Abstract
Serum antibodies from myriad species, particularly birds, can provide key information regarding the transmission and the expansion of the territory of emerging pathogens. Expedient antibody analysis is constrained by a lack of species-specific reagents, a deficiency potentially highlighted by the recent swine-origin influenza A virus (H1N1) outbreak. Available methodologies present difficulties that discourage thorough serologic monitoring of potential disease vectors or hosts. Rapid high-throughput procedures that combined serum amine labeling via biotinylation, contaminant removal, and microsphere-based immunoassays for antibodies to three arboviruses were developed. Agent-specific adaptations of this simple format should facilitate expanded surveillance and diagnostic capabilities regarding pathogens of human and veterinary importance.
Serologic analysis of samples obtained from nondomestic animals, birds in particular, can provide key information regarding the transmission and the expansion of the territory of emerging pathogens (1). Unfortunately, species-specific reagents such as antispecies capture antibodies and detection conjugates, necessary for most rapid diagnostic methods, are frequently unavailable due to a lack of commercial interest. Traditional methods for circumventing species constraints often use live pathogens, are technically challenging, or are so time-consuming that large-scale testing becomes impractical (2). Methods that utilize protein G (4) are able to capture and detect mammalian IgG but cannot be used when the antibodies of interest are IgM and IgY, the former being the first of the Igs to be generated after infection and the latter being the avian and reptilian equivalent of IgG. The results of blocking enzyme-linked immunosorbent assay (ELISAs) (3) are sometimes inconsistent, and test sensitivities are dependent on the relative affinities of the competing monoclonal and serum antibodies to the antigens. Disease surveillance that involves the analysis of antibodies in nondomestic species is thus limited.
The ideal method for the detection of antibodies in a range of species would be rapid, be capable of high throughput, provide a positive signal (as opposed to a signal reduction via competition), and require a small sample volume. Total antibody measurement would be advantageous, because surveillance is usually conducted without knowledge of the timing of infection. Biotin (vitamin H) is well-known to react with free amine groups on proteins (18). We determined that virus-specific serum antibodies independent of the species of origin could be biotinylated and directly detected in microsphere immunoassays (biotin-MIAs) that were modified from established protocols (11). Here we describe the development of two species-independent antibody detection methods for use with arboviruses, which involve animals as vectors or hosts. The first is a duplex procedure for the detection of antibodies to West Nile (WN) and St. Louis encephalitis (SLE) viruses, and the second is a procedure for the identification of anti-eastern equine encephalitis (anti-EEE) virus antibodies.
MATERIALS AND METHODS
Serum samples.
A total of 535 serum samples either were obtained from the diagnostic archives at the DVBID/CDC or remained from previously completed animal studies. The numbers and species used in each portion of the study are indicated separately in the text and figures.
Biotinylation of serum samples.
Serum samples were biotinylated by using a 50 M excess of biotin over the calculated concentration of the amines, as optimized by titration. To 1.25 μl of serum, 4.25 μl of 5.55 mg/ml sulfo-LC-biotin (Pierce, Rockford, IL) and 44.5 μl of phosphate-buffered saline (PBS; pH 7.4) were added. These were incubated for 30 min with mixing at room temperature in the wells of a 100,000-molecular-weight-cutoff filter plate (Acroprep 96 Omega 100K; VWR Scientific, San Francisco, CA) by using a Lab-Line instruments rotary titer plate shaker at 900 rpm (VWR Scientific). Components with molecular masses of <100 kDa, primarily albumin and uncoupled biotin, were removed via vacuum filtration. The retentate, enriched for biotinylated antibodies, was washed with 50 μl PBS and was then vacuum filtered and resuspended in 50 μl PBS, which constituted a 1:40 dilution of the original sample. Candor Low Cross buffer (Boca Scientific, Boca Raton, FL) was used to make further 1:10 dilutions of the samples to achieve a final serum dilution of 1:400, which was determined by initial titration to yield optimal signal-to-noise ratios.
Controls.
Purified monoclonal antibodies (MAbs) served as positive controls and were treated by the same method used for the serum samples. For the WN/SLE virus biotin-MIA, 25 μg of flavivirus group-reactive MAb 6B6C-1, which recognizes all flaviviruses of human medical importance by binding to part of the flaviviral envelope protein fusion peptide, was used (6, 16). For the EEE virus biotin-MIA, 25 μg alphavirus group-reactive MAb 1A4B-6 (17) was used; MAb 1A4B-6 recognizes all medically important alphaviruses, and although it is not proven, it likely recognizes all alphaviruses (J. Roehrig, personal communication). Known antibody-negative sera from representatives of each order of birds, mammals, and reptiles represented in the test sample set were pooled. This was used as a negative control, in which 1.25 μl of the pool was biotinylated in the same way as the samples.
Biotin-MIA.
Two biotin-MIA methods were developed, the WN/SLE virus biotin-MIA and the EEE virus biotin-MIA. The methods were modified versions of existing MIAs (11, 14). Briefly, MicroPlex microsphere sets 32 and 57 (Luminex Corp., Austin, TX) coupled covalently by standard carbodiimide chemistry (19) with MAb 6B6C-1 for the WN and SLE viral antibody tests, respectively, were purchased from Radix Biosolutions (Georgetown, TX). Set 15 coupled to alphavirus group-reactive MAb 2A2C-3 (9) for the EEE viral antibody test was also purchased from Radix Biosolutions. Before the assay was performed, microspheres were reacted with each viral antigen and its corresponding negative control antigen in PBS-1% bovine serum albumin (BSA; Sigma-Aldrich, St. Louis, MO) by mixing at room temperature for 1 h. These reactions were performed for each antigen at a rate of 5,000 microspheres per microliter, as follows: 50 μl set 32-6B6C-1 plus either 40 μl WN virus recombinant antigen (Hennessy Research, Kansas City, MO) (7) or 40 μl negative recombinant antigen (produced by Hennessy Research in a manner identical to that used for the viral antigen but with nontransfected COS-1 cells) plus 410 μl PBS-1% BSA; 50 μl set 57-6B6C-1 plus either 40 μl SLE virus recombinant antigen (CDC) (20) or 40 μl negative recombinant antigen (made with nontransfected COS-1 cells at the CDC) plus 410 μl PBS-1%BSA; and 50 μl set 15-2A2C-3 plus either 2 μl EEE virus suckling mouse brain antigen (CDC) (5) or 2 μl negative suckling mouse brain antigen (made with noninfected suckling mouse brains and processed as described for the preparation of EEE virus at the CDC) plus 448 μl PBS-1% BSA. The preparations were stored at 4°C and were used on the following day or for up to a month thereafter. Freeze-thaw effects were not investigated, since the manufacturer recommends against freezing of the microspheres. For each of 256 samples used to develop the WN/SLE virus biotin-MIA, 2,500 viral antigen-reacted set 32 and 57 microspheres suspended in a total volume of 100 μl PBS-1% BSA (Sigma-Aldrich) were added to a single well of a 96-well filter plate (Millipore Corp., Billerica, MA). Negative antigen-reacted sets 32 and 57 were added to a different well. The microspheres were washed twice with 100 μl PBS-1% BSA on a vacuum manifold. Fifty microliters of prepared sample or controls was added to the wells containing viral or negative antigen-coupled microspheres, and the plates were shaken for 45 min at room temperature on a titer plate shaker at 900 rpm. The wells were washed twice with 100 μl PBS-1% BSA, followed by the addition of 50 μl per well of 4 μg/ml streptavidin-phycoerythrin (Jackson Immunoresearch, West Grove, PA) in PBS-1% BSA. The plates were shaken for 15 min and were then washed twice. The microspheres were resuspended in 100 μl per well of PBS-1% BSA. The reactions were read by using a BioPlex instrument (Bio-Rad Laboratories, Hercules, CA), in which the microsphere sets and the biological reactions associated with them were identified and quantified via a combination of lasers. The raw results were expressed as the median fluorescent intensities (MFIs) of 100 microspheres per sample per set. An identical scheme was used for the EEE virus assay, in which 50 samples were reacted on viral and negative antigen-coupled set 15 microspheres.
PRNTs.
All biotin-MIA results were compared to those of the plaque reduction neutralization test (PRNT) (2), the “gold standard” serologic method in arbovirology, by using 90% plaque reduction to indicate a positive result at a minimum serum dilution of 1:10. The results were recorded as either positive or negative when a single dilution screen had been performed to conserve materials or were reported as titers when the sample was serially diluted.
Determination of cutoffs.
For each of the antigens, the success of the test and the cutoff values that classified the samples as being antibody positive or negative for each virus were determined by comparing the results obtained in the biotin-MIAs to those obtained by PRNT by using receiver operator characteristic (ROC) curves (15). Areas under the ROC curves (AUCs) and the associated 95% confidence intervals (CIs) were calculated (13). Results that were used in the determination of the ROC curves for the WN/SLE virus biotin-MIA pertained only to the infecting virus, as determined by PRNT, to avoid the skewing of the results by the inclusion of apparent false-positive results caused by sera reactive to both flavivirus antigens. Similarly, the rates of false-positive and false-negative results took this into account. Leave-one-out cross-validation (in which the ROC is applied to the whole data set less each observation sequentially, and the average of the prediction error estimates provides a nearly unbiased estimate of the true prediction error of the model) (8) was used to estimate the predictive performance of the cutoff determination procedure.
Cross-reactivity assays.
The cross-reactivities of the antiarboviral antibodies to arboviral antigens is well documented (10). The routine diagnosis of arboviral infection in humans involves screening by an IgM/IgG ELISA, followed by the performance of cross-neutralization tests that include medically important arboviruses from the geographic region of origin of the sample for any sample testing positive by the screening assays. For a virus to be considered the sole infecting agent, the PRNT titer to that virus must be at least four times greater than the titer to any of the other viruses. For the WN/SLE virus biotin-MIA, cross-reactivity was estimated by using 42 archived human diagnostic samples from individuals with confirmed single infections with the flaviviruses Japanese encephalitis (JE) virus and dengue (DEN) virus, alphaviruses EEE virus and Chikungunya (CHIK) virus, and bunyavirus Lacrosse encephalitis (LAC) virus. For the EEE virus biotin-MIA, samples from 10 confirmed human CHIK virus infections and 13 chicken WN virus infections were used to determine cross-reactivity with antibodies to these viruses.
Validation.
Following development, the WN/SLE virus biotin-MIA was validated with 134 avian serum samples from the following species, with the numbers of each species provided in parentheses: Tyto alba (n = 10), Pica pica (n = 1), Falco sparverius (n = 10), Buteo regalis (n = 2), Buteo jamaicensis (n = 7), Megascops asio (n = 4), Buteo swainsoni (n = 8), Asio otus (n = 2), Falco mexicanus (n = 3), Cathartes aura (n = 2), Haliaeetus leucocephalus (n = 1), Buteo lagopus (n = 5), Athene cunicularia (n = 2), Accipiter gentilis (n = 1), Accipiter cooperii (n = 1), Passer domesticus (n = 7), Petrochelidon pyrrhonota (n = 6), Bubo virginianus (n = 10), Corvus brachyrhynchos (n = 10), Sturnus vulgaris (n = 4), Quiscalus quiscula (n = 4), Columba livia (n = 6), Carpodacus mexicanus (n = 4), Phasianus colchicus (n = 3), Agelaius phoeniceus (n = 6), Ardea alba (n = 7), Aardea herodias (n = 4), and Aquila chrysaetos (n = 4). Of these samples, 50 were previously shown to be WN virus positive by PRNT; 84 were virus negative.
RESULTS
For each sample, the MFI value obtained when the samples were reacted against each of the viral antigens (V) was divided by the MFI value obtained for the corresponding negative antigen reaction (N) to yield a V/N value, which was used in further computations. The negative serum control pool gave MFIs of <350 when the samples were reacted against the viral antigens, and positive controls were always >1,000 MFIs when they were reacted against the viral antigens. Negative samples gave MFIs generally in the range of 50 to 350 and positive samples gave MFIs generally in the range of 1,000 to 25,000 when they were reacted against the viral antigens.
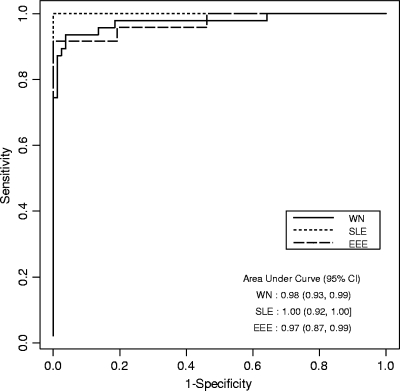
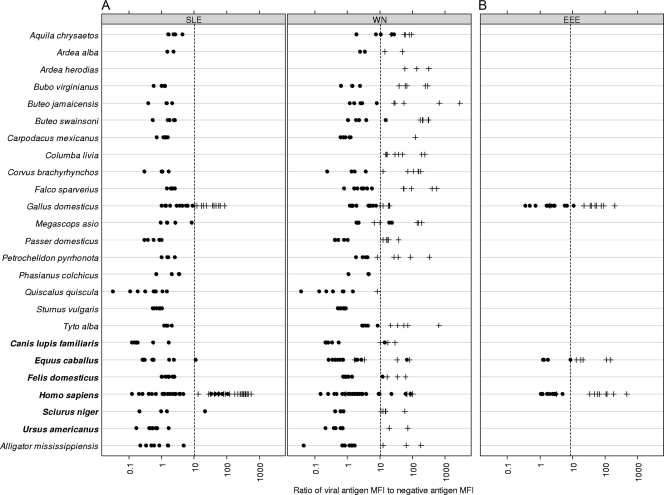
The ROC plots generated for each virus (Fig. 1) gave AUCs of >90%, indicating the good discrimination abilities of the tests. The initial results indicated that a small number of species were susceptible to background reactivity with the negative antigens in the WN/SLE virus biotin-MIA when PBS-1% BSA was used as the final 1:10 diluent. This problem was mitigated by the use of Candor Low Cross buffer and did not alter the results obtained for the other species. For each species exhibiting high background values, the mean MFIs for WN virus- and SLE virus-negative antigens with the original diluent versus the mean MFIs for the WN virus- and SLE virus-negative antigens with the Candor Low Cross buffer is as follows: for Alligator mississippiensis, 486 and 351 versus 46 and 46, respectively; for Ardea alba, 1,556 and 901 versus 8 and 7, respectively; for Equus caballus, 3,917 and 5,175 versus 159 and 151, respectively; for Megascops asio, 929 and 370 versus 230 and 160, respectively; for Petrochelidon pyrrhonota, 950 and 668 versus 50 and 47, respectively; and for Quiscalus quiscula, 1,857 and 1,614 versus 427 and 403, respectively. Therefore, this reagent was adopted as the standard in the protocol. Approximately equal numbers of antibody-positive and -negative samples were used in the development of these tests, with at least half in the WN/SLE virus biotin-MIA originating from wild-caught species. The biotin-MIA results compared to those of PRNT are shown in Fig. 2A for the WN/SLE virus biotin-MIA and Fig. 2B for the EEE virus biotin-MIA.
FIG. 1.
Receiver operator characteristic curves for WN, SLE, and EEE viral antigens in the biotin-MIAs. The sensitivities and the specificities for each antigen were calculated by using the results of PRNT as the gold standard. Areas under the curve and 95% confidence intervals are shown.
FIG. 2.
V/N ratios for samples used to develop the tests are shown. •, PRNT negative; +, PRNT positive; dashed vertical lines, positive V/N cutoff for that antigen. Species are listed in the following order: birds (the normal type above the boldface type). mammals (boldface type), and reptile (the normal type below the boldface type). (A) WN/SLE virus biotin-MIA. Results are shown only for known negative results and positive results for the homologous viral antigens. (B) EEE virus biotin-MIA.
The sensitivity and specificity data for each virus, as plotted on the ROC curves, were used to derive V/N cutoff values above which the samples were deemed positive for the respective viruses. Because all the sensitivity and specificity values were >90%, cutoffs were chosen such that the sensitivity was equal to the specificity. The cutoff values were 10.00 for WN virus, 10.23 for SLE virus, and 8.97 for EEE virus. Figure 2 shows the biotin-MIA results for all viruses species tested compared with those of the PRNTs by application of the calculated cutoffs for each virus. The rates of false-positive and false-negative results computed by using these cutoffs were 5.5% and 5.8%, respectively, for WN virus; 0.0% and 0.0%, respectively, for SLE virus; and 0.0% and 4.0%, respectively, for EEE virus. The overall accuracy of the WN/SLE virus biotin-MIA was 94.4%, and the overall accuracy of the EEE virus biotin-MIA was 98.0%. Cross-validation estimates of the predictive errors mirrored these empirical results, with prediction errors given as overall, false-positive, and false-negative results, as follows: for WN virus, 5.5%, 6.4%, and 4.9%, respectively; for SLE virus, 1.8%, 0.0%, and 4.8%, respectively; and for EEE virus, 6.0%, 8.3%, and 3.8%, respectively. Repeatability was measured to be 100% (95% CI, 80.6 to 100.0%) for 16 samples tested on the same plate at the same time, and reproducibility was measured to be 95.2% (95% CI, 77.3 to 99.8%) for 21 samples tested on different days, on different plates, and on different instruments. The last two parameters were for the WN/SLE virus biotin-MIA only by use of a variety of samples that were known to be WN or SLE virus antibody positive or negative. For the WN/SLE virus biotin-MIA, the samples used comprised 99 samples from WN virus infections, 17 samples from SLE virus infections, and 140 negative samples, as determined by PRNT. Two samples from confirmed WN virus infections gave higher V/N values for the SLE virus antigen, leading to false-positive results for SLE virus. Overall, 27% of all positive samples exhibited reactions to the heterologous antigen in this test.
The 239 WN virus antibody-positive and -negative samples were used to identify a potential equivocal zone surrounding the V/N cutoff value of 10 for the WN virus component of the WN/SLE virus biotin-MIA. Results inconsistent with those of PRNT had V/N values of <5 for 3/131 samples, 5 < 10 for 3/7 samples, 10 < 15 for 3/15 samples, 15 < 25 for 4/19 samples, and >25 for 1/67 samples. For these samples, the total proportion of misclassified results was 5.9%, with 71% of these falling between V/N values of 5 and 25.
Quantified PRNT-positive results were available for 72 of the serum samples taken from individuals with WN virus infection. Of these, 8 were of low titer (1:10/1:20), 18 were of moderate titer (1:40/1:80), and 46 were of high titer (1:160 or above). Positive results for WN virus were obtained in the WN/SLE virus biotin-MIA for 75%, 94%, and 96% of the samples with low, moderate, and high titers, respectively.
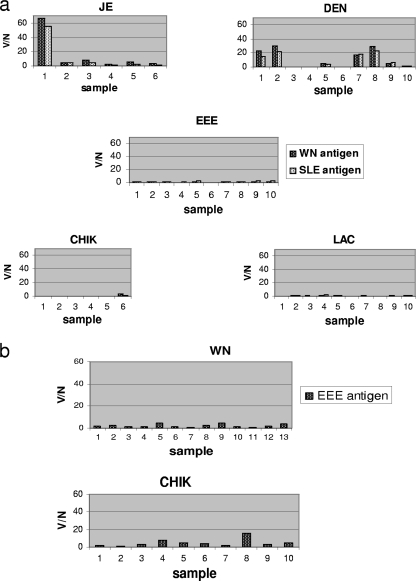
The cross-reactivity results are shown in Fig. 3. Both the WN and SLE viral antigens in the WN/SLE virus biotin-MIA (Fig. 3a) showed significant cross-reactivity against sera from humans with flavivirus infections: for JE virus, 1 of 6 samples, and for DEN virus, 4 of 10 samples. No cross-reactivity was seen with sera with antibodies to the alphaviruses EEE virus (0/10) and CHIK virus (0/6) or against the bunyavirus LAC virus (0/10). Human sera positive for anti-CHIK virus antibodies showed minimal cross-reactivity (1/10), and chicken anti-WN virus antibodies showed no cross-reactivity (0/13) in the EEE virus biotin-MIA (Fig. 3b).
FIG. 3.
Cross-reactivity of the WN/SLE virus biotin-MIA (a) and the EEE virus biotin-MIA (b) with antisera to other confirmed arboviral infections. No dual infections were included in the analyses. The cutoffs for a positive reaction were V/N ratio values of 10.0 for WN virus, 10.23 for SLE virus, and 8.97 for EEE virus.
Validation of the WN virus portion of the WN/SLE virus biotin-MIA was performed with samples that had previously been tested for antibodies to WN virus by PRNT but that were not included in the cutoff determination. Samples were not available to do the same for SLE and EEE virus infections. By using the V/N cutoff value of 10.00 for WN virus, the WN/SLE virus biotin-MIA gave a rate of false-positive results of 0% (95% CI, 95.6 to 100.0%; 84/84 negative samples were correctly identified) and a rate of false-negative results of 10% (95% CI, 78.6 to 95.6%; 45/50 samples positives for WN virus were correctly identified).
A previous study compared specimens from WN virus-infected humans in a WN virus IgM ELISA and an SLE virus IgM ELISA (12). It was consistently found that the optical densities (OD) of the test specimen divided by the OD of the negative control serum sample obtained when it was reacted against the WN viral antigen (positive/negative [P/N] ratio) was at least three times greater than the P/N ratio obtained when the sample was reacted against SLE viral antigen. The reverse was not true of SLE viral infections. A paired t test for the WN/SLE virus biotin-MIA during the development phase showed that the V/N values for the WN viral antigens were consistently twofold or greater than those obtained with the SLE viral antigen when WN virus was the infecting virus (P < 0.001; extrapolated 95% CI, 2.2 to 3.7). In humans, if SLE virus was the infecting virus, a twofold difference in the V/N values was not observed, although the V/N values were consistently higher for SLE virus infections for the limited number of samples tested. For chickens, however, SLE virus infections were distinguishable with a minimum SLE virus-to-WN virus V/N ratio of 6:1 (P < 0.001; extrapolated 95% CI, 8.1 to 11.9).
To confirm that IgM was also detectable by the WN/SLE virus biotin-MIA, 46 human serum samples that were IgM positive, IgG negative, and PRNT positive for WN virus in previous tests were analyzed by the WN/SLE virus biotin-MIA (data not shown). Forty-three were found to be positive (93%), and the remaining three that tested negative were from IgM-equivocal samples.
DISCUSSION
The biotin-MIA is a unique and versatile testing format for a number of reasons. It detects the agent-specific total antibody content in serum samples regardless of the species of origin, requires a minute sample volume, and takes approximately 2 h to perform the test and analyze the results. With the exception of some antigens, most of the reagents are commercially available. While the methods developed here utilized two existing human IgM MIA protocols with minor modifications, clearly there is the potential to multiplex more antigens into a single test. Both the original IgM MIAs combined the serum and anti-human IgM-phycoerythrin conjugate addition steps into a single operation so that these components were coincubated with the antigens. In the biotin-MIAs, coincubation was not possible because of the extremely strong affinity of biotin for streptavidin (21), resulting in a reduction of the signal to a negligible level. As this proof of concept illustrates, the biotin-MIA is potentially adaptable for use with existing MIAs with only minor modifications or for use for the detection of any etiologic agent for which antigens are available for coupling to microspheres, either directly or via a capture antibody. The methods used in the present study incorporated microspheres covalently bound to MAbs as a way to capture antigens. This was to circumvent the need for purified antigens. A simpler approach would be to directly link the antigens to the microspheres when purified products are available.
The results showed that the biotin-MIA is both accurate and sensitive. Unsurprisingly, samples with low PRNT values had the highest rates of false negativity in the WN/SLE virus biotin-MIA, although the sample numbers were limited for this category. A qualitative observation was that only seven samples with true-positive results (6%) gave positive V/N values in conjunction with infecting viral antigen MFIs of <1,000 and six samples with true-negative results (4%) gave false-positive V/N values in conjunction with viral antigen MFIs of <1,000; therefore, any sample with a V/N value greater than the cutoff for that virus but with a viral antigen MFI of <1,000 should be treated as potentially having a false-positive result. Seventy-one percent of the misclassified results had V/N values ranging from 5 to 25. Thus, although a true estimate of an equivocal range can be obtained only by extended experience with a test, this range of V/N values can provide a preliminary working guideline. Overall, samples with equivocal results do not cause a particular problem with this test. For the range suggested, those samples with V/N values falling between 5 and 10 for the WN virus test (1.3% of the samples tested in the present study) may have the greatest risk for misdiagnosis, given that the result for any sample with a positive V/N value would preferably be confirmed. The SLE virus portion of the WN/SLE virus biotin MIA gave rates of false-positive and false-negative results of 0.0%, which could be attributed partly to the relatively small number of samples used and partly to the success of the cutoff placement. A similar situation exists for the EEE virus test, which also gave a rate of false-positive results of 0.0%.
We were initially concerned that irrelevant biotinylated serum proteins would create interference and reduce the sensitivity of the assay. As expected, attempts to adapt this format for ELISA failed due to the overwhelming nonspecific binding of the biotinylated proteins to the microplate surfaces. The key to the success of the biotin-MIA presumably is a combination of factors: (i) the microspheres are relatively inert unless they are specifically coupled, and (ii) because microspheres are assayed inside the BioPlex apparatus, the surface of the assay plate does not interfere with signal detection. In the biotin-MIA, a considerable degree of background interference was noted if a molecular-mass-cutoff plate was not used, most likely due to attachment of the uncoupled biotin (244 Da) to the antigen (data not shown). In addition, while the use of a 10-kDa filter system improved the results, initial experiments showed that use of the 100-kDa-cutoff filter facilitated the achievement of the optimal results, indicating that nonspecific binding of irrelevant biotinylated proteins or other serum components was occurring.
The use of pooled negative samples was a practical way to provide a negative control. While negative results for the individual species being tested would be optimal, this is not always possible in practice. The MFI values for the controls did not figure into the calculation of the results but, rather, provided a means to track whether the tests were running consistently.
The use of microsphere-based assays is expanding domestically and abroad, largely due to the ease of multiplexing and the flexibility of the technology. This assay format is especially suited to the detection of infections in animals that serve as reservoirs for zoonotic pathogens. The ability of the biotin-MIA to simultaneously detect all antibody isotypes is an advantage for surveillance applications because the timing of infection is rarely known. The concentrations of IgG can be assumed to be greater than the concentrations of IgM in many samples, especially those that were not obtained during the acute phase of the disease, and the biotin-MIA gave accurate results with samples collected at random times during infection. Despite the ability of the biotin-MIA to detect IgM, clinical diagnostic applications of this method may not be preferable with species for which individual anti-isotype conjugates are available to create a more informative antibody profile. The biotin-MIA would be particularly useful as a screening tool, with the results for samples with positive results being confirmed by PRNT. The number of samples requiring PRNT would be reduced significantly by using this scheme. Confirmation would be especially pertinent in circumstances in which the biotin-MIA identified disease in a previously unknown species or geographic region.
The biotin-MIA format has the potential to facilitate expanded surveillance and the detection of widespread and important zoonotic pathogens (e.g., additional arboviruses, avian/swine influenza viruses, Francisella tularensis, Borrelia burgdorferi, and Coxiella burnetii). This assay represents the first demonstration of a single platform species-independent rapid test that directly detects the antibodies of interest.
Acknowledgments
We sincerely thank Richard Bowen, Kaci Van Dalen, and Nick Komar for their invaluable help.
Footnotes
Published ahead of print on 18 November 2009.
REFERENCES
- 1.Adrián Diaz, L., N. Komar, A. Visintin, M. J. Dantur Juri, M. Stein, R. Lobo Allende, L. Spinsanti, B. Konigheim, J. Aguilar, M. Laurito, W. Almirón, and M. Contigiani. 2008. West Nile virus in birds, Argentina. Emerg. Infect. Dis. 14:689-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaty, B., C. H. Calisher, and R. E. Shope. 1995. Arboviruses, p. 189-212. In D. Lennette, E. H. Lennette, and E. T. Lennette (ed.), Diagnostic procedures for viral, rickettsial and chlamydial infections, 7th ed. American Public Health Association, Washington, DC.
- 3.Blitvich, B. J., R. A. Bowen, N. L. Marlenee, R. A. Hall, M. L. Bunning, and B. J. Beaty. 2003. Epitope-blocking enzyme-linked immunosorbent assays for detection of West Nile virus antibodies in domestic mammals. J. Clin. Microbiol. 41:2676-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bossart, K. N., J. A. McEachern, A. C. Hickey, V. V. Choudhry, D. S. Dimitrov, B. T. Eaton, and L.-F. Wang. 2007. Neutralization assays for differential henipavirus serology using Bio-Plex protein array systems. J. Virol. Methods 142:29-40. [DOI] [PubMed] [Google Scholar]
- 5.Clarke, D., and J. Casals. 1958. Techniques for hemagglutination and hemagglutination-inhibition with arthropod-borne viruses. Am. J. Trop. Med. Hyg. 7:561-573. [DOI] [PubMed] [Google Scholar]
- 6.Crill, W. D., and G. J. J. Chang. 2004. Localization and characterization of flavivirus envelope glycoprotein cross-reactive epitopes. J. Virol. 78:13975-13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, B. S., G. J. Chang, B. Cropp, J. T. Roehrig, D. A. Martin, C. J. Mitchell, R. Bowen, and M. L. Bunning. 2001. West Nile virus recombinant DNA vaccine protects mouse and horse from virus challenge and expresses in vitro a noninfectious recombinant antigen that can be used in enzyme-linked immunosorbent assays. J. Virol. 75:4040-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davison, A. C., and D. V. Hinkley. 1997. Bootstrap methods and their application. Cambridge University Press, Cambridge, United Kingdom.
- 9.Hunt, A. R., and J. T. Roehrig. 1985. Biochemical and biological characteristics of epitopes on the E1 glycoprotein of western equine encephalitis virus. Virology 142:334-346. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, A. J., D. A. Martin, N. Karabatsos, and J. T. Roehrig. 2000. Detection of anti-arboviral immunoglobulin G by using a monoclonal antibody-based capture enzyme-linked immunosorbent assay. J. Clin. Microbiol. 38:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson, A. J., A. J. Noga, O. Kosoy, R. S. Lanciotti, A. A. Johnson, and B. J. Biggerstaff. 2005. Duplex microsphere-based immunoassay for detection of anti-West Nile virus and anti-St. Louis encephalitis virus immunoglobulin m antibodies. Clin. Diagn. Lab. Immunol. 12:566-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin, D. A., A. Noga, O. Kosoy, A. J. Johnson, L. R. Petersen, and R. S. Lanciotti. 2004. Evaluation of a diagnostic algorithm using immunoglobulin M enzyme-linked immunosorbent assay to differentiate human West Nile Virus and St. Louis encephalitis virus infections during the 2002 West Nile virus epidemic in the United States. Clin. Diagn. Lab. Immunol. 11:1130-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newcombe, R. 2006. Confidence intervals for an effect size measure based on the Mann-Whitney statistic. Part 2. Asymptotic methods and evaluation. Stat. Med. 25:559-573. [DOI] [PubMed] [Google Scholar]
- 14.Noga, A. J., O. L. Kosoy, J. Laven, R. S. Lanciotti, D. A. Martin, and A. J. Johnson. 2005. Detection of eastern equine encephalitis virus IgM antibody by microsphere immunoassay, abstr. TA47. Abstr. 21st Annu. Clin. Virol. Symp.
- 15.Pepe, M. S. 2003. The statistical evaluation of medical tests for classification and prediction. Oxford University Press, New York, NY.
- 16.Roehrig, J., J. Mathews, and D. Trent. 1983. Identification of epitopes on the E-glycoprotein of Saint Louis encephalitis virus using monoclonal antibodies. Virology 128:118-126. [DOI] [PubMed] [Google Scholar]
- 17.Roehrig, J. T., A. R. Hunt, G. J. Chang, B. Sheik, R. A. Bolin, T. F. Tsai, and D. W. Trent. 1990. Identification of monoclonal antibodies capable of differentiating antigenic varieties of eastern equine encephalitis viruses. Am. J. Trop. Med. Hyg. 42:394-398. [DOI] [PubMed] [Google Scholar]
- 18.Schuberth, H. J., A. Kroell, and W. Leibold. 1996. Biotinylation of cell surface MHC molecules: a complementary tool for the study of MHC class II polymorphism in cattle. J. Immunol. Methods 189:89-98. [DOI] [PubMed] [Google Scholar]
- 19.Staros, J. V., R. W. Wright, and D. M. Swingle. 1986. Enhancement by N-hydroxysulfosuccinimide of water-soluble carbodiimide-mediated coupling reactions. Anal. Biochem. 156:220-222. [DOI] [PubMed] [Google Scholar]
- 20.Trainor, N. B., W. D. Crill, J. A. Roberson, and G.-J. J. Chang. 2007. Mutation analysis of the fusion domain region of St. Louis encephalitis virus envelope protein. Virology 360:398-406. [DOI] [PubMed] [Google Scholar]
- 21.Wilchek, M., and E. A. Bayer. 1988. The avidin-biotin complex in bioanalytical applications. Anal. Biochem. 171:1-32. [DOI] [PubMed] [Google Scholar]