Abstract
MRP2 (ABCC2), a member of the ATP binding cassette superfamily of efflux transporters that mediates the apical efflux of organic anions from hepatocytes, enterocytes, and renal epithelial cells, is postulated to undergo post-transcriptional regulation. The MRP2 5′-untranslated region (5′UTR) contains seven upstream start codons and six upstream open reading frames (uORFs). Ribonuclease protection assays in human liver, placenta, kidney, small intestine, and HepG2 cells identified multiple MRP2 transcription initiation sites. We investigated MRP2 5′UTRs [−247 (−247 to −1), −204 (−204 to −1), or −99 (−99 to −1)] for their effects on regulation of gene expression with the use of transient gene expression in HepG2 cells and in vitro translation assays. In HepG2 cells transfected with SV40-MRP2-5′UTR-Luciferase cassettes, luciferase activities of constructs −247 and −204 were significantly lower than that of −99. Disruption of the uORFs at −105 and −74 nucleotides by mutation of ATGs to AAG enhanced luciferase activity significantly without affecting luciferase mRNA expression. The translation efficiencies of T7-5′UTR-Luciferase cassettes determined in vitro were consistent with transfected HepG2 cells and showed that inhibition of translation by the −105 uORF occurred only in the cis configuration and not in the trans configuration and that inhibition of translation by the −105 uORF was independent of the encoded peptide sequence. Characterization of an MRP2 polymorphism, −24C>T, in the MRP2 5′UTR, demonstrated no effect on mRNA expression or downstream ORF translation. These data indicate for the first time that the 5′UTR of MRP2 mRNA transcripts and the uORF at −105 markedly influence MRP2 translation.
The multidrug resistance-associated protein 2 (MRP2; ABCC2) is an important transport protein mediating the efflux of organic anions across the apical domain of hepatocytes, enterocytes, and renal proximal tubule cells. MRP2 mediates the ATP-dependent efflux of glutathione, glucuronide, and sulfate conjugates of endo- and xenobiotics, can mediate the efflux of chemotherapeutic drugs such as methotrexate, and is capable of GSH-stimulated transport of nonconjugated drugs, such as anthracyclines and Vinca alkaloids (Deeley et al., 2006).
Investigation of MRP2 mRNA and protein expression in normal and cancerous kidney cortex found no difference in MRP2 mRNA expression between tumor tissue (clear-cell renal cell cancer) and tumor-free adjacent tissue, but MRP2 protein levels were significantly lower in clear-cell renal cell cancer samples compared with tumor-free adjacent tissue (Haenisch et al., 2007). Furthermore, duodenal MRP2 protein is markedly reduced in patients with inflammatory obstructive cholestasis, whereas MRP2 mRNA is unchanged (Geier et al., 2007). These results indicate that under some conditions, the human MRP2 gene expression undergoes post-transcriptional regulation. We (Mottino et al., 2000; Cao et al., 2001, 2002; Jones et al., 2005) and others (Johnson et al., 2002; Johnson and Klaassen, 2002) have described post-transcriptional regulation of the homologous Mrp2 gene in the rat and have further demonstrated its translational regulation (Zhang et al., 2007).
Post-transcriptional regulation has been less comprehensively characterized compared with transcriptional regulation, although it is often known to be mediated by short sequence RNA elements located in the 5′- or 3′-untranslated regions (UTR) (Pesole et al., 2001). The regulation of translation by the 5′UTR is a major mechanism for post-transcriptional regulation of gene expression; one such element in the 5′UTR is the upstream open reading frame (uORF). uORFs can influence translation of downstream ORFs by regulating the selection or efficiency of the translation start site. Translation of natural eukaryotic mRNA is predicted to initiate at the first ATG encountered by the scanning 40S ribosomal subunit, starting from the 5′m7G cap (Kozak, 1999) and can thus inhibit translation of a downstream ORF. Translation of the downstream ORF by the ribosome occurs either through leaky scanning of uATGs in the 5′UTR when the sequence around the uATG is suboptimal (Kozak, 1986) or through reinitiation when the translation machinery is not dissociated from the mRNA chain after termination of translation of an uORF (Morris and Geballe, 2000). The uORF can also inhibit translation of a downstream ORF if translation of the encoded nascent peptide causes stalling of the ribosome, either at a termination codon or during the elongation process (Hood et al., 2009). Alternatively, there may be internal ribosomal entry sites located in the 5′UTR, such that the translation machinery skips the uATGs and instead uses the internal ribosomal entry sites to initiate translation of the downstream ORF (Le and Maizel, 1997). Whereas some upstream start codons (uATGs) inhibit downstream ORF translation (Meijer et al., 2000; Diba et al., 2001; Kwon et al., 2001; Blaschke et al., 2003; Mihailovich et al., 2007; Song et al., 2007), other uATGs have no such effect (Diba et al., 2001). uORFs can also influence the stability of mRNA by a process termed nonsense-mediated mRNA decay (Hood et al., 2009). In general, uORFs inhibit the translation of the downstream ORF (Morris and Geballe, 2000; Neafsey and Galagan, 2007; Zimmer et al., 2008); inhibition is dependent on the amino-acid sequence of the uORF peptide in some genes (Parola and Kobilka, 1994; Luo and Sachs, 1996; Reynolds et al., 1996; Mize et al., 1998) but not in others (David-Assael et al., 2005).
Although we have investigated the role of rat Mrp2 5′UTR and its uORFs in regulating translation of Mrp2 (Zhang et al., 2007), and the human MRP2 is homologous to the rat Mrp2, the human MRP2 5′UTR seems more complicated, and its role in regulating expression of human MRP2 has not been investigated. Three transcription initiation sites at −247, −204 and −99 nucleotides relative to the start codon ATG of MRP2 have been determined by 5′ rapid amplification of cDNA ends (Tanaka et al., 1999). We here note the presence of seven upstream uATGs and six uORFs in the human MRP2 5′UTR and demonstrate that uATGs in human MRP2 5′UTR inhibit translation efficiency of a downstream ORF and that the uORF at −105 acts as a cis inhibitor in the regulation of MRP2 translation.
Materials and Methods
Vector Construction.
The plasmid pGL3-control vector and Luciferase T7-control DNA vector were purchased from Promega (Madison, WI). MRP2 5′UTRs were amplified by PCR from cDNA that was reverse transcribed from mRNA isolated from HepG2 cells. Gene-specific primers were designed according to the published sequence information (Tanaka et al., 1999) and are shown in Table 1. Figure 1 shows the MRP2 5′UTR sequence. For construction of the pGL3-MRP2 5′UTR-Luciferase vector, two restriction enzyme sites, HindIII and NcoI, were included in each primer to facilitate cloning. The full-length 5′UTR (−247) from the transcription initiation site of −247 to −1 (relative to the MRP2 start codon ATG) was amplified by primers HindIII 247F and NcoI 1R. The fragment of 5′UTR (−204) from −204 to −1 was amplified by primer HindIII 204F and NcoI 1R. The fragment of 5′UTR (−99) from −99 to −1 was amplified by primers HindIII 99F and NcoI 1R. The PCR products were purified with a PCR purification kit from QIAGEN (Valencia, CA), digested with HindIII and NcoI, and then purified on agarose gel. The purified fragment was inserted into the corresponding sites of the pGL3 control vector between the SV40 promoter and the Luciferase gene. For T7-MRP2 5′UTR-Luciferase plasmid construction, we used the restriction enzyme BamHI to replace NcoI in all related primers used for pGL3-MRP2-5′UTR-Luciferase vector construction. For the vector containing MRP2 5′UTR with a mutation of the ATG at −105, −204 was used as a PCR template, and two primers, 204M1F and 204M1R, were used to generate −204M1. For the double mutant with disruption of ATG at both −105 and −74 (−204M2), −204M1 was used as a PCR template together with two primers, 204M2F and 204M2R. For disruption of the ATG at −74 (−99M1), −99 was used as PCR template together with two primers, 204M2F and 204M2R. To determine whether the inhibitory regulation of translation of the −105 ATG is dependent on the amino acid sequence of uORF, the 5′UTR fragment (−111 to −1) and its frame-shifted uORF sequence were cloned into the T7 luciferase control vector. The fragment −111 to −1 was PCR-amplified by a pair of primers hF/hR (Table 1) by using plasmid −204 as template. A HindIII restriction site was designed in each primer to facilitate cloning. The PCR products were column-purified and digested by HindIII, and the fragment was then inserted into the HindIII site of T7 luciferase control vector. We named this plasmid MRP2–22. The plasmid MRP2–22M was constructed by frame-shifting uORF in the plasmid MRP2–22 through 1) deletion of the T at position −101 after ATGG with the primers hFdel/hRdel, 2) insertion of an A before the stop codon TAA at position −39 by using the primers hFins/hRins, and then 3) disruption of the ATG1 at position −74 by mutation to ATA with the use of the primers of hFmut/hRmut. The plasmids −147 and −147M1 were obtained by cloning a spacer sequence from position −147 to −112 between the capped site and −105 ATG in the plasmids MRP2–22 and MRP2–22M, with the use of the pairs of primers hFex/hRex and hFexM/hRexM, respectively. The luciferase ORF and uORF2 were disrupted in the plasmid −147 by a site-directed point mutation with primers MutLucF/MutLucR and h105aagF/h105aagR, respectively, obtaining the plasmids −147M3 and −147M2. All mutagenesis was performed according to the manufacturer's instructions of QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). For analysis of the effect of the −24C>T polymorphism (−99–24C>T), −99 was used as PCR template together with the two primers −24C>TF and −24C>TR. All PCRs were conducted by using Phusion High Fidelity PCR kit from New England BioLabs (Ipswich, MA). All constructs were confirmed by sequencing by Eurofins MWG Operon (Huntsville, AL).
TABLE 1.
Primers
| Primer Name | Primer Sequence |
|---|---|
| HindIII −247F | GCAAGCTTACTTTGGGAACTGGTGAGTC |
| HindIII −204F | GCAAGCTTGTCACATGTCCATCCACTGT |
| HindIII −99F | GCAAGCTTGATAATTCCTGTTCCACTTT |
| T7R | TTAATACGACTCACTATAGGGGATT CCTGGACTGCGTCT |
| NcoI-1R | CGCCATGGGGATTCCTGGACTGCGTCTGG |
| BamHI-1R | GCGGATCCGATTCCTGGACTGCGTCTGG |
| 204M1F | CGGAGAACATCAGAAAGGTAGATAATTCCTGTTCCACTTT |
| 204M1R | AAAGTGGAACAGGAATTATCTACCTTTCTGATGTTCTCCG |
| 204M2F | CCTGTTCCACTTTCTTTGAAGAAACAAGTAAAGA |
| 204M2R | TCTTTACTTGTTTCTTCAAAGAAAGTGGAACAGG |
| 24C>TF | TATTAATAGAAGAGTCTTTGTTCCAGACGCAGTCC |
| 24C>TR | GGACTGCGTCTGGAACAAAGACTCTTCTATTAATA |
| 18SF | CGATTGGATGGTTTAGTGAGG |
| 18SR | AGTTCGACCGTCTTCTCAGC |
| LucF1 | TGAGTACTTCGAAATGTCCGTTC |
| LucR1 | GTATTCAGCCCATATCGTTTCAT |
| SV40F | TCGAGATCTGCGATCTGCATCTCAATTAGT |
| SV40R | TTTACCACATTTGTAGAGGTTTTACTTGCT |
| 74MuF | CCTGTTCCACTTTCTTTGAAGAAACAAGTAAAGA |
| 74MuR | TCTTTACTTGTTTCTTCAAAGAAAGTGGAACAGG |
| T7F | TTAATGCAGCTTAATGCAGCTGGCT |
| LucR2 | AGCTCGCCCCCTCGGAGGATTACAA |
| hF | GGACAAGCTTATCAGAATGGTAGATAATTCCTG |
| hR | GGTAAGCTTGATTCCTGGACTGCGTC |
| hFins | CAACACAATCATATATAATAGAAGAGTCTTCGTTCC |
| hRins | GGAACGAAGACTCTTCTATTATATATGATTGTGTTG |
| hFmut | CCTGTTCCACTTTCTTTGATAAAACAAGTAAAG |
| hRmut | CTTTACTTGTTTTATCAAAGAAAGTGGAACAGG |
| hFdel | CCAAGCTTATCAGAATGGAGATAATTCCTG |
| hRdel | CAGGAATTATCTCCATTCTGATAAGCTTGG |
| hFex | CTCACTATAGGGAGACCCAAGCTTGTTGGGAAAGGTCATCCTTTACGGAGAACATCAGAATGGTAGATAATTCCTGTTC |
| hRex | GAACAGGAATTATCTACCATTCTGATGTTCTCCGTAAAGGATGACCTTTCCCAACAAGCTTGGGTCTCCCTATAGTGAG |
| hFexM | CTCACTATAGGGAGACCCAAGCTTGTTGGGAAAGGTCATCCTTTACGGAGAACATCAGAATGGAGATAATTCCTGTTC |
| hRexM | GAACAGGAATTATCTCCATTCTGATGTTCTCCGTAAAGGATGACCTTTCCCAACAAGCTTGGGTCTCCCTATAGTGAG |
| MutLucF | GCCCGGATCCAAAAGGAAGACGCCAAAAAC |
| MutLucR | GTTTTTGGCGTCTTCCTTTTGGATCCGGGC |
| h105aagF | CCTTTACGGAGAACATCAGAAAGGTAGATAATTCTTGTTCC |
| h105aagR | GGAACAGGAATTATCTACCTTTCTGATGTTCTCCGTAAAGG |
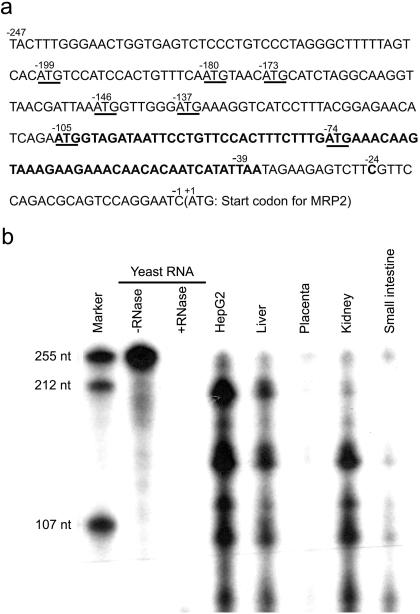
Fig. 1.
The 5′UTR of MRP2 and its transcription start sites. a, cDNA sequence of the full-length 5′UTR of human MRP2. The start codon ATGs in the 5′UTR are underlined. The upstream open reading frame (uORF) beginning at −105 is shown in bold. The numbers are relative to the start codon ATG of MRP2 gene. b, identification of the MRP2 5′UTR transcription initiation sites by ribonuclease protection assay. The [α-32P]UTP-labeled antisense probe of MRP2 5′UTR was synthesized. Total RNA was incubated with 2 × 104 c.p.m. of the radiolabeled probe; after coprecipitation and hybridization, the single-strand RNA was digested by RNase T1. The fragments protected from RNase digestion were identified by electrophoresis on a 6% polyacrylamide denaturing gel. Lane 1, molecular marker; lane 2, 15 μg of yeast RNA without RNase T1 digestion; lane 3, 15 μg of yeast RNA with RNase T1 digestion; lane 4, 10 μg of RNA isolated from HepG2 cells; lane 5, 20 μg of human liver RNA; lane 6, 30 μg of human placenta RNA; lane 7, 30 μg of human kidney RNA; lane 8, 30 μg of human small intestine RNA.
RNAse Protection Assay.
Total RNA of the human liver, placenta, kidney, and small intestine were purchased from Ambion (Austin, TX). Total RNA from HepG2 cells was isolated by using the Magnapure RNA isolation kit from Roche Applied Science (Mannheim, Germany) The DNA templates for synthesis of antisense RNA of MRP2 5′UTR fragments 247, 204, and 99 were obtained by PCR with the use of T7-MRP2 5′UTR-luciferase plasmid DNA as template and three pairs of primers (HindIII −247F and T7 reverse for fragment 247, HindIII −204F, and T7 reverse for fragment 204; HindIII −99F and T7 reverse for fragment 99) (Table 1). The 32P-labeled antisense RNA probes were synthesized by in vitro transcription using MAXIscript kit from Ambion. The 32P-labeled antisense RNA of MRP2 5′UTR fragment −247 was used as the RPA probe. The 32P-labeled antisense RNA of MRP2 5′UTR fragments 247, 204, and 99 were mixed and used as size markers; because of the inclusion of eight nucleotides from the restriction cloning sites, the actual sizes were 255, 212, and 107, respectively. The RPA was carried out with the use of the RPA III kit from Ambion according to the manufacturer's instructions.
HepG2 Cell Culture and Transfection.
The HepG2 cell line was obtained from American Type Culture Collection (Manassas, VA) and was maintained according to their instructions. The SV40 promoter-5′UTR-Luciferase-SV40 polyadenylation signal cassettes were PCR-amplified with the use of two primers, SV40F and SV40R, from individual plasmid DNA. The PCR products were purified with a PCR purification kit (QIAGEN) dissolved in water and quantified by using a UV spectrophotometer. For HepG2 cell transfection, cells were plated in 24 well plates at a density of 0.2 million cells per well 1 day before transfection. The FuGENE HD transfection reagent was used at a FuGENE HD/DNA ratio of 5:2.
In Vitro Translation.
For in vitro translation, the Rabbit Reticulocyte Lysate System (Promega) was used according to the manufacturer's instructions. The DNA template T7-5′UTR-Luciferase cassette was amplified by PCR with the use of two primers, T7F and LucR, with the use of individual plasmid DNA as template. The PCR product was separated on 0.7% agarose gel and purified by using a QIAGEN gel purification column and quantified by UV spectrophotometry. For mRNA synthesis, the mMESSAGE mMACHINE kit from Ambion was used according to the manufacturer's instructions. The mRNA was purified and quantified by UV spectrophotometry. Alternatively, mRNA was also labeled with [α-32P]UTP and quantified by scintillation counting. Free nucleotides in the reaction mixture were removed by using NucAway Spin Columns (Ambion). The luciferase transcripts (0, 2, 4, 10, 20, and 40 ng) were added to the reaction mixture and incubated at 30°C for 60 min. The reactions were terminated on ice.
Luciferase Activity Detection.
The luciferase activity of transfected HepG2 cells or in vitro-translated luciferase was detected by the Luciferase Reporter Assay System from Promega with the use of a luminometer (20/20n; Turner Biosystem, Sunnyvale, CA). The transfected HepG2 cells were directly lysed by the buffer provided, and an aliquot of lysate was used for measurement of the luciferase activity. For in vitro-translated luciferase, the lysis buffer was used to dilute the translation reaction mix, and an aliquot was used to measure luciferase activity.
RNA Isolation and Real-Time RT-PCR Analysis of Gene Expression.
Total RNA from transfected HepG2 cells was isolated by using the GenElute Mammalian Total RNA Miniprep Kit from Sigma-Aldrich (Milwaukee, WI). The cDNA was synthesized by using high-capacity cDNA reverse transcription kits from Applied Biosystems (Foster City, CA) according to the manufacturer's instructions. Primers and UPL probes for real time RT-PCR were designed and ordered from Roche Applied Science (Mannheim, Germany) by using online software (http://www.universalprobelibrary.com), and real-time RT-PCR was determined with the use of a 480 LightCycler (Roche Applied Sciences). For detection of luciferase mRNA, primers lucF1 and lucR1 (Table 1) were used with UPL probe 29. For detection of 18S RNA, primers 18SF and 18SR were used with UPL probe 81. In detail, 1 μg of total RNA was used for cDNA synthesis, and then the synthesized cDNA was diluted to 500 μl. Diluted cDNA (5 μl) was used as template in a 20-μl reaction volume. The target gene expression was normalized by its 18S RNA gene expression.
Data Analysis.
Statistical differences were assessed by using one-way analysis of variance (ANOVA). Post hoc comparisons were performed with the use of Statview software (SAS Institute, Cary, NC).
Results
Sequence analysis of cDNA shows that the 5′UTR of MRP2 contains seven start codon ATGs at −199, −180, −173, −146, −137, −105, and −74 relative to the MRP2 start codon ATG; however, the ATG at −180 is followed immediately by a stop codon (TAA), so there are only six uORF (Fig. 1). The uATG at −105 is flanked by a perfect Kozak motif (AGAATGGTA, where A and G are the flanking sequences at −4 and +3, respectively, that make up a perfect Kozak motif relative to the ATG translation start codon), is in-frame with the MRP2 ORF, terminates at TAA-39, is followed by a second stop codon (TAG), and is predicted to encode a 22-amino acid peptide. There is also an out-of-frame uATG at −74 in the middle of this uORF, and the coding sequence of this uORF extends past the start codon for MRP2 to +234 (Fig. 1). It is noteworthy that the MRP2 ATG (ATCATGC) lacks a perfect Kozak motif. Several transcription initiation sites were detected in the MRP2 5′UTR by using RPA in liver, placenta, kidney, small intestine, and HepG2 cells (Fig. 1). These data confirmed the previously reported three transcription initiation sites at −247, −204, and −99 (Tanaka et al., 1999) and also showed additional transcription initiation sites in HepG2 cells, liver, kidney, and small intestine. MRP2 mRNA transcripts in liver and HepG2 cell are essentially identical but differed from those in placenta, kidney, and small intestines. The major bands in HepG2 cells and liver were detected at −204 and at approximately −150, −120, −99, and −70; a relatively weak band was detected at −247. In placenta, the major band was detected at −247, whereas the kidney showed major bands around −150, −120, −99, and −70 and relatively weak bands at −247 and −204. The small intestine showed major bands around −99 and −70, relatively weak bands around −247 and −150, and no detectable band at −204 (Fig. 1).
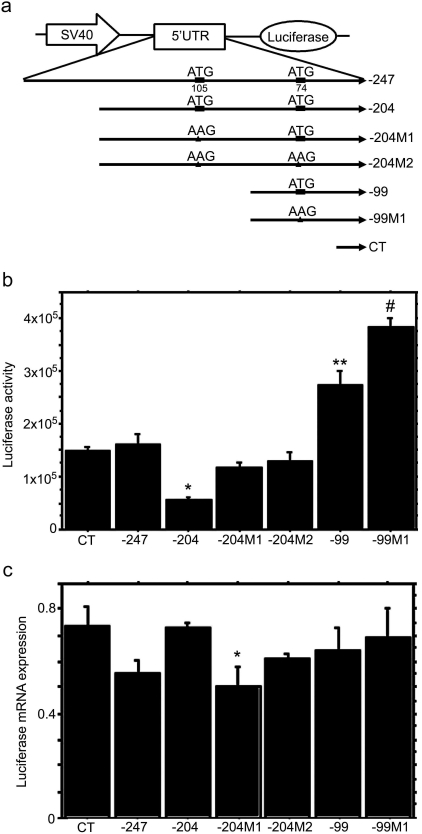
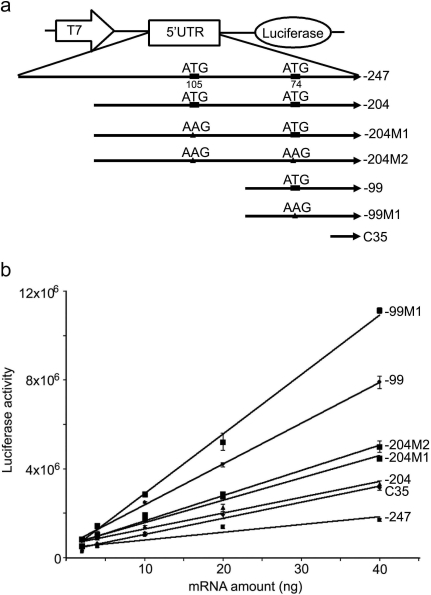
Human MRP2 5′UTRs were obtained by RT-PCR and successfully cloned into the pGL3 control and Luciferase T7 control DNA vectors (Figs. 2a and 3a). To investigate the regulatory role of the MRP2 5′UTR on downstream ORF expression, we inserted MRP2 5′UTRs and the uORF-disrupted mutants between the SV40 promoter and the luciferase gene for HepG2 cell transfection studies (Fig. 2a). In SV40-5′UTR-Luciferase cassette-transfected HepG2 cells, the −99 construct showed a significantly higher luciferase activity compared with the −247, −204, or C35 control constructs (Fig. 2b), despite the absence of any significant differences in the luciferase mRNA expression among these constructs (Fig. 2c). The luciferase activity of −204M1 and −204M2 was significantly higher than that of −204 (Fig. 2b), although the −204M1 luciferase mRNA expression was lower than that of −204 (Fig. 2c). These data suggest that disruption of the uORF by mutation of the start codon ATG at −105 increased the translation efficiency of the downstream ORF. Mutation of the ATG at −74 in construct −99M1 also significantly increased luciferase activity relative to −99, although again there was no difference in their respective mRNA expression levels (Fig. 2c). These data indicated that mutation of these uATGs in the 5′UTR was able to increase translation efficiency of the downstream ORF. To test this directly, in vitro translation assays were conducted for T7-5′UTR-luciferase constructs by using the rabbit reticulocyte lysate system. Results from these in vitro translation assays were consistent with HepG2 cell data, excep that translation of the −204 construct was higher than the −247 construct in the in vitro translation assay (Fig. 3b). Mutation of the −74 ATG to AAG in −204M2 had no further stimulatory effect on translation efficiency than did mutation of the −105 ATG to AAG in −204 M1, indicating that in the longer transcripts, the −105 ATG is the primary regulator of translation efficiency of the ORF. However, mutation of the −74 ATG to AAG further stimulated translation of −99M1, indicating that this uATG could also inhibit translation of the ORF.
Fig. 2.
Role of the MRP2 5′UTR in the regulation of downstream ORF expression. a, schematic representation of the constructs used for HepG2 cell transfection. −247, full length of MRP2 5′UTR, from −247 to −1 relative to MRP2 start codon ATG; −204, fragment of MRP2 5′UTR from −204 to −1; −204M1, −105 ATG was mutated to AAG in −204; −204 M2, −105 ATG and −74 ATG were mutated to AAG in −204; −99, fragment of MRP2 5′UTR from −99 to −1; −99 M1, −74 ATG was mutated to AAG in −99; CT, the fragment between SV40 promoter and luciferase gene in pGL3 control vector. b, effects of MRP2 5′UTR on luciferase activity in transiently transfected HepG2 cells. Two days after transfection, cells were directly lysed and measured for luciferase activity. CT, −247, −204, −204 M1, −204 M2, −99, and −99 M1 are SV40-5′UTR-luciferase cassettes amplified from corresponding vectors as shown in a. Values are mean ± S.E.M. (n = 3). *, p < 0.05 versus −204 M1 and −204M2; **, p < 0.01 versus CT, −247, −204, −204 M1, and −204 M2; #, p < 0.05 versus −99. c, real-time RT-PCR analysis of luciferase mRNA expression in SV40-5′UTR-Luciferase cassette transfected HepG2 cells. After 2 days of transfection, the cells were harvested for RNA isolation. The luciferase mRNA expression was detected by real time RT-PCR and normalized by its 18S expression. Values are mean ± S.E.M. (n = 3). *, p < 0.05 versus −204.
Fig. 3.
Role of MRP2 5′UTR on translation efficiency of the luciferase reporter transcript in in vitro translation assays. a, schematic representation of the constructs used in in vitro translation assays. −247, −204, −204M1, −204M2, −99, and −99M1 are the same as described in Fig. 2a. C35, the fragment between T7 promoter and luciferase gene in T7 control DNA vector. b, in vitro translation assays of 5′UTR-luciferase transcript. The T7-5′UTR-luciferase cassette was amplified by PCR from the corresponding vectors as described in a; the cassettes were used as templates to synthesize the capped, 5′UTR-fused luciferase transcripts. The 5′UTR-luciferase transcripts (0, 2, 4, 10, 20, or 40 ng of mRNA) were added to rabbit reticulocyte lysate mixture. The lines are the best linear fit of the relationship between luciferase activity and mRNA transcript concentration. The slopes represent translation efficiencies. The statistical significance of the difference between the slopes was tested by Prism 4.0 (GraphPad Software, San Diego, CA).
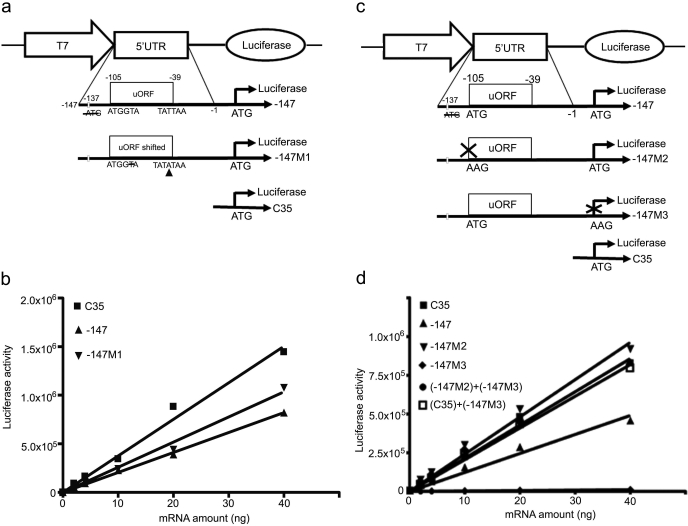
In both HepG2 transient expression experiments and in vitro translational assays, the −204 construct showed a lower translational efficiency than the −204M1 construct, where the −105 ATG was disrupted, consistent with uORF inhibition of translation of the downstream ORF. We further investigated whether this inhibition might be dependent on the uORF-encoded amino acid sequence. To investigate this, the uORF coding frame was shifted by deletion of one nucleotide at −101 and insertion of one nucleotide at −40 in the uORF (Fig. 4a). The site of the mutations ensured that the Kozak motif surrounding the −105 uORF was not disrupted, that the amino acid sequence was maximally altered, and that premature termination codons were not introduced into the uORF. Comparison of the effects of the shifted (−147M1) versus nonshifted (−147) constructs on downstream ORF translation (Fig. 4b) showed that there was no significant difference in inhibition of translation when the amino acid sequence was altered. These data indicated that inhibition of gene translation by the −105 uORF was not dependent on the amino acid sequence of the encoded peptide.
Fig. 4.
Inhibition by uORF at −105 of downstream ORF translation is not dependent on the uORF encoded amino acid sequence and functions as a cis inhibitory element. a, schematic representation of luciferase fusion constructs. −147, the MRP2 fragment from −147 to −1 with mutation of −74 ATG to ATA and deletion of −137 ATG (open bar); −147M1, the uORF was frame-shifted in −147; C35, the same as described in Fig. 3a. b, in vitro translation assays. The capped fusion luciferase transcripts obtained by in vitro transcription (0, 2, 4, 10, 20, and 40 ng of mRNA) were added to the rabbit reticulocyte lysate reaction mixtures and incubated at 30°C for 1 h, terminated on ice, and luciferase activity measured. The lines are the best linear fit of the relationship between luciferase activity and mRNA transcript concentration. The slopes represent translation efficiencies. The statistical significance of the difference between the slopes was tested by GraphPad Prism 4.0. c, schematic representation of the plasmid constructs. −147, the same as described in a; −147M2, the uORF was disrupted in −147; −147M3, the luciferase ORF was disrupted in −147; C35, the same as described in Fig. 3a. d, in vitro translation assays. The transcripts (0, 2, 4, 10, 20, and 40 ng of mRNA) prepared by in vitro transcription were added to the rabbit reticulocyte lysate reaction mixtures and incubated at 30°C for 1 h, terminated on ice, and luciferase activity measured. The lines are the best linear fit of the relationship between luciferase activity and mRNA transcript concentration. The slopes represent translation efficiencies. The statistical significance of the difference between the slopes was tested by GraphPad Prism 4.0.
The inhibition of translation by the −105 uORF raised the question as to whether the uORF-encoded 22 amino acid peptide might inhibit translation as a cis element or act as a trans regulator, where it could regulate translation of an independent gene. We mutated the uORF in construct −147 to create construct −147M2 in which the 22-amino acid peptide would not be translated (Fig. 4c). We also mutated the luciferase ORF in construct −147 to create a new construct, −147M3, in which the luciferase gene would not be translated (Fig. 4c). The −147M3 and −147M2 constructs were mixed in equal amounts in in vitro translation assays. As a control, the −147M3 and C35 constructs were also mixed in equal amounts to test translation efficiency. The translation efficiency of the −147 construct was 57% of that of the C35 construct, whereas the translation efficiency of the −147M2 and C35 constructs were essentially equal, confirming the initial observation that disruption of the −105 uATG increased the translation efficiency 2-fold (p < 0.0001). This also confirmed that the −105 uORF cis inhibited translation of the downstream luciferase mRNA. The translation efficiency of the combination of −147M2 and −147M3 constructs was 89% of that of the −147M2 construct, indicating that the presence of the −147M3 construct did not change the translation efficiency of −147M2 (p = 0.1111). Likewise, the presence of −147M3 did not change the translation efficiency of C35 (p = 0.347) (Fig. 4d). These results show that the −105 uORF regulates translation of a downstream ORF translation as a cis element, and not as a trans-acting factor.
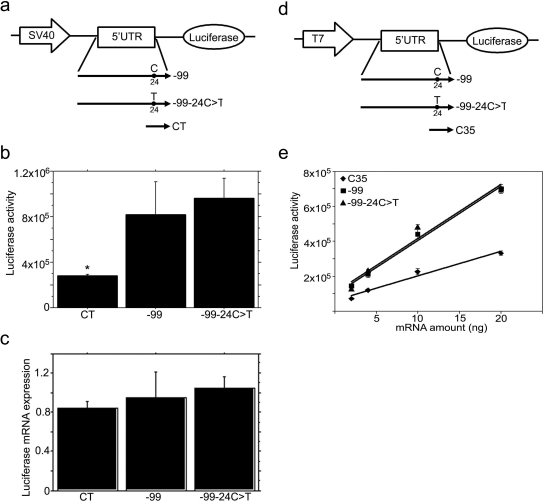
Functional analyses of MRP2 have identified several single nucleotide polymorphisms (SNPs), including −24C>T in the 5′UTR, which have been associated with changes in drug-induced toxicity or pharmacokinetics. The −24C>T SNP represents a synonymous variant in the −74 uORF and could therefore function by influencing MRP2 translation in addition to potential effects on transcription. To determine whether the −24C>T SNP in the 5′UTR might affect expression of the downstream ORF, we made the −24C>T mutant in 5′UTR −99 constructs and tested its effects on regulation of downstream gene expression by using both transient gene expression in HepG2 cells and in vitro translation assays (Fig. 5). Our results show that the −24C>T mutant in these constructs did not affect either mRNA expression or translation of the downstream ORF when characterized in transiently transfected HepG2 cells (Fig. 5, a–c). The same results were obtained in the in vitro translation assay of the −24C>T constructs, confirming that there was no difference in translational efficiency between the wild type and −24C>T mutant (Fig. 5, d and e).
Fig. 5.
The −24C>T polymorphism in the MRP2 5′UTR does not affect mRNA expression or translation of the downstream luciferase gene. a, schematic representation of the constructs used for HepG2 cell transfection. −99 and CT, the same as described in Fig. 2a; −99–24C>T, the −24C was mutated to T in −99. b, luciferase activity in SV40-5′UTR-luciferase cassette transfected HepG2 cells. After 2 days of transfection, the cells were harvested for measurement of luciferase activity. CT, SV40-5′UTR-luciferase cassette amplified from pGL3 control vector; −99, −99–24C>T are SV40-5′UTR-luciferase cassettes amplified from corresponding vectors as described in a. Values are mean ± S.E.M. (n = 3). *, p < 0.001 versus −99 or −99–24C>T. c, real-time RT-PCR analysis of luciferase mRNA in SV40-5′UTR-Luciferase cassette transfected HepG2 cells. Two days after transfection, RNA was isolated, and luciferase mRNA expression detected by real time RT-PCR and normalized by 18S expression. Values are mean ± S.E.M. (n = 3). d, schematic representation of the constructs used in in vitro translation assays. −99 and −99–24C>T, the same as described in a; C35, the same as described in Fig. 3 a. e, in vitro translation assays of 5′UTR-luciferase transcript. The T7-5′UTR-luciferase cassettes were amplified by PCR from corresponding vectors as described in c; the cassettes were used as templates to synthesize the capped, 5′UTR-fused luciferase transcripts. The 5′UTR-luciferase transcripts (0, 2, 4, 10, or 20 ng of mRNA) were added to rabbit reticulocyte lysate mixture. The lines are the best linear fit of the relationship between luciferase activity and mRNA transcript concentration. The slopes represent translation efficiencies. The statistical significance of the difference between the slopes was tested by GraphPad Prism 4.0.
Discussion
The present studies demonstrate for the first time the importance of the 5′UTRs of human MRP2 in the translational regulation of MRP2 protein expression, and the importance of the uORF −105 in inhibiting translation. The 5′UTR of the human MRP2 gene contains seven uATGs and six uORFs. We focused on uORF −105 because this is the only uATG that is flanked by a perfect Kozak motif and also because its location in the 5′UTR is similar to that in the rat Mrp2 5′UTR, where an uATG at −109 is flanked by a perfect Kozak motif and inhibits translation of the downstream ORF (Zhang et al., 2007). The −105 uORF in human MRP2 encodes a 22-amino acid peptide and is in-frame with the MRP2 ORF, but two stop codons at −39 (TAA) and −36 (TAG) almost certainly ensure that an N-terminal-extended form of MRP2 is not generated. The presence of an in-frame uORF is unusual, in that statistical analyses of genome organization show that the in-frame uATGs are more strongly suppressed than out-of-frame uATGs (Iacono et al., 2005). It is noteworthy that the 5′UTR of the murine Mrp2 (GenBank accession number NM_013806) also contains a putative uORF with a perfect Kozak motif beginning at −110 nucleotides; further work is needed to identify the murine Mrp2 transcription start sites and the role such an uORF might have in regulating Mrp2 translation. The conservation of the uORFs at −109, −105, and −110 nucleotides in the rat, human, and mouse, respectively, implies an important and shared regulatory mechanism. The amino acid sequence of the peptides encoded by these uORFs for the rat, human, and mouse Mrp2/MRP2 are not conserved, consistent with genome statistical analyses showing that there is greater conservation of the location of such uORFs than of the encoded amino acid sequences (Neafsey and Galagan, 2007).
Mutation of uATG −105 significantly increased the translation efficiency in HepG2 cells (Fig. 2) and in vitro (Fig. 3), indicating that uATG −105 was recognized by the ribosome and inhibited the translation of the downstream ORF. These findings are consistent with most other findings that uATGs inhibit downstream ORF translation (Meijer et al., 2000; Diba et al., 2001; Kwon et al., 2001; Blaschke et al., 2003; Mihailovich et al., 2007; Song et al., 2007). Further studies showed that the amino acid sequence of the peptide encoded by uORF-105 was not important in regulating translation and that uORF-105 acted only as a cis element to inhibit downstream ORF translation. These data indicate that translation of the downstream ORF (luciferase) occurred either by leaky scanning (i.e., some ribosomes bypassed uORF-105 and continued scanning the 5′UTR and translated the ORF) or by reinitiation of translation of the downstream ORF after termination of translation of the uORF-105. Mutation of the uATG −74 in the −204M1 construct did not further stimulate translation in HepG2 cells (Fig. 2) or in vitro (Fig. 3), implying that this uORF does not play a major role in regulating translation of the downstream ORF, at least in the longer 5′UTR transcripts. This suggests, but does not prove, that most of the scanning ribosomes translate the downstream ORF by reinitiating translation, rather than by leaky scanning of the longer 5′UTR transcripts. Because uORF −74 terminates inside the MRP2 coding region at +234, initiation of translation of uORF −74 would markedly inhibit translation of the downstream ORF. The presence of the uORF −105 with a perfect Kozak motif might therefore prevent initiation of translation at uORF-74 in the longer 5′UTR transcripts. However, mutation of uATG-74 to AAG in the shorter −99 construct further increased translation of the downstream ORF, clearly indicating its inhibitory potential.
The MRP2 5′UTR −99 showed a much higher translational efficiency than −247 or −204 (Figs. 2 and 3). Because the regulation of translation by 5′UTR is independent of the length of the 5′UTR (Anant et al., 2002), it is likely that the presence of the additional uATGs in the −247 and −204 5′UTRs is responsible for their lower translational efficiency. There are seven uATGs in −247 or −204, but only one uATG in −99. At present, we cannot exclude other mechanisms that might inhibit translation in the longer 5′UTRs; although the secondary structure of the −247 or −204 5′UTRs might affect translation efficiency, the fact that the translational efficiency of the −204 fragment is similar to that of the control (C35) fragment (Fig. 2), argues against this possibility. Alternatively, a translation suppressor may bind to the 5′ end of the longer 5′UTR and suppress translation, or a translational active element between −99 and −1 may increase its translational efficiency. We are currently investigating the mechanism for the higher translational efficiency of the −99 MRP2 5′UTR.
Human MRP2 gene transcription initiation sites −247, −204, and −99 have been identified by 5′ rapid amplification of cDNA ends analysis of a human placenta library, and the −247 site is described as the “major” site (Tanaka et al., 1999); analysis of placenta RNA by RPA demonstrated the predominant band at −247, consistent with these results. The RPA analyses detected at least eight putative transcription initiation sites, including the identified three transcription initiation sites −247, −204, and −99 in HepG2 cells, liver, and kidney (Fig. 1). The presence of multiple transcription initiation sites for MRP2 is consistent with the absence of a functionally relevant TATA box in the MRP2 gene (Scotto, 2003). These results indicate that like rat Mrp2, human MRP2 uses different transcription start sites and therefore different 5′UTRs as a potential mechanism to control gene expression at the translational level. In the rat, the liver and kidney use different Mrp2 transcripts, such that in kidney, −132 is the predominant transcription start site and includes the inhibitory −109 uORF, whereas in liver, the transcript beginning at −98 predominates, avoiding the inhibitory −109 uORF (Zhang et al., 2007). MRP2 expression in human placenta increases with gestational age, with a 4-fold increase in MRP2 mRNA versus a 1.6-fold increase in MRP2 protein expression from early preterm (<32 weeks of gestation) to term (>37 weeks of gestation) (Meyer zu Schwabedissen et al., 2005). We do not know the gestational age of the purchased placental RNA; however, it could be useful to determine whether the transcription start sites differ with gestational age and whether such differences might explain the differential induction of MRP2 mRNA versus protein. In patients with obstructive cholestasis, upper intestinal MRP2 protein was reduced to 27% compared with MRP2 protein expression in age- and gender-matched control patients with mild gastritis, esophagitis, or acid reflux; however, there were no detectable difference in mRNA levels in these cholestatic versus control patients (Dietrich et al., 2004). The extent of down-regulation of MRP2 protein expression seemed to correlate with the duration of cholestasis, and MRP2 protein expression increased in those subjects in which bile flow was restored. In the purchased intestinal RNA (the section of the intestine was not known), shorter MRP2 mRNA transcripts predominated compared with liver, implying more efficient translation. Further studies are needed to determine whether changes in the transcription start sites of MRP2 mRNA could mediate changes in MRP2 protein expression in the upper intestine in patients with obstructive cholestasis. Finally, even in the longer MRP2 mRNA transcripts, the selection of the translation start site by the scanning ribosome could greatly influence the translation efficiency. Thus, if the ribosome were to bypass the −105 uORF and instead translate the −74 uORF, translation of the MRP2 ORF would be markedly reduced. The best-studied example of how uORFs act as translational regulators is in the GCN4 gene of Saccharomyces cerevisiae, where the GCN4 protein transcriptionally activates amino acid biosynthetic genes (Hinnebusch, 2005). Translation of GCN4 mRNA is derepressed in amino acid-deprived cells; derepression is the orchestrated effect of four short uORFs (named uORF1–4) in GCN4 mRNA, together with the phosphorylation status of eukaryotic initiation factor 2. In an amino acid-rich medium, ribosomes translate uORF1 and reinitiate primarily at uORF4 but are therefore unable to access the GCN4 ATG. In amino acid-deprived cells, ribosomes still translate uORF1, but bypass uORF2 to uORF4 and reinitiate at GCN4 ATG. In this case, uORF 4 and GCN4 ORF are competitors for the scanning ribosomes after translation of uORF1; the level of the active form of eukaryotic initiation factor 2 determines whether uORF4 or GCN4 captures the ribosomes more efficiently. Clearly, much more work is needed to understand the factors that regulate MPR2 transcription, the use of MRP2 transcription start sites, and the translation start sites.
It is apparent that regulation of expression of functional MRP2 occurs at multiple levels and time frames: increased transcription and selection of transcription start site for long-term regulation, selection of translational start site for short-term regulation, and localization of MRP2 within the cell, either in the apical domain, or in endocytic vesicles (Mottino et al., 2002), for immediate regulation. This array of regulatory mechanisms should thus allow the cell to respond to different stimuli and needs in various ways.
The MRP2 −24C>T variant occurs with a relatively high allelic frequency (18%) in Japanese subjects (Suzuki and Sugiyama, 2002) and is associated with changes in MRP2 expression or drug-induced toxicity or pharmacokinetics. MRP2 −24C>T is associated with higher methotrexate plasma levels in female pediatric patients treated for acute lymphoblastic leukemia (Gradhand and Kim, 2008), with increased susceptibility to toxic liver injury when a component of an MRP2 haplotype (Choi et al., 2007), and with decreased MRP2 mRNA expression in noncancerous kidney cortex (Haenisch et al., 2007). Although there has been some suggestion that the −24C>T SNP might affect the binding of transcription factors or RNA stability, we found no effect of this SNP on MRP2 mRNA expression or translational efficiency. In the present studies, the −24C>T mutation was tested by using MRP2 5′UTR fragment −99, which was inserted between the SV40 promoter and luciferase reporter. Because the SV40 promoter drove the transcription of the luciferase gene, the lack of effect on luciferase mRNA expression probably reflects the lack of effect of this mutation on mRNA stability. Clearly, the −24C>T mutation had no effect on the translational efficiency of the downstream ORF (Fig. 5). However, we cannot rule out a potential effect of the −24C>T mutation on differential transcriptional regulation of MRP2.
In summary, the present studies provide the first clear evidence of the role of the 5′UTR of MRP2 in regulating its translation and provide a potential mechanism to explain the observed post-transcriptional regulation of MRP2. The uORF −105, containing a perfect Kozak motif, was shown to inhibit translation of the main ORF, most likely by initiating translation of the encoded peptide, followed by reinitiation of translation of MRP2. Because inhibition of downstream ORF translation is not dependent on the uORF encoded peptide sequence, the conserved uORF in rat and human Mrp2/MRP2 and the putative uORF in murine Mrp2, suggest that this uORF is an important feature in translational regulation of this key efflux transporter.
Acknowledgments
We thank Dr. Robert Rhoads (Louisiana State University, Shreveport, LA) for his generous help and advice in these studies.
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grant GM55343] and the National Institutes of Health National Institute of Child Health & Human Development [Grant HD58299].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
doi:10.1124/mol.109.058982.
- MRP
- multidrug resistance-associated protein
- UTR
- untranslated region
- uORF
- upstream open reading frame
- uATG
- upstream start codon
- PCR
- polymerase chain reaction
- F
- forward
- R
- reverse
- SV40
- simian virus 40
- RPA
- RNAse protection assay
- RT-PCR
- reverse-transcription polymerase chain reaction
- UPL
- universal probe library
- SNP
- single nucleotide polymorphism.
References
- Anant S, Mukhopadhyay D, Hirano K, Brasitus TA, Davidson NO. (2002) Apobec-1 transcription in rat colon cancer: decreased apobec-1 protein production through alterations in polysome distribution and mRNA translation associated with upstream AUGs. Biochim Biophys Acta 1575: 54–62 [DOI] [PubMed] [Google Scholar]
- Blaschke RJ, Töpfer C, Marchini A, Steinbeisser H, Janssen JW, Rappold GA. (2003) Transcriptional and translational regulation of the Leri-Weill and Turner syndrome homeobox gene SHOX. J Biol Chem 278: 47820–47826 [DOI] [PubMed] [Google Scholar]
- Cao J, Huang L, Liu Y, Hoffman T, Stieger B, Meier PJ, Vore M. (2001) Differential regulation of hepatic bile salt and organic anion transporters in pregnant and postpartum rats and the role of prolactin. Hepatology 33: 140–147 [DOI] [PubMed] [Google Scholar]
- Cao J, Stieger B, Meier PJ, Vore M. (2002) Expression of rat hepatic multidrug resistance-associated proteins and organic anion transporters in pregnancy. Am J Physiol Gastrointest Liver Physiol 283: G757–G766 [DOI] [PubMed] [Google Scholar]
- Choi JH, Ahn BM, Yi J, Lee JH, Lee JH, Nam SW, Chon CY, Han KH, Ahn SH, Jang IJ, Cho JY, Suh Y, Cho MO, Lee JE, Kim KH, Lee MG. (2007) MRP2 haplotypes confer differential susceptibility to toxic liver injury. Pharmacogenet Genomics 17: 403–415 [DOI] [PubMed] [Google Scholar]
- David-Assael O, Saul H, Saul V, Mizrachy-Dagri T, Berezin I, Brook E, Shaul O. (2005) Expression of AtMHX, an Arabidopsis vacuolar metal transporter, is repressed by the 5′ untranslated region of its gene. J Exp Bot 56: 1039–1047 [DOI] [PubMed] [Google Scholar]
- Deeley RG, Westlake C, Cole SP. (2006) Transmembrane transport of endo- and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiol Rev 86: 849–899 [DOI] [PubMed] [Google Scholar]
- Diba F, Watson CS, Gametchu B. (2001) 5′UTR sequences of the glucocorticoid receptor 1A transcript encode a peptide associated with translational regulation of the glucocorticoid receptor. J Cell Biochem 81: 149–161 [PubMed] [Google Scholar]
- Dietrich CG, Geier A, Salein N, Lammert F, Roeb E, Oude Elferink RP, Matern S, Gartung C. (2004) Consequences of bile duct obstruction on intestinal expression and function of multidrug resistance-associated protein 2. Gastroenterology 126: 1044–1053 [DOI] [PubMed] [Google Scholar]
- Geier A, Wagner M, Dietrich CG, Trauner M. (2007) Principles of hepatic organic anion transporter regulation during cholestasis, inflammation and liver regeneration. Biochim Biophys Acta 1773: 283–308 [DOI] [PubMed] [Google Scholar]
- Gradhand U, Kim RB. (2008) Pharmacogenomics of MRP transporters (ABCC1–5) and BCRP (ABCG2). Drug Metab Rev 40: 317–354 [DOI] [PubMed] [Google Scholar]
- Haenisch S, Zimmermann U, Dazert E, Wruck CJ, Dazert P, Siegmund W, Siegmund S, Kroemer HK, Warzok RW, Cascorbi I. (2007) Influence of polymorphisms of ABCB1 and ABCC2 on mRNA and protein expression in normal and cancerous kidney cortex. Pharmacogenomics J 7: 56–65 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol 59: 407–450 [DOI] [PubMed] [Google Scholar]
- Hood HM, Neafsey DE, Galagan J, Sachs MS. (2009) Evolutionary role of upstream open reading frames in mediating gene regulation in fungi. Annu Rev Microbiol 63: 385–409 [DOI] [PubMed] [Google Scholar]
- Iacono M, Mignone F, Pesole G. (2005) uAUG and uORFs in human and rodent 5′untranslated mRNAs. Gene 349: 97–105 [DOI] [PubMed] [Google Scholar]
- Johnson DR, Guo GL, Klaassen CD. (2002) Expression of rat Multidrug Resistance Protein 2 (Mrp2) in male and female rats during normal and pregnenolone-16alpha-carbonitrile (PCN)-induced postnatal ontogeny. Toxicology 178: 209–219 [DOI] [PubMed] [Google Scholar]
- Johnson DR, Klaassen CD. (2002) Regulation of rat multidrug resistance protein 2 by classes of prototypical microsomal enzyme inducers that activate distinct transcription pathways. Toxicol Sci 67: 182–189 [DOI] [PubMed] [Google Scholar]
- Jones BR, Li W, Cao J, Hoffman TA, Gerk PM, Vore M. (2005) The role of protein synthesis and degradation in the post-transcriptional regulation of rat multidrug resistance-associated protein 2 (Mrp2, Abcc2). Mol Pharmacol 68: 701–710 [DOI] [PubMed] [Google Scholar]
- Kozak M. (1986) Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44: 283–292 [DOI] [PubMed] [Google Scholar]
- Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene 234: 187–208 [DOI] [PubMed] [Google Scholar]
- Kwon HS, Lee DK, Lee JJ, Edenberg HJ, Ahn YH, Hur MW. (2001) Posttranscriptional regulation of human ADH5/FDH and Myf6 gene expression by upstream AUG codons. Arch Biochem Biophys 386: 163–171 [DOI] [PubMed] [Google Scholar]
- Le SY, Maizel JV., Jr (1997) A common RNA structural motif involved in the internal initiation of translation of cellular mRNAs. Nucleic Acids Res 25: 362–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Sachs MS. (1996) Role of an upstream open reading frame in mediating arginine-specific translational control in Neurospora crassa. J Bacteriol 178: 2172–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HA, Dictus WJ, Keuning ED, Thomas AA. (2000) Translational control of the Xenopus laevis connexin-41 5′-untranslated region by three upstream open reading frames. J Biol Chem 275: 30787–30793 [DOI] [PubMed] [Google Scholar]
- Meyer zu Schwabedissen HE, Jedlitschky G, Gratz M, Haenisch S, Linnemann K, Fusch C, Cascorbi I, Kroemer HK. (2005) Variable expression of MRP2 (ABCC2) in human placenta: influence of gestational age and cellular differentiation. Drug Metab Dispos 33: 896–904 [DOI] [PubMed] [Google Scholar]
- Mihailovich M, Thermann R, Grohovaz F, Hentze MW, Zacchetti D. (2007) Complex translational regulation of BACE1 involves upstream AUGs and stimulatory elements within the 5′ untranslated region. Nucleic Acids Res 35: 2975–2985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mize GJ, Ruan H, Low JJ, Morris DR. (1998) The inhibitory upstream open reading frame from mammalian S-adenosylmethionine decarboxylase mRNA has a strict sequence specificity in critical positions. J Biol Chem 273: 32500–32505 [DOI] [PubMed] [Google Scholar]
- Morris DR, Geballe AP. (2000) Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol 20: 8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottino AD, Cao J, Veggi LM, Crocenzi F, Roma MG, Vore M. (2002) Altered localization and activity of canalicular Mrp2 in estradiol-17beta-d-glucuronide-induced cholestasis. Hepatology 35: 1409–1419 [DOI] [PubMed] [Google Scholar]
- Mottino AD, Hoffman T, Jennes L, Vore M. (2000) Expression and localization of multidrug resistant protein mrp2 in rat small intestine. J Pharmacol Exp Ther 293: 717–723 [PubMed] [Google Scholar]
- Neafsey DE, Galagan JE. (2007) Dual modes of natural selection on upstream open reading frames. Mol Biol Evol 24: 1744–1751 [DOI] [PubMed] [Google Scholar]
- Parola AL, Kobilka BK. (1994) The peptide product of a 5′ leader cistron in the beta 2 adrenergic receptor mRNA inhibits receptor synthesis. J Biol Chem 269: 4497–4505 [PubMed] [Google Scholar]
- Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, Liuni S. (2001) Structural and functional features of eukaryotic mRNA untranslated regions. Gene 276: 73–81 [DOI] [PubMed] [Google Scholar]
- Reynolds K, Zimmer AM, Zimmer A. (1996) Regulation of RAR beta 2 mRNA expression: evidence for an inhibitory peptide encoded in the 5′-untranslated region. J Cell Biol 134: 827–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotto KW. (2003) Transcriptional regulation of ABC drug transporters. Oncogene 22: 7496–7511 [DOI] [PubMed] [Google Scholar]
- Song KY, Hwang CK, Kim CS, Choi HS, Law PY, Wei LN, Loh HH. (2007) Translational repression of mouse mu opioid receptor expression via leaky scanning. Nucleic Acids Res 35: 1501–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Sugiyama Y. (2002) Single nucleotide polymorphisms in multidrug resistance associated protein 2 (MRP2/ABCC2): its impact on drug disposition. Adv Drug Deliv Rev 54: 1311–1331 [DOI] [PubMed] [Google Scholar]
- Tanaka T, Uchiumi T, Hinoshita E, Inokuchi A, Toh S, Wada M, Takano H, Kohno K, Kuwano M. (1999) The human multidrug resistance protein 2 gene: functional characterization of the 5′-flanking region and expression in hepatic cells. Hepatology 30: 1507–1512 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li W, Vore M. (2007) Translational regulation of rat multidrug resistance-associated protein 2 expression is mediated by upstream open reading frames in the 5′ untranslated region. Mol Pharmacol 71: 377–383 [DOI] [PubMed] [Google Scholar]
- Zimmer M, Ebert BL, Neil C, Brenner K, Papaioannou I, Melas A, Tolliday N, Lamb J, Pantopoulos K, Golub T, Iliopoulos O. (2008) Small-molecule inhibitors of HIF-2a translation link its 5′UTR iron-responsive element to oxygen sensing. Mol Cell 32: 838–848 [DOI] [PMC free article] [PubMed] [Google Scholar]