Abstract
The cytochrome P450 26 family is believed to be responsible for all-trans-retinoic acid (atRA) metabolism and elimination in the human fetus and adults. CYP26A1 and CYP26B1 mRNA is expressed in a tissue-specific manner, and mice in which the CPY26 isoform has been knocked out show distinct malformations and lethality. The aim of this study was to determine differences in CYP26A1 and CYP26B1 regulation and expression. Analysis of CYP26A1 and CYP26B1 expression in a panel of 57 human livers showed CYP26A1 to be the major CYP26 isoform present in the liver, and its expression to be subject to large interindividual variability between donors. CYP26A1 and retinoic acid receptor (RAR) β were found to be greatly inducible by atRA in HepG2 cells, whereas CYP26B1, RARα, and RARγ were induced to a much lesser extent. Based on treatments with RAR isoform-selective ligands, RARα is the major isoform responsible for CYP26A1 and RARβ induction in HepG2 cells. Classic cytochrome P450 inducers did not affect CYP26 transcription, whereas the peroxisome proliferator-activated receptor (PPAR) γ agonists pioglitazone and rosiglitazone up-regulated CYP26B1 transcription by as much as 209- ± 80-fold and CYP26A1 by 10-fold. RARβ was also up-regulated by pioglitazone and rosiglitazone. CYP26B1 induction by PPARγ agonists was abolished by the irreversible PPARγ antagonist 2-chloro-5-nitrobenzanilide (GW9662), whereas RARβ and CYP26A1 induction was unaffected by GW9662. Overall, the results of this study suggest that CYP26B1 and CYP26A1 are regulated by different nuclear receptors, resulting in tissue-specific expression patterns. The fact that drugs can alter the expression of CYP26 enzymes may have toxicological and therapeutic importance.
Vitamin A is essential for many biological functions including maintenance of epithelia, the immune system, regulation of apoptosis, embryonic development and osteogenesis (Blomhoff and Blomhoff, 2006). Most biological effects of atRA are mediated by all-trans-retinoic acid (atRA) binding to nuclear retinoic acid receptors (RAR), which heterodimerize with retinoid X receptors (RXR) and regulate the transcription of an array of target genes. Because of its multiple biological effects, the in vivo concentrations of atRA, the active form of vitamin A (Lampen et al., 2001a), are tightly controlled (Bendich and Langseth, 1989; Hathcock et al., 1990; Clagett-Dame and DeLuca, 2002). It has been proposed that cellular concentrations of atRA are regulated by complex systems that include synthesis of atRA from retinal by retinaldehyde dehydrogenases and elimination of atRA by metabolic enzymes, among which the CYP26 family seems most crucial (Napoli, 1999; Ross, 2003; Duester, 2008).
Cytochrome P450 (P450) family 26 has three human isoforms: CYP26A1, CYP26B1, and CYP26C1. All three are known to metabolize atRA (White et al., 1997, 2000; Taimi et al., 2004). Based on knockout mice data, CYP26A1 and CYP26B1 are both essential for development, whereas CYP26C1 is functionally redundant with CYP26A1 (Abu-Abed et al., 2001; Sakai et al., 2001; Yashiro et al., 2004; Uehara et al., 2007). Based on mRNA data in mouse, chick, and zebrafish embryos, the expression of CYP26 isoforms is very cell- and tissue-specific, and the different isoforms are rarely expressed simultaneously in the same tissue during development (MacLean et al., 2001; Sakai et al., 2001; Abu-Abed et al., 2002; Reijntjes et al., 2004; Yashiro et al., 2004; Hernandez et al., 2007). RNA expression data from adult human tissues has also indicated that CYP26 enzymes are expressed in a tissue-specific manner (White et al., 2000; Xi and Yang, 2008).
There is compelling evidence that CYP26A1 expression is induced by atRA, and two distinct RAREs have been characterized in the CYP26A1 promoter (Loudig et al., 2000; Ozpolat et al., 2005; Zolfaghari et al., 2007), but other processes have also been shown to contribute to CYP26 regulation. Organochlorine pesticides have been shown to activate RARs and strongly induce CYP26A1 in HepG2 cells (Lemaire et al., 2005). The tumor suppressor adenomatous polyposis coli, signaling via WNT-independent and WNT-dependent pathways up-regulate CYP26A1 expression in human and mouse adenomas and in the intestine of apcmcr mutant zebrafish embryos (Shelton et al., 2006). Sex hormones such as gestagens up-regulate CYP26A1 expression in mouse uterus (Fritzsche et al., 2007), whereas lipopolysaccharide-induced inflammation suppresses atRA-induced CYP26A1 and CYP26B1 expression in rat liver (Zolfaghari et al., 2007). These studies suggest that complex cross-talk exists in pathways that regulate CYP26 expression. However, no studies have been published that would have compared the regulation of CYP26A1 and CYP26B1 in a specific cell system, and mechanisms that control CYP26B1 transcription are largely not established.
We hypothesized that the biological, phenotypic differences between CYP26A1 and CYP26B1 are due to differences in the regulation of these enzymes. The aim of this study was to determine whether different mechanisms are responsible for regulation of CYP26A1 and CYP26B1 transcription in human liver and to test whether xenobiotics can affect CYP26 transcription.
Materials and Methods
Materials.
The hepatocarcinoma HepG2 cell line was a gift from Dr. Kenneth E. Thummel (University of Washington, Seattle, WA), and hepatocytes were purchased from CellzDirect (Durham, NC). Actinomycin D, all-trans-atRA, AM580, clofibrate, dexamethasone, 5,6-dichlorobenzimidazole 1-β-d-ribofuranoside (DRB), estradiol, GW9662, L- 165,041, phenobarbital, phenytoin, progesterone, and rifampicin were purchased from Sigma-Aldrich (St. Louis, MO). Pioglitazone was purchased from Altan Biochemicals (Orange, CT), and rosiglitazone and troglitazone were from Cayman Chemical (Ann Arbor, MI). TTNPB and AC55649 were purchased from Tocris (Ellisville, MO). Stock solutions were prepared in either DMSO or ethanol and stored at −20°C. TaqMan real-time Universal PCR Master Mix and PCR primers and fluorescent probes were obtained from Applied Biosystems (Foster City, CA). Probes were labeled with the 5′ reporter dye 5-carboxyfluorescein and a nonfluorescent black hole quencher on the 3′ end. Primer and probe pairs used include: CYP26A1 (Hs00175627_m1), CYP26B1 (Hs00219866_m1), CYP4A11 (Hs00167961_m1), GAPDH (Hs99999905_m1), peroxisome proliferator-activated receptor (PPAR) α (Hs00947539_m1), PPARδ (Hs00602622_m1), PPARγ (Hs01115512_m1), RARα (Hs00940446_m1), RARβ (Hs00233407_m1), and RARγ (Hs00171273_m1).
Cell Culture.
HepG2 cells were maintained in 5% carbon dioxide in a humidified incubator at 37°C. The growth medium used was Dulbecco's modified Eagle's medium (Invitrogen, Carlsbad, CA) supplemented with 10% (v/v) fetal bovine serum, 1% nonessential amino acids, 1% sodium pyruvate, and 50 mg/liter penicillin-streptomycin. Cells were plated at 106 cells per well in six-well plastic tissue culture plates and were given 24 h to adhere before treatment began. All treatments were added as 0.1% DMSO diluted in either growth medium or differentiation medium (DM). The differentiation medium contained growth medium with the addition of 5 nM atRA. DMSO (0.1%) or ethanol vehicle treatment was used as control.
RNA Extraction and Quantitative PCR.
All HepG2 cells and hepatocytes were harvested, and total RNA was isolated from each well using TRI reagent (Invitrogen) according to the manufacturer's recommendations. Total RNA was quantified using the Quant-iT RiboGreen RNA Assay Kit (Invitrogen), and RNA quality was confirmed via gel electrophoresis. cDNA was generated by reverse transcription using the TaqMan reverse transcription reagents kit (Applied Biosystems) and 1 μg of total RNA according to the manufacturer's recommendations. Quantitative real-time PCR was conducted using relevant TaqMan primers and probes on a StepOnePlus Real-Time PCR instrument (Applied Biosystems) using one holding stage cycle of 50°C for 2 min, then 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min.
Absolute Quantification of CYP26A1 and CYP26B1.
CYP26B1 cDNA was obtained from Origene (Rockville, MD). Full-length CYP26A1 mRNA was extracted from human embryonic kidney 293 cells, and cDNA was synthesized using reverse transcription-PCR. The CYP26A1 cDNA was cloned into pCRblunt-II TOPO vector (Invitrogen), and the sequence was verified to be identical with that of GenBank accession number NM000783. CYP26A1 cDNA was obtained by PCR amplification of this plasmid (forward primer, 5′-gctcgagatggggctcccggcgctgc-3′ and reverse primer: 5′-cgcatgctcagatttccccatggaaatg-3′) and the resulting product was purified and quantified. Linear CYP26B1 cDNA was generated in a similar way using forward primer 5′- gctcgagatgctctttgagggcttgg-3′ and reverse primer 5′- cgcatgcttagactgtggcgctcagcatg-3′. The linear cDNA copy number was determined using the Quant-iT dsDNA Assay Kit Broad Range (Invitrogen). Serial dilutions of the DNA ranging from 4 copies/μl to 4.2 × 106 copies/μl were prepared and amplified using the real-time PCR and PCR conditions as stated above. The linearity and amplification efficiency of the CYP26A1 and CYP26B1 real-time assays were validated using CYP26A1 and CYP26B1 linearized plasmid cDNA. A linear relationship between CT values and log-transformed cDNA copy numbers was observed between 420 and 4.2 × 106 copies (y = −1.44ln(x)+45.46, r2 = 0.9957) for CYP26A1 and between 5000 and 2.5 × 106 copies (y = −1.51ln(x)+45.98,2 = 0.9999) for CYP26B1. The amplification efficiency of the two CYP26 isoforms was similar, although the CYP26A1 assay allowed accurate quantification of lower copy numbers. We could accurately quantify CYP26A1 above 500 copies per well (CT ≈ 36) and CYP26B1 above 5000 copies (CT ≈ 34) of the cDNA. The relationship between lower amounts of cDNA copies and threshold values was not linear; hence, CT values higher than 36 for CYP26A1 and 34 for CYP26B1 were not used in any of the subsequent analyses.
Human Liver CYP26 Expression.
CYP26A1 and CYP26B1 RNA were quantified from 57 human livers from the University of Washington human liver bank. The liver tissue in the bank is from anonymized donors; donor age, gender, cause of death, ICU medications, home medications, and liver pathology were recorded for all donors. RNA was extracted and cDNA was synthesized from 1 μg of mRNA using the method described previously for HepG2 cells under Cell Culture. 10% of the total cDNA synthesized was used for each real-time PCR reaction. Absolute RNA quantification from each human sample was done using real-time PCR, and the standard curve was obtained with linearized cDNA. Livers that had GAPDH values more than 2 S.D. from the mean (n = 3) were excluded from analysis. For included livers, GAPDH CT values had a standard deviation of ±1. Livers that had CYP26 RNA copy numbers lower than the limit of quantification obtained from the standard curve were excluded from further analysis.
Seven human liver microsome samples were selected based on CYP26A1 mRNA levels to represent high and low CYP26A1 mRNA expression, and CYP26A1 protein expression was measured using Western blotting. Microsomal preparations were diluted in sample buffer to yield a final concentration of 4 μg/μl. The diluted microsomal preparations were boiled (3 min), loaded onto 0.25% SDS-10% polyacrylamide gels (8 × 15 cm), and the proteins were separated by electrophoresis. The proteins were transferred for 1 h at 100 V and 1.5 A to polyvinylidene difluoride membranes (Millipore, Billerica, MA), after which the membranes were placed in blocking buffer [50% Odyssey block (LI-COR Biosciences, Lincoln, NE) and 50% PBS] for 1 h at room temperature. Tween 20 (final concentration, 0.1%) was added together with the primary antibodies. The membranes were incubated with rabbit anti-CYP26A1 antibody (Lutz et al., 2009) at a 1:50,000 dilution overnight, after which the membrane was rinsed four times with PBS-Tween 20 and incubated for 1 h with the secondary Alexa Fluor 680 (Invitrogen) anti-rabbit antibody mixture (1:4000) in 1:1 mixture of Odyssey blocking buffer and PBS-0.1% Tween 20. The membrane was rinsed again with PBS-0.1% Tween 20 and stored in PBS at 4°C until imaged. CYP26A1 was visualized by fluorescence using Odyssey infrared imaging system (LI-COR Biosciences).
Induction of CYP26 by atRA.
CYP26 induction by atRA was studied in HepG2 cells grown in growth medium. The time course of CYP26 induction was determined with 100 nM atRA treatments, and cells were harvested at time points between 4 and 72 h of treatment. Media was changed every 24 h to maintain the presence of RA. After a peak induction time of 24 h was detected, the EC50 of CYP26 induction by atRA was determined by treating cells with seven different atRA concentrations between 1 nM and 1 μM.
CYP26 RNA Half-Life.
To determine the half-life of CYP26A1 and CYP26B1 mRNA, we used the RNA synthesis inhibitors DRB and actinomycin D. HepG2 cells were pretreated with 500 nM atRA, and after 24 h, the cells were washed with PBS and then treated with either 80 μM DRB or 200 nM actinomycin D. Cells were harvested at four time points after DRB or actinomycin D treatment (0, 6, 12, and 24 h), and RNA was extracted. Half-life of the mRNA was calculated from a log-linear fit of RNA copy number as percentage of time 0 h versus time.
Effect of RAR Isoforms on CYP26 Induction.
To investigate the different effects of the individual RAR isoforms on CYP26 regulation in HepG2 cells, we used three selective RAR agonists: AM580 for RARα (Delescluse et al., 1991), AC55649 for RARβ (Lund et al., 2005), and TTNPB as a pan-RAR agonist (Nagpal et al., 1995). Cells were treated for 24 h with the RAR agonists at a concentration of 10 nM, and cells were harvested at the end of this time period. For these agonists, a concentration of 10 nM has been shown to be selective for the target RARs, and the relevant RAR(s) have Kd values less than 10 nM (Delescluse et al., 1991; Nagpal et al., 1995; Lund et al., 2005). The relative expression levels of the individual RAR isoforms, as indicated by mRNA levels, were quantified using real-time PCR in the HepG2 cells at baseline and after atRA treatment.
Screen of Xenobiotics for CYP26A1 or CYP26B1 Induction.
A panel of 11 xenobiotics was screened for their possible inductive effects on CYP26A1 and CYP26B1. The xenobiotics were selected based on their known regulatory effects on P450 expression. This panel included carbamazepine (20 μM), clofibrate (10 and 100 μM), dexamethasone (10 μM), estradiol (10 μM), L-165,041 (10 and 100 μM), phenobarbital (1 mM), phenytoin (20 μM), progesterone (10 μM), rifampin (20 μM), rosiglitazone (10 and 100 μM), and troglitazone (20 μM). HepG2 cells were treated with each of these compounds in growth medium for 48 h after plating, and total RNA was extracted and analyzed.
Effect of PPAR Agonist Concentrations on CYP26 Transcription.
HepG2 cells were plated and pretreated with DM for 24 h before treatment began. First cells were treated with 100 μM clofibrate (PPARα), L-165,041 (PPARβ/δ), and rosiglitazone and pioglitazone (PPARγ) in DM, and the time course of CYP26 induction by these compounds was determined. Treatment medium was changed, and cells were harvested every 24 h until 120 h. Total RNA was extracted and target transcripts were quantified by real-time PCR. Then, cells were treated for 72 h with clofibrate, L-165,041, rosiglitazone, and pioglitazone at four concentrations between 0.5 and 100 μM. A separate experiment was conducted to determine the EC50 value for CYP26B1 induction by pioglitazone using six concentrations between 0.5 and 250 μM pioglitazone.
Effect of GW9662, a PPARγ Antagonist, on CYP26 Induction by Pioglitazone and Rosiglitazone.
After 24 h pretreatment with DM, DMSO vehicle or 10 μM GW9662 (irreversible PPARγ antagonist) was added to cells and allowed to equilibrate for 2 h before adding 50 μM rosiglitazone or pioglitazone. Cells were redosed with fresh media and drugs (either GW9662 or vehicle and rosiglitazone/pioglitazone) every 24 h until they were harvested at 72 h for mRNA quantification.
Regulation of CYP26A1 and CYP26B1 in Human Hepatocytes.
Hepatocytes were maintained in 5% carbon dioxide in a humidified incubator at 37°C, and Williams E maintenance media was the growth medium (CellzDirect). Forty-eight-well plated fresh hepatocytes from two donors (Hu1076 and Hu1078) were purchased from CellzDirect. Hepatocytes were revived according to supplier's recommendation, and maintained with the use of Williams E maintenance media. Cells were either 1) treated with 100 nM atRA or 10 μM AM580 for 48 h or 2) pretreated with 5 nM atRA for 24 h and then treated with pioglitazone and 5 nM RA for 48 h with fresh pioglitazone plus atRA media added at 24 h after drug treatment began. All treatments were added as less than 0.1% DMSO diluted into Williams E maintenance media and DMSO, or DMSO with 5 nM atRA was used as the vehicle treatment. Cryopreserved hepatocytes from one donor (Hu4100) were thawed and plated according to manufacturer's recommendations at a density of 6 × 104 cells per well in a 96-well plate. The cells were given 24 h to adhere before treatment began. Cells were collected at 6, 24, and 48 h for analysis with fresh maintenance media, and drug was changed every 24 h. All treatments were added as less than 0.1% DMSO diluted into Williams E maintenance media, and DMSO was used as the vehicle treatment.
Data Analysis.
Each treatment was done as three biological replicates and the means and S.D. are reported. Treatments were compared with controls grown in the presence of vehicle for the same length of time. For all cell culture experiments, the relative quantification (-fold change) was calculated with the ΔΔCT method and GAPDH as a housekeeping gene (according to the manufacturer's instructions). Real-time analysis was done in duplicate, and the average between the two was used in calculations. EC50 and Emax values were determined using WinNonlin 5.2 (Pharsight, Cary, NC) and the equation E = E0 + (((Emax − E0) × Cγ)/(Cγ + EC50γ)), in which E0 is the expression level in the absence of the inducer, Emax is the maximum fold-induction, γ is the Hill coefficient (slope factor), C is the concentration of the inducer, and EC50 is the concentration of the inducer needed to obtain 50% of the maximum induction. Significant differences between treatments were evaluated using Student's t test with Bonferroni adjustment for multiple comparison, resulting in p < 0.01 being considered significant. Correlation between CYP26A1 and CYP26B1 mRNA as well as between CYP26 and RARα and PPARγ mRNA in human liver tissues was tested with linear regression. Differences in CYP26A1 and CYP26B1 expression levels between different liver pathologies were tested using Student's t test, with p < 0.05 considered significant.
Results
CYP26A1 and CYP26B1 Expression in Human Livers.
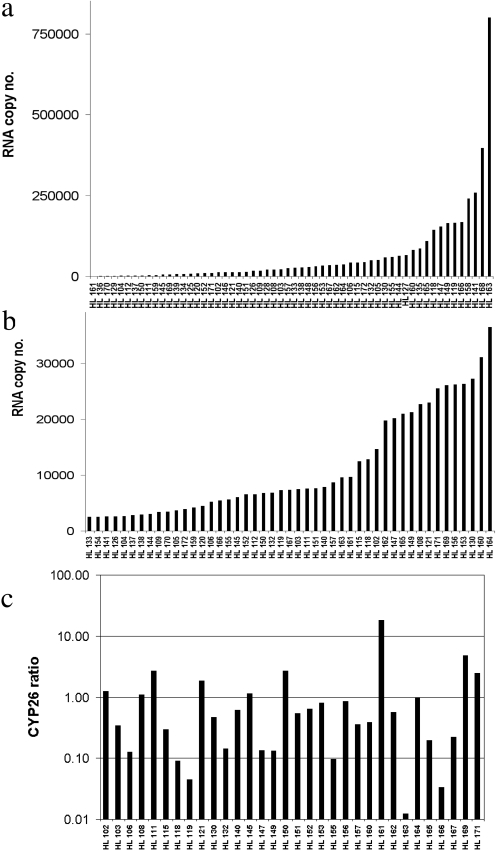
CYP26A1 and CYP26B1 mRNA was quantified in 57 human livers (Fig. 1, a and b). CYP26A1 and CYP26B1 were quantifiable in 95 and 54% of the livers analyzed, respectively. Based on absolute quantification, CYP26A1 transcripts were on average 6-fold higher than CYP26B1 mRNA. The mean amount of transcripts in the livers with quantifiable mRNA was 65,212 ± 124,544 and 14,815 ± 9149 for CYP26A1 and CYP26B1, respectively. However, in nine livers, CYP26B1 was more abundant than CYP26A1. The ratio between CYP26B1 and CYP26A1 transcripts is shown in Fig. 1c. No specific drug treatment or liver pathology was associated with the high CYP26B1 transcripts in these livers.
Fig. 1.
Absolute quantification of CYP26A1 (a) and CYP26B1 (b) mRNA in a panel of human livers. Absolute quantification of mRNA expression was determined by real-time PCR in 57 human livers using CYP26A1 and CYP26B1 cDNA as calibrators. The ratio between CYP26B1 and CYP26A1 mRNA expression is shown in c. In a and b, livers are presented in the order of increasing CYP26 transcript amounts, and the donor number is listed in the x-axis.
Large interindividual variability in CYP26A1 and CYP26B1 expression was observed. Within the livers of detectable transcripts, the variation for CYP26A1 expression was 300- and 474-fold within 1 and 2 S.D. of the mean copy number, respectively, and CYP26B1 copy numbers varied between 4- and 6-fold, within 1 and 2 S.D. of the mean copy number. No correlation between CYP26A1 and CYP26B1 transcripts was observed (R2 = 0.00006), indicating that CYP26A1 and CYP26B1 are not coregulated in this tissue.
The donor histories were investigated to determine factors affecting CYP26 regulation. Table 1 shows the analysis of the effect of fatty liver, liver hypoxia, smoking, and alcohol use in CYP26A1 and CYP26B1 transcripts. CYP26B1 transcripts were significantly (p < 0.05) higher in fatty livers than in nonfatty livers, whereas CYP26A1 transcripts were significantly lower (p < 0.05) in ischemic livers than in nonischemic livers. Other donor factors, such as drug treatment, alcohol, or tobacco use did not significantly affect CYP26 transcription, and none of the donors was treated with PPAR agonists such as rosiglitazone, pioglitazone, or clofibrate. There was no correlation between donor age and CYP26 transcription for either CYP26A1 or CYP26B1 (p > 0.05).
TABLE 1.
Effect of donor history on CYP26A1 and CYP26B1 expression
| mRNA | Alcohol |
Fatty liver |
Smoker |
Ischemia |
||||
|---|---|---|---|---|---|---|---|---|
| + | − | + | − | + | − | + | − | |
| × 103 | ||||||||
| CYP26A1 | 53 ± 73 | 73 ± 141 | 75 ± 161 | 60 ± 89 | 54 ± 71 | 74 ± 145 | 24 ± 25 | 87 ± 147 |
| P value | 0.01 | |||||||
| CYP26B1 | 17 ± 9 | 14 ± 9 | 19 ± 9 | 12 ± 9 | 16 ± 9 | 14 ± 9 | 13 ± 12 | 16 ± 8 |
| P value | 0.04 | |||||||
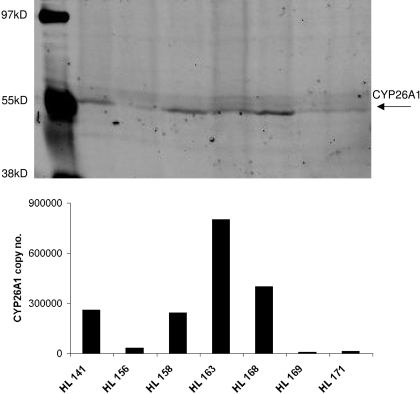
To test whether mRNA levels correlate with CYP26A1 protein, Western blotting was conducted from a set of seven human livers. Figure 2 shows the CYP26A1 protein detected in correlation with the mRNA levels in these livers. Expression of CYP26A1 could be predicted from the mRNA amount in the livers; in livers with low mRNA expression, no corresponding CYP26A1 protein was detectable, whereas in the livers with high mRNA quantification, CYP26A1 protein was detectable.
Fig. 2.
Expression of CYP26A1 and CYP26B1 protein in human liver microsomes. CYP26A1 expression was measured by Western blotting (top) and the expression level compared with mRNA quantification in the same livers (bottom). The bars in the bottom graph correspond to the same liver preparations run on the Western blot.
atRA and CYP26A1 Transcription in HepG2 Cells.
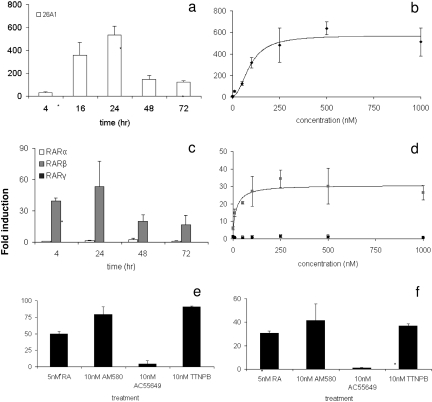
To investigate the differences in regulation of CYP26A1 and CYP26B1, we used HepG2 cells to first test the induction of CYP26 by atRA. Basal expression of CYP26B1 in the HepG2 cells was below our limit of quantification with an average CT value of 38, whereas CYP26A1 could be quantified in the absence of RA treatment with an average CT value of 36. Overall, CYP26A1 was more responsive to atRA treatment than CYP26B1. When HepG2 cells were treated with atRA, CYP26A1 transcription increased significantly (Fig. 3a). CYP26A1 induction peaked at 24 h after treatment (533- ± 80-fold induction) and then steadily declined to 123- ± 10-fold at 72 h of treatment. A dose response analysis of CYP26A1 induction by atRA after 24-h treatment yielded an EC50 value of 93 ± 19 nM and an Emax value of 572- ± 48-fold (Fig. 3b). CYP26B1 transcripts could not be accurately quantified at any time point of atRA treatment at 100 nM, and basal CYP26B1 expression was undetectable in the time-course experiment. In the dose-response experiment, however, CYP26B1 transcripts were quantifiable at atRA concentrations above 50 nM. It is noteworthy that increasing concentrations of atRA did not cause a detectable increase in CYP26B1 transcription.
Fig. 3.
Induction of CYP26 and RAR transcription by retinoic acid (atRA) and synthetic RAR agonists. CYP26A1 (a) and RAR isoforms (c) depict the time course of induction in HepG2 cells after treatment with 100 nM atRA. CYP26A1 (b) and RAR (d) show concentration-response curves for CYP26A1 and RARβ induction after 24 h of atRA treatment, reported as -fold induction compared with untreated control. d, light gray squares depict RARβ and closed squares depict RARα. Induction of CYP26A1 (e) and RARβ (f) by 10 nM AM580, an RARα agonist; 10 nM AC55649, an RARβ agonist; and 10 nM TTNPB, a RAR pan-agonist, compared with atRA. Data are presented as means ± S.D. (n = 3), and all -fold induction values are relative to vehicle control matched for culture time. *, significant (P < 0.01) difference between treated cells and untreated control.
To better explain the time course of CYP26A1 induction, the mRNA half-life of CYP26A1 and CYP26B1 was measured using DRB and actinomycin D treatments. The CYP26A1 mRNA half-life was 7 h after DRB treatment and 9 h after actinomycin D treatment, whereas CYP26B1 half-life was 7 h after both treatments (data not shown). Based on these half-life estimates after treatment with a direct inducer, a new steady state of CYP26 transcripts is predicted to be reached within 24 h of treatment. However, any indirect induction or negative feedback would prolong the time required to reach steady state.
Role of RAR Isoforms in CYP26 Induction.
To test whether the different extent of induction of CYP26A1 and CYP26B1 by atRA in the HepG2 cells was due to discrete involvement of RAR isoforms, two sets of experiments were conducted. First, the basal expression of RARα, RARβ, and RARγ in the HepG2 cells was measured by analyzing transcripts from control cells and after the various atRA treatments (Fig. 3, c and d). Second, the induction of CYP26A1, CYP26B1, and RAR isoforms was measured after treatment with isoform-selective RAR agonists (Fig. 3, e and f). Basal expression levels of RARα, RARβ, and RARγ were low in HepG2 cells. RARα was the most abundant isoform (CT 30–32), but RARβ could also be detected in all experiments (CT 34–38). The expression of RARγ was very low (CT between 35 and undetermined) suggesting a lack of corresponding protein in HepG2 cells. It is noteworthy that when we analyzed basal expression in human livers, the abundance rank order of the RAR isoforms was the same as in HepG2 cells, with RARα being the most prevalent (CT 28–30), followed by RARβ (CT 30–32), and finally by RARγ (CT 32–36). In the human liver samples, no correlation was observed between RARα mRNA and CYP26A1 or CYP26B1 mRNA values (p > 0.05). In HepG2 cells, RARβ but not RARα was inducible by 100 nM atRA, and the induction followed a similar time course as seen for CYP26A1 (Fig. 3c). The EC50 value for induction of RARβ by atRA was 21 ± 17 nM, and the corresponding Emax was 31- ± 4-fold. Transcription of RARα and RARγ was not induced even at 1 μM atRA (Fig. 3d).
The synthetic RARα-selective agonist AM580 and RAR pan-agonist TTNPB up-regulated CYP26A1 and CYP26B1 transcription as well as RARβ transcripts, at 10 nM, whereas the selective RARβ agonist AC55649 had no effect on CYP26 or RAR transcription (Fig. 3, e and f). Consistent with the data from atRA treatments, CYP26A1 induction was quantifiable (85- ± 7-fold) after RAR activation, whereas CYP26B1 induction could not be quantified because of the lack of baseline expression. After AM580 treatment, CYP26B1 transcripts were quantifiable (CT = 32), indicating induction by RARα. It is noteworthy that neither CYP26A1 nor CYP26B1 was induced after RARβ activation, but RARβ was up-regulated by AM580 and TTNPB (42- ± 14-fold and 37- ± 2-fold, respectively) similar to CYP26A1 (Fig. 3f). AC55649, the RARβ agonist, had no effect on RARβ transcription either.
Screen for Xenobiotic Inducers of CYP26A1 and CYP26B1.
Eleven xenobiotics, including carbamazepine, clofibrate, dexamethasone, estradiol, L-165,041, phenobarbital, phenytoin, progesterone, rifampin, rosiglitazone, and troglitazone, were tested for their ability to induce CYP26A1 and CYP26B1 transcription. These xenobiotics were specifically chosen to cover a mechanistically diverse group of compounds associated with P450 induction. In the absence of atRA pretreatment, CYP26B1 could not be quantified reliably, and no CYP26A1 induction was observed after any of the treatments. In the presence of 5 nM atRA, rosiglitazone and L-165,041 induced CYP26A1, whereas clofibrate down-regulated CYP26A1, and <10-fold induction was seen with estradiol (data not shown). We also saw an induction of CYP26B1 by L-165,041 and rosiglitazone, but -fold change could not be accurately quantified. Based on the poor expression of CYP26 in the absence of atRA, a pretreatment and cotreatment with 5 nM atRA was adopted for reliable basal detection of CYP26A1 and CYP26B1 in subsequent studies.
Induction of CYP26B1 and RARβ by PPARγ Agonists.
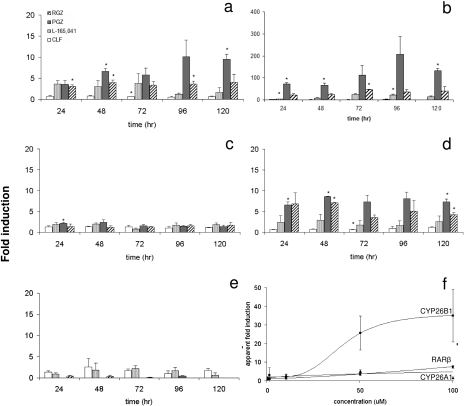
As a result of the potent effects of PPAR agonists on CYP26 transcripts in the initial screen, PPAR agonists were chosen for further characterization. In the presence of 5 nM atRA, CYP26A1 and CYP26B1 were significantly induced by rosiglitazone and pioglitazone (100 μM) in multiple time points, whereas induction by L-165,041 was detected only in one time point (Fig. 4, a and b). Clofibrate down-regulated CYP26A1 at one time point, but the down-regulation was not consistent. It is noteworthy that the overall magnitude of CYP26B1 induction was much greater (∼4–200-fold) than that of CYP26A1 (∼2–10-fold). No clear time point for maximum CYP26A1 and CYP26B1 induction was observed between 24 and 120 h of treatment, suggesting that maximum induction was already reached at 24 h. To test whether induction of RARβ or PPARγ contributed to CYP26 induction in the atRA cotreated cells, transcripts of these genes were measured. We found that RARβ was inducible by rosiglitazone and pioglitazone, whereas PPARγ transcripts did not change consistently after treatment with any of the compounds tested despite significant induction by pioglitazone after 24 h of treatment (Fig. 4, c and d). It is noteworthy that PPARγ was abundant (CT ∼27) in the HepG2 cells and in human livers (CT ∼30). However, there was no correlation between PPARγ and CYP26 transcripts in the human liver bank (p > 0.05). As a control of PPARα activation by clofibrate, CYP4A11 transcripts were measured. As expected, CYP4A11 was up-regulated by clofibrate and L-165,041 (0.6–2.5-fold) and down-regulated by rosiglitazone and pioglitazone (undetectable to 0.4-fold). CYP4A11 was also down-regulated by atRA (data not shown).
Fig. 4.
Effect of PPAR agonists on CYP26 and RAR transcription. The time course of induction of CYP26A1 (a), CYP26B1 (b), PPARγ (c), RARβ (d), and CYP4A11 (e), after treatment with 100 μM PPAR agonists is shown. The PPAR subtype-selective agonists used were clofibrate (CLF; PPARα), L-165,041 (PPARβ/δ), rosiglitazone and pioglitazone (RGZ and PGZ; PPARγ). f, dose response curves for induction of CYP26B1, RARβ, and CYP26A1 mRNA expression after treatment with PGZ for 72 h. Data are presented as means ± S.D. (n = 3), and all -fold induction is calculated relative to vehicle control. *, significant (P < 0.01) difference between drug-treated and untreated control.
The effect of the selective PPAR agonists on CYP26B1 (Table 2) and CYP26A1 (Table 3) transcripts was concentration-dependent. Ligand concentrations greater than 100 μM were not tested because of the potential for nonselective effects and lack of ligand solubility. The magnitude of CYP26B1 induction increased with agonist concentration for PPARγ and PPARβ/δ, whereas corresponding induction for CYP26A1 was not observed. PPARα agonist clofibrate down-regulated both CYP26A1 and CYP26B1 in a dose-dependent manner (Tables 2 and 3). The greatest inductive effect was observed with 100 μM piogliotazone, with a 54.2-fold induction of CYP26B1. In a separate experiment, an EC50 value of 40 ± 4 μM and an Emax value of 36- ± 1-fold were obtained for CYP26B1 induction by pioglitazone. EC50 and Emax values for RARβ induction by pioglitazone could not be estimated due to lack of sufficiently high concentrations to see saturating effects (EC50 >100 μM; Fig. 4f).
TABLE 2.
Dose-dependent induction of CYP26B1 by 5 nM atRA and PPAR ligands
HepG2 cells were pretreated with 5 nM atRA and then cotreated with 5 nM atRA and PPAR ligands for 72 h.
| Concentration | RGZ | PGZ | L-165,041 | CLF |
|---|---|---|---|---|
| 0.5 μM | 1.2 ± 0.8 | 1.3 ± 0.5 | 1.4 ± 0.2 | 1.1 ± 0.4 |
| 1 μM | 1.1 ± 0.2 | 1.5 ± 0.3 | 1.4 ± 0.4 | 1.6 ± 0.7 |
| 10 μM | 2.6 ± 0.7 | 3.9 ± 1.0 | 2.3 ± 0.5 | 0.7 ± 0.2 |
| 100 μM | 35.8 ± 10.8 | 54.2 ± 11.2 | 28.2 ± 15.7 | 0.3 ± 0.01 |
RGZ, rosiglitazone; PGZ, pioglitazone; CLF, clofibrate.
TABLE 3.
Dose-dependent induction of CYP26A1 by 5 nM atRA and PPAR ligands
HepG2 cells were pretreated with 5 nM atRA and then cotreated with 5 nM atRA and PPAR ligands for 72 h.
| Concentration | RGZ | PGZ | L-165,041 | CLF |
|---|---|---|---|---|
| 0.5 μM | 1.0 ± 0.2 | 1.7 ± 0.5 | 1.3 ± 0.2 | 1.1 ± 0.2 |
| 1 μM | 1.1 ± 0.1 | 1.5 ± 0.1 | 1.3 ± 0.01 | 1.1 ± 0.1 |
| 10 μM | 1.4 ± 0.2 | 2.3 ± 0.5 | 1.3 ± 0.3 | 0.9 ± 0.2 |
| 100 μM | 3.4 ± 1.2 | 2.0 ± 0.6 | 1.3 ± 0.9 | 0.3 ± 0.02 |
RGZ, rosiglitazone; PGZ, pioglitazone; CLF, clofibrate.
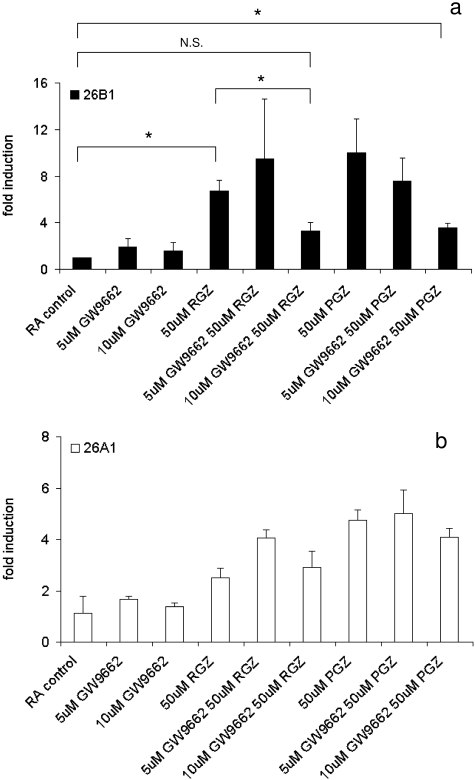
Effects of a PPARγ Antagonist, GW9662, on CYP26 Induction by Rosiglitazone and Pioglitazone.
To confirm the role of PPARγ in CYP26B1 induction, we used an irreversible PPARγ antagonist (GW9662) to block pioglitazone and rosiglitazone from binding to the PPARγ receptor. GW9662 decreased CYP26B1 induction by pioglitazone in a concentration-dependent manner supporting the role of PPARγ in CYP26B1 regulation (Fig. 5a). Ten micromolar GW9662, but not 5 μM, reduced CYP26B1 induction by rosiglitazone from 6.7- to 3.3-fold (p < 0.01). Likewise, the induction of CYP26B1 by pioglitazone was reduced from 10- to 3.6-fold by 10 μM GW9662. On the other hand, GW9662 had no effect on CYP26A1 induction by pioglitazone and rosiglitazone under the same conditions, suggesting that the induction of CYP26A1 by pioglitazone and rosiglitazone was not mediated by PPARγ.
Fig. 5.
The irreversible PPARγ antagonist GW9662 blocks CYP26B1 induction by rosiglitazone and pioglitazone. a, concentration-dependent effect of 72 h treatment with GW9662 (5 and 10 μM) in the induction of CYP26B1 transcription by rosiglitazone (RGZ) and pioglitazone (PGZ). b, absence of effect of GW9662 in CYP26A1 induction by rosiglitazone and pioglitazone. HepG2 cells were treated with 50 μM PGZ or RGZ in the presence or absence of 5 or 10 μM GW9662 as described under Materials and Methods. Data are presented as means ± S.D. (n = 3), and -fold induction relative to vehicle control. *, significant (P < 0.01) difference between the indicated treatments. N.S., not significant (P < 0.01).
Induction of CYP26A1 and CYP26B1 in Human Hepatocytes.
To test whether the CYP26 induction pattern was a cell-line-specific phenomenon, we treated human hepatocytes from three donors with atRA, pioglitazone, and AM580. Similar to the data obtained in human liver bank tissue, the baseline expression of CYP26A1 and CYP26B1 was variable among donors, and the response to the treatments varied greatly among donors. The CT values for CYP26A1 and CYP26B1, respectively, were 38 and 33 in donor Hu1076, 32 and 32 in donor Hu1078, and 38 and undetermined in donor Hu4100. In Hu4100, 10 nM treatment with atRA induced CYP26A1 transcripts 162- ± 16-fold and 7- ± 3-fold after 6 and 48 h of treatment, reflecting similar time course as observed in HepG2 cells. CYP26B1 was not quantifiable in this donor after atRA treatment. CYP26A1 was induced 773- ± 43-fold in Hu1076 after 100 nM treatment of atRA, whereas CYP26B1 induction by atRA was inconsistent in this donor and could not be quantified. In Hu1078, no induction of CYP26A1 or CYP26B1 by atRA (100 nM) was detected, perhaps because of the high baseline expression of both enzymes, which could result in atRA depletion.
Similar to the results in HepG2 cells, AM580, an RARα agonist, induced CYP26A1 and CYP26B1 transcripts in both Hu1076 and Hu1078, but the extent of induction was greater for CYP26A1 than CYP26B1 in both donors. In Hu1076, AM580 induced CYP26A1 26,000- ± 3000-fold and CYP26B1 404 ± 39-fold, whereas in Hu1078, 27-fold induction of CYP26A1 and 17-fold induction of CYP26B1 was observed.
Induction of CYP26 by pioglitazone was tested in all donors after cotreatment with 5 nM atRA. In Hu1078, CYP26A1 and CYP26B1 were induced in a pioglitazone concentration-dependent manner, 27- and 4-fold after 20 μM pioglitazone and 64- and 12-fold after 50 μM pioglitazone for CYP26A1 and CYP26B1, respectively. Only CYP26A1 induction by pioglitazone was quantifiable in Hu1076. A 5- and 3-fold induction of CYP26A1 was detected after 20 and 50 μM pioglitazone treatment, respectively, compared with 5 nM atRA alone. Finally, in Hu4100, pioglitazone down-regulated CYP26A1 to 0.5-fold compared with treatment with 5 nM atRA alone.
Discussion
Based on the mRNA data, CYP26A1 is the major CYP26 enzyme present in human liver. The presence of CYP26A1 protein in human liver was confirmed by Western blotting. The predominant expression of CYP26A1 in the liver is in agreement with previous reports of tissue distribution of CYP26 mRNA in adult humans (Ray et al., 1997; Xi and Yang, 2008) and rodents (Yamamoto et al., 2000; Wang et al., 2002). The caveat of previous human studies is that relative expression was determined in only a single donor. The importance of analyzing multiple donors for CYP26 expression is illustrated by the significant interindividual variability of CYP26 mRNA and protein observed among human livers. Because of interindividual variability, results from a single donor may be misleading regarding the general abundance of CYP26 in that tissue. The variability in CYP26A1 mRNA is not unexpected, because CYP26A1 is strongly regulated by atRA in the rodent liver and in human cell lines (Ray et al., 1997; White et al., 1997; Yamamoto et al., 2000; Wang et al., 2002; Loudig et al., 2005; Ozpolat et al., 2005), and dietary status of the donor is expected to alter CYP26A1 transcription in the liver.
Several factors were identified in donor histories that affected CYP26 transcripts. CYP26A1 transcripts were significantly lower in ischemic livers than in nonischemic livers, and CYP26B1 transcripts were significantly higher in fatty livers than in nonfatty livers, suggesting different roles of CYP26A1 and CYP26B1 in maintaining tissue health. In contrast to previous observations in the rat (Yamamoto et al., 2000), no correlation between donor age and CYP26 expression was observed, perhaps due to the generally more variable donor pathology.
The basal transcript levels of CYP26A1 and CYP26B1 in HepG2 cells followed a pattern similar to that observed in human livers: CYP26B1 was virtually undetermined and CYP26A1 was low. When treated with atRA, CYP26A1 was significantly induced, whereas CYP26B1 induction was weak and variable. CYP26A1 expression and induction profile in HepG2 cells is in good agreement with previously published work using this cell line (White et al., 1997; Ozpolat et al., 2005), whereas the weak CYP26B1 induction in this atRA-responsive cell line was unexpected, because both CYP26A1 and CYP26B1 have been shown to be inducible by atRA in MCF-7 cells and in rats (Loudig et al., 2000; White et al., 2000; Zolfaghari et al., 2007). No correlation between CYP26A1 and CYP26B1 transcription was found in the HepG2 cells or in human livers.
Based on the different expression pattern of CYP26A1 and CYP26B1 and reported induction of both enzymes by atRA, we hypothesized that different RAR isoforms are responsible for distinct CYP26A1 and CYP26B1 regulation. To test this, we analyzed the presence of RAR isoform mRNA in a subset of the livers (n = 14) and in the HepG2 cells. In both sample sets, RARα was most abundant, closely followed by RARβ, and RARγ was not quantifiable. The expression of RARα and RARβ and lack of RARγ in human liver and HepG2 cells is in agreement with previous reports of spatially and temporally specific expression of RAR isoforms (de The et al., 1989; Krust et al., 1989; Zelent et al., 1989). No correlation between RARα and CYP26A1 or CYP26B1 mRNA in the human liver bank was observed and RARα transcripts had minimal variability between donors. Because the activation of RARα is highly dependent on ligand concentrations, these data suggest that basal RARα expression is sufficient to accomplish CYP26A1 induction.
The requirement of specific RAR isoform in CYP26 induction was tested using selective RAR agonists. CYP26A1 and RARβ were induced by AM580 (RARα agonist) and TTNPB (RAR pan-agonist), whereas no clear induction of CYP26B1 was detected. AC55649 (RARβ agonist) had no effect on CYP26A1, CYP26B1, or RAR transcription. Hence, RARα activation seems to be responsible for CYP26A1 and RARβ induction by atRA in HepG2 cells and hepatocytes. The role of RARα in CYP26A1 activation has been demonstrated previously in promyelocytic leukemia cells, embryocarcinoma cells, and intestinal cells (Lampen et al., 2001a; Ozpolat et al., 2002; Idres et al., 2005; Pozzi et al., 2006). In contrast, in mouse F9 cells, RARγ was shown to regulate CYP26A1 gene expression, because CYP26A1 induction was lost in RARγ(−/−) cell lines (Abu-Abed et al., 1998).
The correlation between RARβ and CYP26A1 induction in our studies is striking. The time course and dose response of RARβ induction by atRA closely followed that of CYP26A1. It has been shown that CYP26A1-RARE and RARβ2-RARE are similar (Loudig et al., 2000) and this conservation is likely to lead to the related regulation pattern. RARα and RARγ share the DR5 regions of CYP26A1 and RARβ RAREs but differ in the identity of the nucleotides separating the direct repeats (Loudig et al., 2000), and as shown by us and others, are not induced by atRA in the HepG2 cells (de The et al., 1989). Whether the subtle differences in their RAREs explain the different induction of RARs is unknown. It is also possible that differences in induction are related to different populations of transcriptional activators and repressors. In contrast, no RARE has been reported in the CYP26B1 promoter, although CYP26B1 has been shown to be inducible by atRA in HeLa and MCF-7 cells (White et al., 2000). Based on our data, CYP26B1 requires atRA to be expressed in the liver or liver-derived cell lines, but other mechanisms are needed for induction.
Some of the biological effects of atRA may also be due to its ability to bind to PPARs instead of RAR (Shaw et al., 2003; Schug et al., 2007). Cell fate (apoptosis or proliferation) can be determined by whether atRA binds to RARs or PPARs (Schug et al., 2007). It is not clear, however, which atRA target genes respond to PPAR activation and which respond to RAR activation. We investigated whether PPAR activation in the presence and absence of atRA could induce CYP26 expression. We also screened an additional eight compounds that target a variety of nuclear receptors. Both CYP26A1 and CYP26B1 were susceptible to induction by various xenobiotics, but the magnitude of induction was lower than that observed after atRA, suggesting that a significant effect of these drugs in vivo is unlikely. This supports previous findings of a 1.2-fold induction of CYP26B1 mRNA after phenobarbital treatment in human hepatocytes (Finkelstein et al., 2006). It is also possible that low expression of some nuclear receptors in HepG2 cells is responsible for the low CYP26 induction by xenobiotics.
We report for the first time that PPARγ receptor activation specifically induces CYP26B1 and that the magnitude of induction by PPAR agonists is comparable with that observed after atRA treatment. In a previous study, phytanic acid and docosahexaenoic acid, which are PPARα activators and RXRα ligands, were shown to induce an unspecified CYP26 transcription and atRA metabolism when combined with atRA in Caco-2 cells (Lampen et al., 2001b). In our study, clofibrate, a PPARα agonist, down-regulated CYP26A1 and CYP26B1 transcripts in the presence of atRA. The greatest magnitude of CYP26B1 induction was obtained with PPARγ agonists, although the PPARβ/δ agonist L-165,041 also induced CYP26 transcripts. The selective induction by PPARγ agonists cannot be explained by the relative abundance of this PPAR isoform as in the HepG2 cells PPARα and PPARγ had similar abundance. All three PPAR isoforms were also abundant (CT 25–30) in human livers tested (n = 16), although no correlation between PPARγ and CYP26A1 or CYP26B1 mRNA was observed. Instead, CYP26B1 but not CYP26A1 transcripts were significantly higher in fatty versus nonfatty livers, a result that could be explained by PPARγ activation in fatty livers and warrant further investigation.
Our data suggest that the induction of CYP26B1 by rosiglitazone and pioglitazone is a direct PPARγ-mediated effect. The irreversible PPARγ antagonist GW9662 abolished CYP26B1 induction in a dose-dependent manner. Based on the fact that GW9662 did not diminish CYP26A1 or RARβ induction, the induction of CYP26A1 and RARβ by pioglitazone and rosiglitazone is most likely not due to PPARγ activation, although PPARγ-RXR heterodimers have been shown to bind to the RARβ promoter and induce RARβ transcription (James et al., 2003). Based on the half-life for CYP26B1 mRNA (7–10 h), induced steady-state mRNA levels would be reached between 21 and 40 h, as was observed. If CYP26B1 induction required increased protein synthesis of an intermediate signaling factor, the time course of induction should be slower. Unfortunately, cycloheximide induced CYP26 expression on its own (data not shown); hence, we could not test the dependence of CYP26B1 induction on new protein synthesis.
Considerable interindividual variability was observed in human hepatocytes in their response to the tested inducers. Although the hepatocyte experiments supported the findings of regulation of CYP26A1 and CYP26B1 in HepG2 cells, the magnitude of induction seemed dependent on the baseline expression of the CYP26 enzymes as well as other donor-specific factors. The fact that pioglitazone and AM580 induced CYP26 in human hepatocytes as well as in HepG2 cells suggests that the results of this study are relevant to the in vivo situation. Because rosiglitazone and pioglitazone induce CYP26B1 at therapeutic concentrations in vitro, one would predict that CYP26B1 induction would occur also in vivo.
In conclusion, the data presented here suggest that distinct mechanisms and multiple transcriptional elements are responsible for regulating CYP26A1 and CYP26B1 transcripts, and these mechanisms may be responsible for tissue- and time-specific differences in CYP26 expression. In the human liver, CYP26A1 expression seems to be primarily regulated by RARα and cellular atRA concentrations, whereas CYP26B1, if present, is induced by PPARγ-mediated pathways.
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grants R01-GM081596, P01-GM32165].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.109.059071.
- atRA
- all-trans-retinoic acid
- RAR
- retinoic acid receptor
- RXR
- retinoid X receptor
- P450
- cytochrome P450
- AM580
- 4-[(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)carboxamido]benzoic acid
- DRB
- 5,6-dichlorobenzimidazole 1-β-d-ribofuranoside
- GW9662
- 2-chloro-5-nitrobenzanilide
- L-165,041
- [4-]3-(4-acetyl-3-hydroxy-2-propylphenoxy)propoxy]phenoxy]acetic acid
- TTNPB
- 4-[(E)-2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl]benzoic acid
- AC55649
- 4′-octyl-[1,1′-biphenyl]-4-carboxylic acid
- DMSO
- dimethyl sulfoxide
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- PPAR
- peroxisome proliferator-activated receptor
- DM
- differentiation medium
- PCR
- polymerase chain reaction
- PBS
- phosphate-buffered saline
- RARE
- retinoic acid response element.
References
- Abu-Abed S, Dollé P, Metzger D, Beckett B, Chambon P, Petkovich M. (2001) The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes Dev 15: 226–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Abed S, MacLean G, Fraulob V, Chambon P, Petkovich M, Dollé P. (2002) Differential expression of the retinoic acid-metabolizing enzymes CYP26A1 and CYP26B1 during murine organogenesis. Mech Dev 110: 173–177 [DOI] [PubMed] [Google Scholar]
- Abu-Abed SS, Beckett BR, Chiba H, Chithalen JV, Jones G, Metzger D, Chambon P, Petkovich M. (1998) Mouse P450RAI (CYP26) expression and retinoic acid-inducible retinoic acid metabolism in F9 cells are regulated by retinoic acid receptor gamma and retinoid X receptor alpha. J Biol Chem 273: 2409–2415 [DOI] [PubMed] [Google Scholar]
- Bendich A, Langseth L. (1989) Safety of vitamin A. Am J Clin Nutr 49: 358–371 [DOI] [PubMed] [Google Scholar]
- Blomhoff R, Blomhoff HK. (2006) Overview of retinoid metabolism and function. J Neurobiol 66: 606–630 [DOI] [PubMed] [Google Scholar]
- Clagett-Dame M, DeLuca HF. (2002) The role of vitamin A in mammalian reproduction and embryonic development. Annu Rev Nutr 22: 347–381 [DOI] [PubMed] [Google Scholar]
- de The H, Marchio A, Tiollais P, Dejean A. (1989) Differential expression and ligand regulation of the retinoic acid receptor alpha and beta genes. EMBO J 8: 429–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delescluse C, Cavey MT, Martin B, Bernard BA, Reichert U, Maignan J, Darmon M, Shroot B. (1991) Selective high affinity retinoic acid receptor alpha or beta-gamma ligands. Mol Pharmacol 40: 556–562 [PubMed] [Google Scholar]
- Duester G. (2008) Retinoic acid synthesis and signaling during early organogenesis. Cell 134: 921–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein D, Lamba V, Assem M, Rengelshausen J, Yasuda K, Strom S, Schuetz E. (2006) ADME transcriptome in Hispanic versus White donor livers: evidence of a globally enhanced NR1I3 (CAR, constitutive androstane receptor) gene signature in Hispanics. Xenobiotica 36: 989–1012 [DOI] [PubMed] [Google Scholar]
- Fritzsche B, Vermot J, Neumann U, Schmidt A, Schweigert FJ, Dollé P, Rühl R. (2007) Regulation of expression of the retinoic acid metabolizing enzyme CYP26A1 in uteri of ovariectomized mice after treatment with ovarian steroid hormones. Mol Reprod Dev 74: 258–264 [DOI] [PubMed] [Google Scholar]
- Hathcock JN, Hattan DG, Jenkins MY, McDonald JT, Sundaresan PR, Wilkening VL. (1990) Evaluation of vitamin A toxicity. Am J Clin Nutr 52: 183–202 [DOI] [PubMed] [Google Scholar]
- Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. (2007) Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development 134: 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idres N, Marill J, Chabot GG. (2005) Regulation of CYP26A1 expression by selective RAR and RXR agonists in human NB4 promyelocytic leukemia cells. Biochem Pharmacol 69: 1595–1601 [DOI] [PubMed] [Google Scholar]
- James SY, Lin F, Kolluri SK, Dawson MI, Zhang XK. (2003) Regulation of retinoic acid receptor beta expression by peroxisome proliferator-activated receptor gamma ligands in cancer cells. Cancer Res 63: 3531–3538 [PubMed] [Google Scholar]
- Krust A, Kastner P, Petkovich M, Zelent A, Chambon P. (1989) A third human retinoic acid receptor, hRAR-gamma. Proc Natl Acad Sci U S A 86: 5310–5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampen A, Meyer S, Nau H. (2001a) Effects of receptor-selective retinoids on CYP26 gene expression and metabolism of all-trans-retinoic acid in intestinal cells. Drug Metab Dispos 29: 742–747 [PubMed] [Google Scholar]
- Lampen A, Meyer S, Nau H. (2001b) Phytanic acid and docosahexaenoic acid increase the metabolism of all-trans-retinoic acid and CYP26 gene expression in intestinal cells. Biochim Biophys Acta 1521: 97–106 [DOI] [PubMed] [Google Scholar]
- Lemaire G, Balaguer P, Michel S, Rahmani R. (2005) Activation of retinoic acid receptor-dependent transcription by organochlorine pesticides. Toxicol Appl Pharmacol 202: 38–49 [DOI] [PubMed] [Google Scholar]
- Loudig O, Babichuk C, White J, Abu-Abed S, Mueller C, Petkovich M. (2000) Cytochrome P450RAI(CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol Endocrinol 14: 1483–1497 [DOI] [PubMed] [Google Scholar]
- Loudig O, Maclean GA, Dore NL, Luu L, Petkovich M. (2005) Transcriptional co-operativity between distant retinoic acid response elements in regulation of Cyp26A1 inducibility. Biochem J 392: 241–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund BW, Piu F, Gauthier NK, Eeg A, Currier E, Sherbukhin V, Brann MR, Hacksell U, Olsson R. (2005) Discovery of a potent, orally available, and isoform-selective retinoic acid beta2 receptor agonist. J Med Chem 48: 7517–7519 [DOI] [PubMed] [Google Scholar]
- Lutz JD, Dixit V, Yeung CK, Dickmann LJ, Zelter A, Thatcher JE, Nelson WL, Isoherranen N. (2009) Expression and functional characterization of cytochrome P450 26A1, a retinoic acid hydroxylase. Biochem Pharmacol. 77: 258–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean G, Abu-Abed S, Dollé P, Tahayato A, Chambon P, Petkovich M. (2001) Cloning of a novel retinoic-acid metabolizing cytochrome P450, Cyp26B1, and comparative expression analysis with Cyp26A1 during early murine development. Mech Dev 107: 195–201 [DOI] [PubMed] [Google Scholar]
- Nagpal S, Athanikar J, Chandraratna RA. (1995) Separation of transactivation and AP1 antagonism functions of retinoic acid receptor alpha. J Biol Chem 270: 923–927 [DOI] [PubMed] [Google Scholar]
- Napoli JL. (1999) Interactions of retinoid binding proteins and enzymes in retinoid metabolism. Biochim Biophys Acta 1440: 139–162 [DOI] [PubMed] [Google Scholar]
- Ozpolat B, Mehta K, Lopez-Berestein G. (2005) Regulation of a highly specific retinoic acid-4-hydroxylase (CYP26A1) enzyme and all-trans-retinoic acid metabolism in human intestinal, liver, endothelial, and acute promyelocytic leukemia cells. Leuk Lymphoma 46: 1497–1506 [DOI] [PubMed] [Google Scholar]
- Ozpolat B, Mehta K, Tari AM, Lopez-Berestein G. (2002) all-trans-Retinoic acid-induced expression and regulation of retinoic acid 4-hydroxylase (CYP26) in human promyelocytic leukemia. Am J Hematol 70: 39–47 [DOI] [PubMed] [Google Scholar]
- Pozzi S, Rossetti S, Bistulfi G, Sacchi N. (2006) RAR-mediated epigenetic control of the cytochrome P450 Cyp26a1 in embryocarcinoma cells. Oncogene 25: 1400–1407 [DOI] [PubMed] [Google Scholar]
- Ray WJ, Bain G, Yao M, Gottlieb DI. (1997) CYP26, a novel mammalian cytochrome P450, is induced by retinoic acid and defines a new family. J Biol Chem 272: 18702–18708 [DOI] [PubMed] [Google Scholar]
- Reijntjes S, Gale E, Maden M. (2004) Generating gradients of retinoic acid in the chick embryo: Cyp26C1 expression and a comparative analysis of the Cyp26 enzymes. Dev Dyn 230: 509–517 [DOI] [PubMed] [Google Scholar]
- Ross AC. (2003) Retinoid production and catabolism: role of diet in regulating retinol esterification and retinoic Acid oxidation. J Nutr 133: 291S–296S [DOI] [PubMed] [Google Scholar]
- Sakai Y, Meno C, Fujii H, Nishino J, Shiratori H, Saijoh Y, Rossant J, Hamada H. (2001) The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes Dev 15: 213–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. (2007) Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell 129: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw N, Elholm M, Noy N. (2003) Retinoic acid is a high affinity selective ligand for the peroxisome proliferator-activated receptor beta/delta. J Biol Chem 278: 41589–41592 [DOI] [PubMed] [Google Scholar]
- Shelton DN, Sandoval IT, Eisinger A, Chidester S, Ratnayake A, Ireland CM, Jones DA. (2006) Up-regulation of CYP26A1 in adenomatous polyposis coli-deficient vertebrates via a WNT-dependent mechanism: implications for intestinal cell differentiation and colon tumor development. Cancer Res 66: 7571–7577 [DOI] [PubMed] [Google Scholar]
- Taimi M, Helvig C, Wisniewski J, Ramshaw H, White J, Amad M, Korczak B, Petkovich M. (2004) A novel human cytochrome P450, CYP26C1, involved in metabolism of 9-cis and all-trans isomers of retinoic acid. J Biol Chem 279: 77–85 [DOI] [PubMed] [Google Scholar]
- Uehara M, Yashiro K, Mamiya S, Nishino J, Chambon P, Dolle P, Sakai Y. (2007) CYP26A1 and CYP26C1 cooperatively regulate anterior-posterior patterning of the developing brain and the production of migratory cranial neural crest cells in the mouse. Dev Biol 302: 399–411 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zolfaghari R, Ross AC. (2002) Cloning of rat cytochrome P450RAI (CYP26) cDNA and regulation of its gene expression by all-trans-retinoic acid in vivo. Arch Biochem Biophys 401: 235–243 [DOI] [PubMed] [Google Scholar]
- White JA, Beckett-Jones B, Guo YD, Dilworth FJ, Bonasoro J, Jones G, Petkovich M. (1997) cDNA cloning of human retinoic acid-metabolizing enzyme (hP450RAI) identifies a novel family of cytochromes P450. J Biol Chem 272: 18538–18541 [DOI] [PubMed] [Google Scholar]
- White JA, Ramshaw H, Taimi M, Stangle W, Zhang A, Everingham S, Creighton S, Tam SP, Jones G, Petkovich M. (2000) Identification of the human cytochrome P450, P450RAI-2, which is predominantly expressed in the adult cerebellum and is responsible for all-trans-retinoic acid metabolism. Proc Natl Acad Sci U S A 97: 6403–6408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi J, Yang Z. (2008) Expression of RALDHs (ALDH1As) and CYP26s in human tissues and during the neural differentiation of P19 embryonal carcinoma stem cell. Gene Expr Patterns 8: 438–442 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Zolfaghari R, Ross AC. (2000) Regulation of CYP26 (cytochrome P450RAI) mRNA expression and retinoic acid metabolism by retinoids and dietary vitamin A in liver of mice and rats. Faseb J 14: 2119–2127 [DOI] [PubMed] [Google Scholar]
- Yashiro K, Zhao X, Uehara M, Yamashita K, Nishijima M, Nishino J, Saijoh Y, Sakai Y, Hamada H. (2004) Regulation of retinoic acid distribution is required for proximodistal patterning and outgrowth of the developing mouse limb. Dev Cell 6: 411–422 [DOI] [PubMed] [Google Scholar]
- Zelent A, Krust A, Petkovich M, Kastner P, Chambon P. (1989) Cloning of murine alpha and beta retinoic acid receptors and a novel receptor gamma predominantly expressed in skin. Nature 339: 714–717 [DOI] [PubMed] [Google Scholar]
- Zolfaghari R, Cifelli CJ, Lieu SO, Chen Q, Li NQ, Ross AC. (2007) Lipopolysaccharide opposes the induction of CYP26A1 and CYP26B1 gene expression by retinoic acid in the rat liver in vivo. Am J Physiol Gastrointest Liver Physiol 292: G1029–G1036 [DOI] [PMC free article] [PubMed] [Google Scholar]