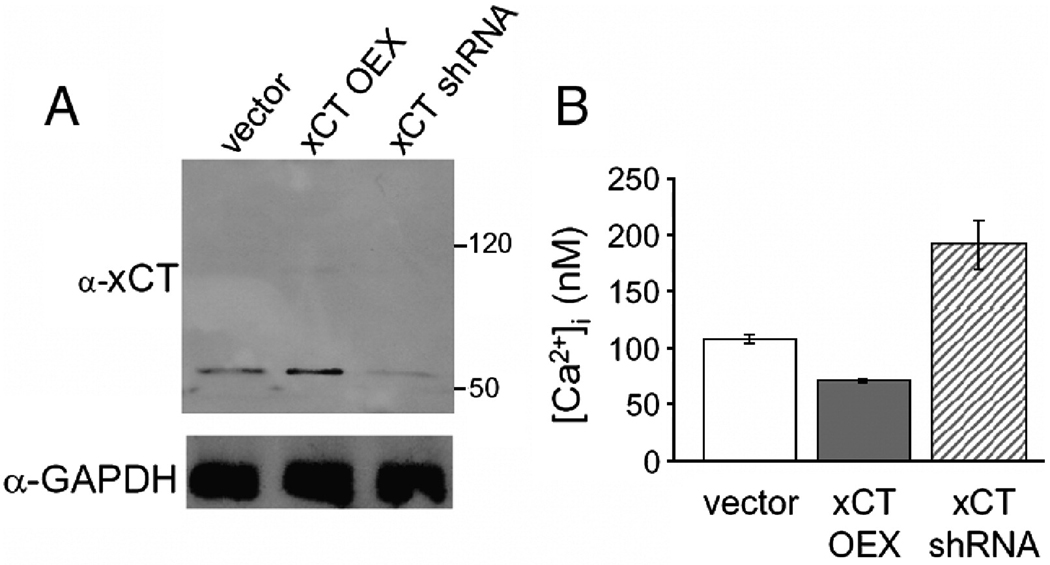
Additional evidence for the effect of xCT on calcium levels came from RNAi experiments which indicated that the effects of xCT were dose-dependent. Cells stably expressing shRNAs targeting xCT contained less xCT protein (Figure 5A) and were more sensitive to CoCl2 and dihydropyridine calcium channel blockers (data not shown). Yet, these cells contained higher Ca2+ concentrations and displayed increased Ca2+ signaling in response to PDGF (Figure 5 B). Taken together these results indicate that xCT overexpression bypasses the requirement of calcium signaling in G1/S progression. Rather than having a positive effect on calcium-based signaling per se, xCT overexpression has a general effect decreasing cytoplasmic calcium levels in cells under resting and stimulated conditions. This is likely due to the effect of GSH on a variety of calcium handling systems (see [39] for review, and Discussion).
Figure 5.
The effect of xCT on intracellular Ca2+ is dose-dependent. An shRNA targeting xCT affects resting calcium levels in Swiss 3T3 fibroblasts. A. Immunoblots probed with anti-xCT antibody raised to the C-terminal 8 amino acids of human xCT. B. expression of an shRNA targeting xCT (hatched bars) increases resting [Ca2+]I compared to cells overexpressing xCT (solid bars) and HM IV vector control cells (open bars). Measurements were made as in Figure 4.
Cells overexpressing xCT exhibit increased AP-1 activity
To identify cell cycle regulatory mechanisms involved in the xCT-mediated bypass of calcium antagonist imposed G1 arrest, we examined the effect of xCT overexpression on the activity of signaling pathways associated with cell cycle progression and proliferation. Growth factor (PDGF)-induced cell cycle progression results from activation of the Ras/Mek/ Erk MAPk and the PI3kinase/AKT pathways which are involved in accumulation and stabilization of cyclin D1 proteins and in the elimination of the cell cycle inhibitor, p27 [40]. Immunoblot analysis indicated that both vector control cells and cells overexpressing xCT have identical expression patterns of p42/44 ERK, AKT and p~AKT and cyclin D1 in the absence of calcium channel antagonists (data not shown). No differences in the expression patterns for AKT, p~AKT and cyclin D1 were observed between vector control and xCT cells in the presence of calcium channel antagonists (data not shown). However, in the presence of CoCl2 and nifedipine, xCT cells produce p42/44 ERK expression patterns that reflect cell cycle progression, while vector control cells produce chronic upregulation expression patterns indicative of senescence/quiescence (Supplementary Figure 3) [41, 42] [43]. This is not surprising since xCT cells are actively proliferating under these conditons.
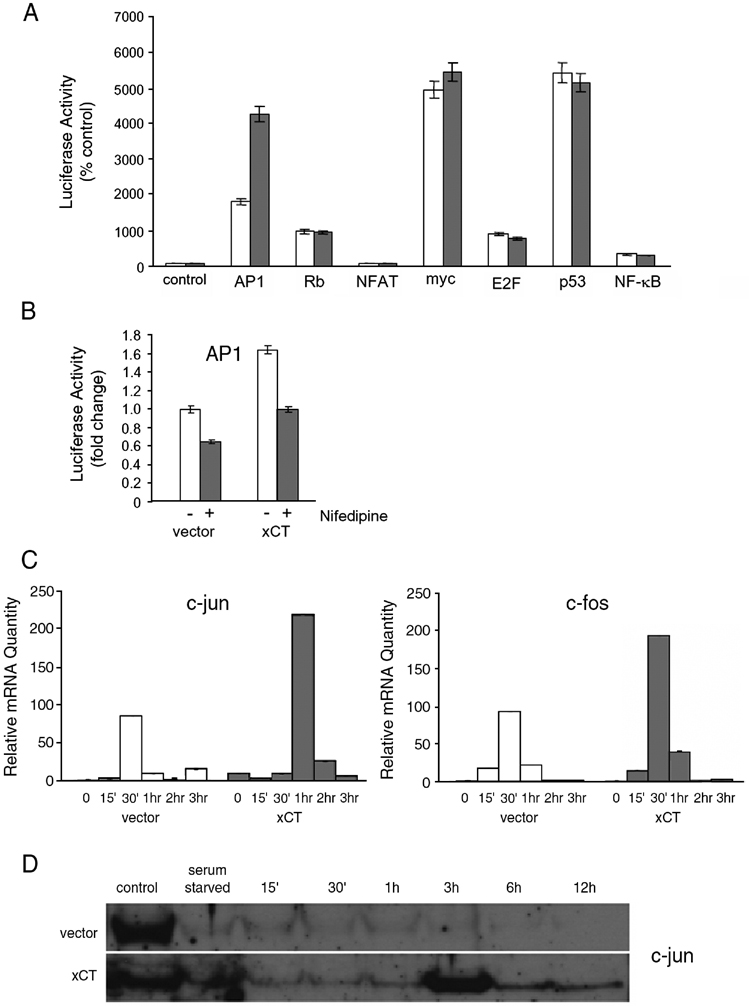
We also measured the activities of several transcription factors that regulate proliferation using luciferase reporter assays in cells overexpressing xCT. No difference in the activities of c-Myc, NFAT, NF-κB, E2F and p53 between vector control and cells overexpressing xCT was observed. However, xCT cells displayed a 2.4-fold increase in AP-1 activity over control cells in the absence of Ca2+ antagonists (Figure 6A). In its most widely accepted forms AP-1 consists of heterodimers of Jun and Fos family members which dimerize using leucine zipper domains [44]. Interestingly, c-Jun was isolated in the screen as a gene whose overexpression conferred resistance to calcium channel antagonists (Table I). Together these results suggested that the effect of xCT on AP-1 activity might be responsible for the ability of these cells to proliferate in the presence of Ca2+ antagonists. As shown in Figure 6B, nifedipine decreased AP-1 activity in vector control cells to 65% of normal. Activity was also decreased in xCT overexpressing cells to 60% of untreated, however, even in the presence of 50µM nifedipine, reporter levels were approximately the same as untreated vector controls. This near normal amount of AP-1 activity that was retained in the presence of drugs likely accounts for the observed channel blocker resistant phenotype.
Figure 6.
A. Cells overexpressing xCT display increased AP-1 transcription factor activity even in the presence of nifedipine. NIH 3T3 cells overexpressing xCT (solid bars) and HM IV vector control cells (open bars) were transiently transfected with a control luciferase reporter plasmid, TA, and luciferase reporter plasmids for AP-1, Rb, NFAT, myc, E2F, p53, and NF-κB. B. Cells were transiently co-transfected with an AP-1 firefly luciferase reporter and a control Renilla luciferase plasmid and grown for 24 hours. Cells were then grown for an additional 24 hours with or without 50 µM nifedipine and subsequently lysed for analysis of luciferase activity. C. Cells overexpressing xCT possess higher mRNA levels of AP-1 component transcription factors Jun and Fos. NIH 3T3 cells were serum starved for 48 hours and stimulated with 10ng/ml PDGF. Total RNA was extracted at the indicated times reverse transcribed, and qPCR was performed. D. xCT increases c-jun protein levels. NIH 3T3 fibroblasts overexpressing xCT or a vector control were serum starved for 48 hours then stimulated with serum. Cell lysates were collected at the indicated times and expression of c-jun was analyzed by immunoblotting.
AP-1 activity is determined by levels of its constituent subunits as well as by several regulatory mechanisms that respond to a variety of stimuli (for review [44]). C-Jun and c-Fos, being immediate early genes, are among the first genes expressed after mitogenic stimulation (reviewed by [45]). Each posseses TRE elements in their promoter regions and have been shown to regulate their own transcription[46]. As shown in Figure 6C, cells that overexpress xCT have increased levels of both c-fos (2.05 fold increase) and c-Jun (2.56 fold increase) messages. Both peak within the first hour after stimulation. Since c-Jun was isolated in the original screen, we tested levels of c-Jun protein and found that basal levels were increased (Figure 6D) and that a large peak in c-Jun protein was found reproducibly at 3 hrs post-growth factor stimulation in xCT overexpressing cells. The peak at this time fits well with the G1 blockade by the calcium antagonists and the ability of xCT cells to progress to S-phase. Taken together, these results indicate that the effect of xCT overexpression on AP-1 activity is due to increased levels and activity of jun and fos caused by increased intracellular thiol reducing equivalents which are sufficient to allow cell cycle progression even in the presence of calcium channel antagonists. It is worth noting that other transcription factors that we tested share some of these regulatory attributes with AP-1 e.g. NF-κB is highly redox regulated, NFAT is regulated by calcium and ectopic expression of Myc can bypass the requirement for mitogenic signals for entry into S phase [47, 48]. However, since no difference in activity between control and xCT cells for these transcription factors was observed, indicates that AP-1 is a specific target of xCT overexpression in these cells.
DISCUSSION
Cell cycle arrest in late G1 by cobalt and dihydropyridine-type calcium channel blockers is a well established phenomenon, though pharmacological approaches have failed to identify the mechanisms involved [13, 16, 23–26]. This growth inhibition is at the heart of several therapies proposed to counteract undesirable cell proliferation in sensitive cell types[49]. The genetic screens presented here represent an unbiased approach to the identification of genes involved in calcium regulation of cell cycle progression. Although genes with known links to calcium signaling were isolated in these screens, the gene that had the greatest effect on bypassing the drug-induced cell cycle arrest was the cystine-glutamate exchanger. Our results show that xCT acts as a dosage dependent suppressor of the cell cycle arrest induced by calcium channel blockers through its effects on cellular antioxidant production. Expression levels of xCT directly affect cellular cystine uptake and in turn GSH levels in mammalian cells. Our results indicate that increased GSH due to xCT overexpression directly affects Ca2+ physiology, AP-1 activation and cell cycle progression.
xCT is a member of the CD98 light chain family [50]. These variable substrate specificity transporters all share a 12-transmembrane segment topology and association with a single transmembrane domain CD98 heavy chain protein. The CD98 heavy chain is a surface glycoprotein that was initially isolated on T lymphocytes and has subsequently been found on a number of rapidly proliferating cells[51]. This protein has been implicated in integrin function [52], HIV gp160 induced cell fusion [53], and oncogenic transformation[54]. Although poorly understood at present, these attributes suggest a dynamic role for the heterodimers in linking solute transport to proliferation-related aspects of the immediate cellular environment. Correlative studies have indicated that GSH levels are linked to cystine transport in a number of cells including endothelial cells [35, 55], smooth muscle cells [56], macrophages [57], fibroblasts [58], brain cells [40, 59] and pancreatic acinar and islet cells [60]. Although xCT is generally thought to impact chemotherapeutic resistance[33, 37], our results show that it directly impacts cell proliferation signaling through AP-1 by increasing GSH levels in mouse fibroblasts. This is of note as xCT and its associated heavy chain, CD98, are frequently overexpressed in tumors [33, 34].
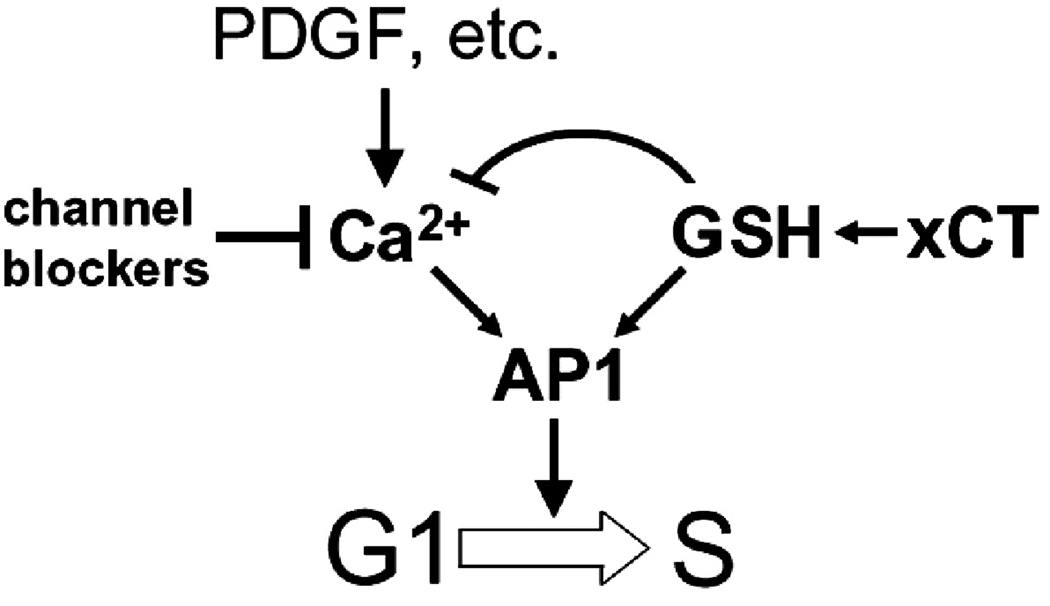
c-Jun was also isolated as a gene whose overexpression allowed cells to progress through the cell cycle in the presence of calcium channel antagonists, albeit to a lesser degree than did xCT (Table I). C-Jun expression has been shown to promote entry into S-phase in the absence of mitogenic stimulation [48, 61] and substantial evidence shows that it is redox regulated (reviewed by [62]. Figure 4 shows that cells that overexpress xCT have increased signaling through AP-1 even in the presence of calcium channel antagonists. A model explaining these relationships and which is consistent with our results is shown in Figure 7. Overexpression of xCT increases reduced intracellular thiols or GSH levels within cells which in turn effectively bypasses the requirement for Ca2+ signaling in G1. Since xCT overexpression did not lead to increased proliferation rates and entered quiescence normally upon serum starvation, we infer that cytokine signaling is still required for proliferation and that AP-1 may be a critical target of calcium signaling in G1. Calcium regulation of AP-1 has been found in basic calcium phosphate induced activation of human fibroblasts[63], in arachidonic acid signaling in stromal cells[64], hypoxic stress response in lung cancer cells [65] and in electrical signaling in neurons[66]. It is possible that other targets of GSH exist and since xCT overexpression was found to have the largest effect on bypassing the G1 arrest, it may be that it is the intracellular redox environment that is most important for the onset of DNA synthesis. GSH is increasingly being recognized as a general regulatory molecule whose modification of proteins is part of normal cell physiology and signaling [67–70] and recent studies have shown that intracellular reduction is the key requirement for allowing DNA synthesis to occur[71].
Figure 7.
Model depicting how Ca2+ signaling and the cellular redox environment may regulate cell cycle progression to S phase. Overexpression of xCT increases GSH levels within cells, thus bypassing the requirement for growth factor-induced Ca2+ influx. The resultant increase in cellular GSH levels has a negative feedback effect on cytoplasmic free Ca2+ concentrations.
The data also reinforce the notion that redox physiology and calcium metabolism are tightly linked in non-excitable cells [72]. Cells that overexpress xCT have lower cytoplasmic free calcium levels and the addition of GSH to control cells phenocopies this effect (Figure 5). Numerous studies have shown direct effects of cellular redox status on proteins that regulate intracellular Ca2+ levels [39]. Activity of the sarco/endoplasmic reticulum Ca2+ATPase (SERCA), which is required for progression through G1/S [73], is inhibited by oxidation [74, 75] as is the activity of the plasma membrane Ca2+ ATPase (PMCA) [76]. Similarly, oxidized GSH, (GSSG) has been shown to directly effect the depletion of IP3-sensitive Ca2+ stores [77] and, the activity of RYR is redox regulated [78, 79]. Although this implies that Ca2+-dependent processes may not be required for cell cycle progression, it is possible that these processes are activated at lower Ca2+ concentrations. Glutathionylation of a number of calcium binding proteins is known to occur [80–82], some of which display an significantly increased affinity for Ca2+ when modified in this way [82]. Those proteins that are activated by Ca2+-calmodulin, or directly by binding calcium could then effectively become activated without the requirement for normal calcium signaling per se. In any event the effects of GSH on Ca2+ levels are consistent with the seemingly paradoxical result of cells with the ability to overcome calcium channel antagonist-induced cell cycle arrest actually having less measurable calcium.
CONCLUSIONS
xCT acts as a dosage dependent suppressor of the cell cycle arrest induced by calcium channel blockers through its effects on cellular antioxidant production.
Expression levels of xCT directly affect cellular cystine uptake and in turn GSH levels in mammalian cells.
Increased GSH levels due to xCT overexpression directly affect Ca2+ physiology, generally decreasing calcium levels under a variety of conditions.
AP-1 activity is increased by xCT overexpression allowing cell cycle progression to occur.
Supplementary Material
Supplementary Figure 1. Overexpression of xCT does not confer resistance to several other drugs that inhibit proliferation. Growth of 3T3 cells infected with empty HM IV retrovirus or HM IV-xCT. Cells at a density of 104 cells/well were treated as indicated and growth after 5 days was visualized by crystal violet staining.
Supplementary Figure 2. Overexpression of the multidrug resistance gene (MDR1) does not confer resistance to nifedipine. NIH3T3 cells infected with retroviruses directing the expression of the p-glycoprotein MDR1, xCT and a control (empty) vector were exposed to vinblastine and nifedipine at the indicated concentrations. Growth after 5 days was measured by crystal violet staining.
Supplementary Figure 3. ERK expression patterns in vector control cells were indicative of quiescence while cells overexpressing xCT continued to display a proliferative pattern in the presence of nifedipine. Swiss 3T3 fibroblasts were serum starved for 24 hours then stimulated with 20 ng/ml PDGF in the absence (top two panels from a single blot) or presence of 50 µM nifedipine (bottom two panels from a single blot). Cell lysates were collected at the indicated times and expression of p42/44 ERK analyzed by immunoblotting. No difference in ERK expression was observed between the cell lines in the absence of nifedipine.
ACKNOWLEDGEMENTS
The authors would like to thank Drs. Nancy Thompson, Francois Verrey, Jeff Leiden, Lukas Kuhn, Igor Roninson for reagents and Xiaocun Chen for technical assistance. Cheryl Eifert and members of the Aguirre-Ghiso and Begley labs are thanked for helpful suggestions. Supported in part by NIH R43 GM59496-01 and U.S. Army Medical Research Acquisition Activity W8IWXH- 04-1-0474 grants to DSC.
REFERENCES
- 1.Kahl CR, Means AR. Endocr Rev. 2003;24(6):719–736. doi: 10.1210/er.2003-0008. [DOI] [PubMed] [Google Scholar]
- 2.Takuwa N, Iwamoto A, Kumada M, Yamashita K, Takuwa Y. J Biol Chem. 1991;266(3):1403–1409. [PubMed] [Google Scholar]
- 3.Barbiero G, Munaron L, Antoniotti S, Baccino FM, Bonelli G, Lovisolo D. Cell Calcium. 1995;18(6):542–556. doi: 10.1016/0143-4160(95)90016-0. [DOI] [PubMed] [Google Scholar]
- 4.Munaron L, Antoniotti S, Lovisolo D. J Cell Mol Med. 2004;8(2):161–168. doi: 10.1111/j.1582-4934.2004.tb00271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitfield JF, Boynton AL, MacManus JP, Sikorska M, Tsang BK. Mol Cell Biochem. 1979;27(3):155–179. doi: 10.1007/BF00215364. [DOI] [PubMed] [Google Scholar]
- 6.Hazelton B, Mitchell B, Tupper J. J Cell Biol. 1979;83(2 Pt 1):487–498. doi: 10.1083/jcb.83.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boynton AL, Whitfield JF. In Vitro. 1976;12(7):479–484. doi: 10.1007/BF02796490. [DOI] [PubMed] [Google Scholar]
- 8.Boynton AL, Whitfield JF, Isaacs RJ. J Cell Physiol. 1976;87(1):25–32. doi: 10.1002/jcp.1040870105. [DOI] [PubMed] [Google Scholar]
- 9.Boynton AL, Whitfield JF, Isaacs RJ. In Vitro. 1976;12(2):120–123. doi: 10.1007/BF02796358. [DOI] [PubMed] [Google Scholar]
- 10.Takuwa N, Zhou W, Kumada M, Takuwa Y. FEBS Lett. 1992;306(2–3):173–175. doi: 10.1016/0014-5793(92)80993-q. [DOI] [PubMed] [Google Scholar]
- 11.Miyakawa T, Kojima M, Ui M. Biochem J. 1998;329(Pt 1):107–114. doi: 10.1042/bj3290107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber KR, Schmidt WF, Thompson EA, Forsthoefel AM, Neuberg RW, Ettinger RS. Br J Cancer. 1989;59(5):714–718. doi: 10.1038/bjc.1989.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nilsson J, Sjolund M, Palmberg L, Von Euler AM, Jonzon B, Thyberg J. Atherosclerosis. 1985;58(1–3):109–122. doi: 10.1016/0021-9150(85)90059-0. [DOI] [PubMed] [Google Scholar]
- 14.Grier CE, 3rd, Mastro AM. J Cell Physiol. 1985;124(1):131–136. doi: 10.1002/jcp.1041240121. [DOI] [PubMed] [Google Scholar]
- 15.Zeitler H, Ko Y, Glodny B, Totzke G, Appenheimer M, Sachinidis A, Vetter H. Cancer Detect Prev. 1997;21(4):332–339. [PubMed] [Google Scholar]
- 16.Stepien O, Gogusev J, Zhu DL, Iouzalen L, Herembert T, Drueke TB, Marche P. J Cardiovasc Pharmacol. 1998;31(5):786–793. doi: 10.1097/00005344-199805000-00019. [DOI] [PubMed] [Google Scholar]
- 17.Munaron L, Antoniotti S, Fiorio Pla A, Lovisolo D. Curr Med Chem. 2004;11(12):1533–1543. doi: 10.2174/0929867043365008. [DOI] [PubMed] [Google Scholar]
- 18.Hannon GJ, Sun P, Carnero A, Xie LY, Maestro R, Conklin DS, Beach D. Science. 1999;283(5405):1129–1130. doi: 10.1126/science.283.5405.1129. [DOI] [PubMed] [Google Scholar]
- 19.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, Sheth N, Bradshaw J, Burchard J, Kulkarni A, Cavet G, Sachidanandam R, McCombie WR, Cleary MA, Elledge SJ, Hannon GJ. Nat Genet. 2005;37(11):1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 20.Sato H, Tamba M, Ishii T, Bannai S. J Biol Chem. 1999;274(17):11455–11458. doi: 10.1074/jbc.274.17.11455. [DOI] [PubMed] [Google Scholar]
- 21.Grynkiewicz G, Poenie M, Tsien RY. J Biol Chem. 1985;260(6):3440–3450. [PubMed] [Google Scholar]
- 22.Laemmli UK, Quittner SF. Virology. 1974;62(2):483–499. doi: 10.1016/0042-6822(74)90409-7. [DOI] [PubMed] [Google Scholar]
- 23.Lee YS, Sayeed MM, Wurster RD. Mol Chem Neuropathol. 1994;22(2):81–95. doi: 10.1007/BF03160097. [DOI] [PubMed] [Google Scholar]
- 24.Ko Y, Totzke G, Graack GH, Heidgen FJ, Meyer zu Brickwedde MK, Dusing R, Vetter H, Sachinidis A. J Hypertens. 1993;11(11):1171–1178. [PubMed] [Google Scholar]
- 25.Kataoka S, Alam R, Dash PK, Yatsu FM. Stroke. 1997;28(2):364–369. doi: 10.1161/01.str.28.2.364. [DOI] [PubMed] [Google Scholar]
- 26.Wang HX, Tao L, Rao MR. Zhongguo Yao Li Xue Bao. 1997;18(2):136–139. [PubMed] [Google Scholar]
- 27.Sun P, Dong P, Dai K, Hannon GJ, Beach D. Science. 1998;282(5397):2270–2272. doi: 10.1126/science.282.5397.2270. [DOI] [PubMed] [Google Scholar]
- 28.Brown R, Marshall CJ, Pennie SG, Hall A. Embo J. 1984;3(6):1321–1326. doi: 10.1002/j.1460-2075.1984.tb01970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi K, Frommel TO, Stern RK, Perez CF, Kriegler M, Tsuruo T, Roninson IB. Proc Natl Acad Sci U S A. 1991;88(16):7386–7390. doi: 10.1073/pnas.88.16.7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura E, Sato M, Yang H, Miyagawa F, Harasaki M, Tomita K, Matsuoka S, Noma A, Iwai K, Minato N. J Biol Chem. 1999;274(5):3009–3016. doi: 10.1074/jbc.274.5.3009. [DOI] [PubMed] [Google Scholar]
- 31.Pfeiffer R, Spindler B, Loffing J, Skelly PJ, Shoemaker CB, Verrey F. FEBS Lett. 1998;439(1–2):157–162. doi: 10.1016/s0014-5793(98)01359-3. [DOI] [PubMed] [Google Scholar]
- 32.Dun Y, Mysona B, Van Ells T, Amarnath L, Shamsul Ola M, Ganapathy V, Smith SB. Cell Tissue Res. 2006;324(2):189–202. doi: 10.1007/s00441-005-0116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, Dai Z, Barbacioru C, Sadee W. Cancer Res. 2005;65(16):7446–7454. doi: 10.1158/0008-5472.CAN-04-4267. [DOI] [PubMed] [Google Scholar]
- 34.Hara K, Kudoh H, Enomoto T, Hashimoto Y, Masuko T. Oncogene. 2000;19(54):6209–6215. doi: 10.1038/sj.onc.1204019. [DOI] [PubMed] [Google Scholar]
- 35.Miura K, Ishii T, Sugita Y, Bannai S. Am J Physiol. 1992;262(1 Pt 1):C50–C58. doi: 10.1152/ajpcell.1992.262.1.C50. [DOI] [PubMed] [Google Scholar]
- 36.Warr O, Takahashi M, Attwell D. J Physiol. 1999;514(Pt 3):783–793. doi: 10.1111/j.1469-7793.1999.783ad.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okuno S, Sato H, Kuriyama-Matsumura K, Tamba M, Wang H, Sohda S, Hamada H, Yoshikawa H, Kondo T, Bannai S. Br J Cancer. 2003;88(6):951–956. doi: 10.1038/sj.bjc.6600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bannai S, Ishii T. J Cell Physiol. 1982;112(2):265–272. doi: 10.1002/jcp.1041120216. [DOI] [PubMed] [Google Scholar]
- 39.Zima AV, Blatter LA. Cardiovasc Res. 2006;71(2):310–321. doi: 10.1016/j.cardiores.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 40.Kato S, Negishi K, Mawatari K, Kuo CH. Neuroscience. 1992;48(4):903–914. doi: 10.1016/0306-4522(92)90278-a. [DOI] [PubMed] [Google Scholar]
- 41.Weber JD, Raben DM, Phillips PJ, Baldassare JJ. Biochem J. 1997;326(Pt 1):61–68. doi: 10.1042/bj3260061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tombes RM, Auer KL, Mikkelsen R, Valerie K, Wymann MP, Marshall CJ, McMahon M, Dent P. Biochem J. 1998;330(Pt 3):1451–1460. doi: 10.1042/bj3301451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cook SJ, Aziz N, McMahon M. Mol Cell Biol. 1999;19(1):330–341. doi: 10.1128/mcb.19.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaulian E, Karin M. Oncogene. 2001;20(19):2390–2400. doi: 10.1038/sj.onc.1204383. [DOI] [PubMed] [Google Scholar]
- 45.Eferl R, Wagner EF. Nat Rev Cancer. 2003;3(11):859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 46.Lamph WW, Wamsley P, Sassone-Corsi P, Verma IM. Nature. 1988;334(6183):629–631. doi: 10.1038/334629a0. [DOI] [PubMed] [Google Scholar]
- 47.Cole MD, Nikiforov MA. Curr Top Microbiol Immunol. 2006;302:33–50. doi: 10.1007/3-540-32952-8_2. [DOI] [PubMed] [Google Scholar]
- 48.Clark W, Black EJ, MacLaren A, Kruse U, LaThangue N, Vogt PK, Gillespie DA. Mol Cell Biol. 2000;20(7):2529–2542. doi: 10.1128/mcb.20.7.2529-2542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schachter M. Int J Cardiol. 1997;62 Suppl 2:S9–S15. doi: 10.1016/s0167-5273(97)00236-2. [DOI] [PubMed] [Google Scholar]
- 50.Verrey F, Jack DL, Paulsen IT, Saier MH, Jr, Pfeiffer R. J Membr Biol. 1999;172(3):181–192. doi: 10.1007/s002329900595. [DOI] [PubMed] [Google Scholar]
- 51.Diaz LA, Jr, Fox DA. J Biol Regul Homeost Agents. 1998;12(1–2):25–32. [PubMed] [Google Scholar]
- 52.Fenczik CA, Sethi T, Ramos JW, Hughes PE, Ginsberg MH. Nature. 1997;390(6655):81–85. doi: 10.1038/36349. [DOI] [PubMed] [Google Scholar]
- 53.Ohgimoto S, Tabata N, Suga S, Nishio M, Ohta H, Tsurudome M, Komada H, Kawano M, Watanabe N, Ito Y. J Immunol. 1995;155(7):3585–3592. [PubMed] [Google Scholar]
- 54.Hara K, Kudoh H, Enomoto T, Hashimoto Y, Masuko T. Biochem Biophys Res Commun. 1999;262(3):720–725. doi: 10.1006/bbrc.1999.1051. [DOI] [PubMed] [Google Scholar]
- 55.Susanto I, Wright SE, Lawson RS, Williams CE, Deneke SM. Am J Physiol. 1998;274(2 Pt 1):L296–L300. doi: 10.1152/ajplung.1998.274.2.L296. [DOI] [PubMed] [Google Scholar]
- 56.Siow RC, Ishii T, Sato H, Taketani S, Leake DS, Sweiry JH, Pearson JD, Bannai S, Mann GE. FEBS Lett. 1995;368(2):239–242. doi: 10.1016/0014-5793(95)00650-x. [DOI] [PubMed] [Google Scholar]
- 57.Sato H, Fujiwara K, Sagara J, Bannai S. Biochem J. 1990;310(Pt 2):547–551. doi: 10.1042/bj3100547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bannai S. J Biol Chem. 1984;259(4):2435–2440. [PubMed] [Google Scholar]
- 59.Murphy TH, Miyamoto M, Sastre A, Schnaar RL, Coyle JT. Neuron. 1989;2(6):1547–1558. doi: 10.1016/0896-6273(89)90043-3. [DOI] [PubMed] [Google Scholar]
- 60.Sato H, Kuriyama-Matsumura K, Siow RC, Ishii T, Bannai S, Mann GE. Biochim Biophys Acta. 1998;1414(1–2):85–94. doi: 10.1016/s0005-2736(98)00159-x. [DOI] [PubMed] [Google Scholar]
- 61.Clark W, Gillespie DA. Cell Growth Differ. 1997;8(4):371–380. [PubMed] [Google Scholar]
- 62.Gius D, Botero A, Shah S, Curry HA. Toxicol Lett. 1999;106(2–3):93–106. doi: 10.1016/s0378-4274(99)00024-7. [DOI] [PubMed] [Google Scholar]
- 63.McCarthy GM, Augustine JA, Baldwin AS, Christopherson PA, Cheung HS, Westfall PR, Scheinman RI. J Biol Chem. 1998;273(52):35161–35169. doi: 10.1074/jbc.273.52.35161. [DOI] [PubMed] [Google Scholar]
- 64.Rizzo MT, Leaver AH, Yu WM, Kovacs RJ. Prostaglandins Leukot Essent Fatty Acids. 1999;60(3):187–198. doi: 10.1054/plef.1999.0024. [DOI] [PubMed] [Google Scholar]
- 65.Salnikow K, Kluz T, Costa M, Piquemal D, Demidenko ZN, Xie K, Blagosklonny MV. Mol Cell Biol. 2002;22(6):1734–1741. doi: 10.1128/MCB.22.6.1734-1741.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cruzalegui FH, Hardingham GE, Bading H. Embo J. 1999;18(5):1335–1344. doi: 10.1093/emboj/18.5.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rigacci S, Iantomasi T, Marraccini P, Berti A, Vincenzini MT, Ramponi G. Biochem J. 1997;324(Pt 3):791–796. doi: 10.1042/bj3240791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gomez LD, Noctor G, Knight MR, Foyer CH. J Exp Bot. 2004;55(404):1851–1859. doi: 10.1093/jxb/erh202. [DOI] [PubMed] [Google Scholar]
- 69.Nakamura H, Nakamura K, Yodoi J. Annu Rev Immunol. 1997;15:351–369. doi: 10.1146/annurev.immunol.15.1.351. [DOI] [PubMed] [Google Scholar]
- 70.Shackelford RE, Heinloth AN, Heard SC, Paules RS. Antioxid Redox Signal. 2005;7(7–8):940–950. doi: 10.1089/ars.2005.7.940. [DOI] [PubMed] [Google Scholar]
- 71.Chen Z, Odstrcil EA, Tu BP, McKnight SL. Science. 2007;316(5833):1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- 72.Reed DJ. Semin Liver Dis. 1990;10(4):285–292. doi: 10.1055/s-2008-1040484. [DOI] [PubMed] [Google Scholar]
- 73.Simon VR, Moran MF. Cell Prolif. 2001;34(1):15–30. doi: 10.1046/j.1365-2184.2001.00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grover AK, Samson SE, Fomin VP, Werstiuk ES. Am J Physiol. 1995;269(3 Pt 1):C546–C553. doi: 10.1152/ajpcell.1995.269.3.C546. [DOI] [PubMed] [Google Scholar]
- 75.Barnes KA, Samson SE, Grover AK. Mol Cell Biochem. 2000;203(1–2):17–21. doi: 10.1023/a:1007053802481. [DOI] [PubMed] [Google Scholar]
- 76.Chen B, Mayer MU, Squier TC. Biochemistry. 2005;44(12):4737–4747. doi: 10.1021/bi0474113. [DOI] [PubMed] [Google Scholar]
- 77.Renard-Rooney DC, Joseph SK, Seitz MB, Thomas AP. Biochem J. 1995;310(Pt 1):185–192. doi: 10.1042/bj3100185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saimi Y, Kung C. Annu Rev Physiol. 2002;64:289–311. doi: 10.1146/annurev.physiol.64.100301.111649. [DOI] [PubMed] [Google Scholar]
- 79.Balshaw DM, Yamaguchi N, Meissner G. J Membr Biol. 2002;185(1):1–8. doi: 10.1007/s00232-001-0111-4. [DOI] [PubMed] [Google Scholar]
- 80.Zhukova L, Zhukov I, Bal W, Wyslouch-Cieszynska A. Biochim Biophys Acta. 2004;1742(1–3):191–201. doi: 10.1016/j.bbamcr.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 81.Cedervall T, Berggard T, Borek V, Thulin E, Linse S, Akerfeldt KS. Biochemistry. 2005;44(2):684–693. doi: 10.1021/bi049232r. [DOI] [PubMed] [Google Scholar]
- 82.Goch G, Vdovenko S, Kozlowska H, Bierzynski A. Febs J. 2005;272(10):2557–2565. doi: 10.1111/j.1742-4658.2005.04680.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Overexpression of xCT does not confer resistance to several other drugs that inhibit proliferation. Growth of 3T3 cells infected with empty HM IV retrovirus or HM IV-xCT. Cells at a density of 104 cells/well were treated as indicated and growth after 5 days was visualized by crystal violet staining.
Supplementary Figure 2. Overexpression of the multidrug resistance gene (MDR1) does not confer resistance to nifedipine. NIH3T3 cells infected with retroviruses directing the expression of the p-glycoprotein MDR1, xCT and a control (empty) vector were exposed to vinblastine and nifedipine at the indicated concentrations. Growth after 5 days was measured by crystal violet staining.
Supplementary Figure 3. ERK expression patterns in vector control cells were indicative of quiescence while cells overexpressing xCT continued to display a proliferative pattern in the presence of nifedipine. Swiss 3T3 fibroblasts were serum starved for 24 hours then stimulated with 20 ng/ml PDGF in the absence (top two panels from a single blot) or presence of 50 µM nifedipine (bottom two panels from a single blot). Cell lysates were collected at the indicated times and expression of p42/44 ERK analyzed by immunoblotting. No difference in ERK expression was observed between the cell lines in the absence of nifedipine.