Abstract
Noroviruses (NoVs) resembling human NoV genotype GIV (Alphatron-like) have recently been detected in carnivores. By using an enzyme-linked immunosorbent assay based on baculovirus-expressed capsid protein VP1 of lion strain GGIV.2/Pistoia/387/06/ITA, NoV-specific antibodies were detected in cats (16.11%) and dogs (4.8%), demonstrating that these animals are exposed to infections caused by NoVs.
Noroviruses (NoVs) have been identified as the most common cause of viral gastroenteritis in humans. NoV infections affect persons of all age groups (13) and are predominantly transmitted through the fecal-oral route, either indirectly through contaminated food, water, or surfaces or directly from person to person (4). NoVs are small, rounded, nonenveloped, single-stranded RNA viruses belonging to the family Caliciviridae. On the basis of the VP1 capsid gene sequence, NoVs have been classified into five genogroups (genogroups GI to GV) and at least 32 genotypes (11, 16, 18); but only GI, GII, and GIV are known to infect humans, with GII viruses accounting for the majority of NoV infections (15).
NoVs genetically and antigenically closely related to human NoVs have been discovered in cows and pigs (14, 16). Recently, NoVs have been detected in the stools of a captive lion cub (strain Pistoia/387/06/ITA) (11) and a dog (strain Bari/170/07/ITA) (10) with enteric signs. Upon sequence analysis, the lion and canine NoVs were found to share 90.1% amino acid (aa) identity in the capsid protein and were closely related to human NoV GIV (Alphatron-like) (69.3% to 70.1% aa identity). According to widely accepted classification criteria (18), the lion and canine NoVs were classified as having a novel genotype, genotype GIV.2, within genogroup GIV (10, 11). However, whether these novel caliciviruses are common in domestic carnivores or whether these were detected serendipitously remains unclear. In the study described here, the prevalence of antibodies specific for NoV GIV in carnivores was assessed by using virus-like particles (VLPs).
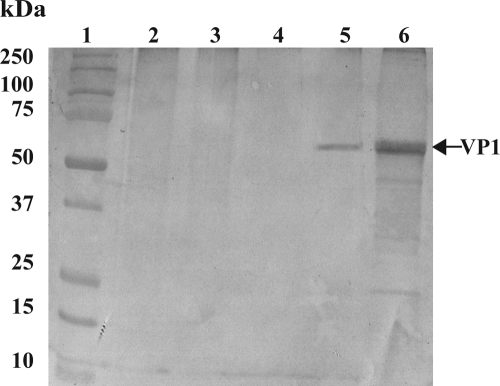
VLPs were developed with the RNA of the prototype lion NoV GIV strain Pistoia/387/06/ITA (GenBank accession number EF450827) (11). Briefly, the full-length VP1 (open reading frame 2) was cloned into the vector pRN16 (kindly provided by Polly Roy), and transfection was performed with triple-cut linearized Autograph californica multiple nucleopolyhedrosis virus DNA. The recombinant baculovirus was selected and plaque purified on Spodoptera frugiperda (Sf21) cells, as described previously (2). For the large-scale production of the VLPs, a 100-ml Sf9 cell suspension culture was infected with the recombinant baculovirus at a multiplicity of infection of 3 PFU/cell. The recombinant protein was isolated from the culture medium at 48 h postinfection. After purification by rate-zonal centrifugation on a discontinuous sucrose gradient (60, 40, 30, 20%), the recombinant VP1 and the assembled VLPs were analyzed by electrophoresis on a 12% SDS-polyacrylamide gel and by electron microscopy. Hyperimmune serum against the purified lion VLPs was raised in two rabbits. The specificity of the serum was tested by Western blotting (WB), with the lion GIV VLPs and dog GIV strain Bari/170/07/ITA being used as positive controls and wild-type baculovirus and vaccine FCV strain F9 being used as negative controls (Fig. 1).
FIG. 1.
Western blotting analysis of lion GIV VLPs using rabbit hyperimmune serum. Lane 1, Precision Plus protein standards (Bio-Rad, Italy); lane 2, mock-infected Sf9 cells; lane 3, wild-type baculovirus Sf9 insect cells; lane 4, FCV strain F9 purified from the supernatant of CrFK cells; lane 5, dog GIV strain Bari/170/07/IT purified from a fecal sample suspension; lane 6, lion GIV VLPs purified from the supernatants of Sf9 insect cells.
For the development of the enzyme-linked immunosorbent assay (ELISA), purified VLPs were coated onto 96-well enzyme immunoassay plates (Costar, Italy) at 100 μl per well (final concentration, 8 μg/ml) in carbonate-bicarbonate buffer (0.05 M, pH 9.6), and the plates were incubated at 4°C overnight. After the plates were blocked with 1% bovine serum albumin in phosphate-buffered saline (PBS) buffer at room temperature (RT) for 2 h, the VLP-coated microplates were incubated with 100 μl of dog and cat serum samples diluted to 1:50 in PBS at 37°C for 1 h. The plates were washed three times in PBS with 0.1% Tween 20 (PBST) and were then incubated with goat anti-cat IgG (1:1,000) and anti-dog IgG (1:2,000) conjugated with horseradish peroxidase (Sigma-Aldrich, Italy) for 1 h at 37°C. The plates were washed three times in PBST prior to the addition of 2,2′-azino-di-(3-ethylbenzthiazoline-6-sulfonate) (ABTS) substrate. Each reaction was completed by incubation at room temperature for 20 min, and the absorbance was measured at 405 nm. Wild-type baculovirus Sf9 insect cells were used to obtain a positive/negative ratio (optical density of the GIV VLPs/optical density of the wild-type baculovirus Sf9 insect cells) to evaluate the background binding. In order to establish the cutoff value, 25 cat serum samples negative for the lion GIV VLPs by WB assay and a rabbit negative control serum sample were tested. A mean with a standard deviation (SD) was calculated. The cutoff value was established as the mean value plus 3 SDs. A total of 211 serum samples collected from adult cats (ages, >1 year) from several geographical settings in Italy were tested. Ninety-six serum samples were collected from private veterinary clinics in Teramo, Italy; 44 were from rescue colonies in Reggio Emilia, Italy; 34 were from the clinic of the Faculty of Veterinary Medicine of Bari (Bari, Italy); and 37 were from stray cats living in the Rome, Italy, Biopark. In addition, 103 serum samples from adult dogs (ages, >1 year) collected in Teramo from 2006 to 2008 were tested.
The overall prevalence of lion NoV GIV-specific antibodies in cats was 16.1% (34/211), with a higher seroprevalence rate (32.0%) being detected in stray cats living in the Rome Biopark than in the other cats (14.6% to 6.8%). The difference in the estimated prevalence between the two groups was statistically significant (χ2 = 8.8393, P > 0.01). Five of 103 (4.8%) serum samples from dogs were also positive for antibodies against the lion NoV GIV.2 (Table 1).
TABLE 1.
Results of serological investigation by the ELISA with feline and canine sera
| Origin | Feline sera |
Canine sera |
||
|---|---|---|---|---|
| No. of serum samples | No. (%) positive | No. of serum samples | No. (%) positive | |
| Teramo | 96 | 14 (14.6) | 103 | 5 (4.8) |
| Reggio Emilia | 44 | 3 (6.8) | ||
| Rome Biopark | 37 | 12 (32.0) | ||
| Bari | 34 | 5 (14.1) | ||
| Total | 211 | 34 (16.1) | 103 | 5 (4.8) |
With the exception of murine norovirus (GV) (17), no reproducible cell culture system has been described for NoV. Therefore, epidemiological studies of NoVs necessarily rely on the expression of synthetic antigens, and the baculovirus system appears to be particularly adequate, since the baculovirus-expressed full-length VP1 of NoV tends to assemble into virus-like particles whose structures and features resemble the native structure and the antigenic features of wild-type NoVs (7). ELISAs based on VLPs have successfully been used to gather information on the epidemiology of NoVs in humans and animals (3, 6). In this study, using baculovirus-generated VLPs based on lion NoV GIV.2, NoV-specific antibodies were detected in feline and canine sera by ELISA. These findings clearly indicate that domestic carnivores are susceptible to infection with NoV GIV. Human NoV GIV (Alphatron-like) has been identified only sporadically in human patients, although epidemiological studies based on the analysis of sewage and wastewater in Japan and Italy have unexpectedly revealed high prevalence rates, thus suggesting that these enteric pathogens are more common than was previously believed in human populations and the environment (8, 9).
Also, cross-reactions with antibodies raised to NoV non-GIV.2 strains cannot be ruled out, as limited antigenic cross-reactivity between NoVs of different genogroups may occur, as evidenced by the use of VLPs and antisera generated from panels of NoV genotypes of genogroups I and II (5, 16).
A higher prevalence of NoV-specific antibodies was detected in stray cats, while the prevalence rates were lower in family cats and dogs. The high prevalence rate found in stray cats living within the Rome Biopark can reasonably be explained to be the result of social interactions among free-ranging cats or as exposure to other sources of infection represented by other animal species. In contrast, we observed a low prevalence in dogs. Since the rabbit hyperimmune serum against the lion NoV VLPs reacted strongly with dog NoV strain Bari/170/07/ITA, a lack of cross-reactivity between the two strains was ruled out. Moreover, several studies have demonstrated that antibody ELISAs are generally more broadly reactive than antigen ELISAs (6). Finally, it is possible that strains genetically/antigenically diverse circulate in carnivores and that their distribution varies markedly, thus requiring the construction of panels of VLPs.
To our knowledge, this is the first study showing that NoV may circulate among domestic carnivores. Dogs are regarded as vectors of viral, bacterial, and parasitic zoonoses in people of all ages (13); but the risks linked to the transmission of enteric viruses has not been studied. However, evidence of the exposure of young children to infection by rotavirus strains of canine and feline origin has been collected (1). Also, in a seroepidemiological study conducted in rural Mexico, the presence of dogs in or near a home was recognized as a risk factor for the acquisition of NoV-specific IgA antibodies in infants (12). The availability of ELISAs specific for NoV GIV.2 could be useful to assess better whether these novel viruses may pose a zoonotic risk.
Acknowledgments
We thank Camillo Di Rocco and Paradies Paola for assistance in the laboratory and the collection of serum samples.
This work was financed by grants from the University of Teramo, Teramo, Italy, and from the Italian Ministry of University and Research.
Footnotes
Published ahead of print on 18 November 2009.
REFERENCES
- 1.De Grazia, S., V. Martella, G. M. Giammanco, M. Iturriza-Gomara, S. Ramirez, A Cascio, C. Colomba, and A. Serenella. 2007. Canine-origin G3P[3] rotavirus strain in child with acute gastroenteritis. Emerg. Infect. Dis. 13:1091-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Martino, B., F. Marsilio, and P. Roy. 2007. Assembly of feline calicivirus-like particle and its immunogenicity. Vet. Microbiol. 120:173-178. [DOI] [PubMed] [Google Scholar]
- 3.Farkas, T., S. Nakajima, M. Sugieda, X. Deng, W. M. Zhong, and X. Jiang. 2005. Seroprevalence of noroviruses in swine. J. Clin. Microbiol. 43:657-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glass, R. I., J. Noel, T. Ando, R. Fankhouser, G. Belliot, A. Mounts, U. D. Parashar, J. S. Bresee, and S. S. Monroe. 2000. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J. Infect. Dis. 181:S254-S261. [DOI] [PubMed] [Google Scholar]
- 5.Hansman, G. S., K. Natori, H. Shirato-Horikoshi, S. Ogawa, T. Oka, K. Katayama, T. Tanaka, T. Miyoshi, K. Sakae, S. Kobayashi, M. Shinohara, K. Uchida, N. Sakurai, K. Shinozaki, M. Okada, Y. Seto, K. Kamata, N. Nagata, K. Tanaka, T. Miyamura, and N. Takeda. 2006. Genetic and antigenic diversity among noroviruses. J. Gen. Virol. 87:909-919. [DOI] [PubMed] [Google Scholar]
- 6.Jiang, X., N. Wilton, W. M. Zhong, T. Farkas, P. W. Huang, E. Barrett, M. Guerrero, G. Ruiz-Palacios, K. Y. Green, J. Green, A. D. Hale, M. K. Estes, L. K. Pickering, and D. O. Matson. 2000. Diagnosis of human calicivirus by use of enzyme immunoassays. J. Infect. Dis. 181:S349-S359. [DOI] [PubMed] [Google Scholar]
- 7.Jiang, X., W. M. Zhong, T. Farkas, P. W. Huang, N. Wilton, E. Barret, D. Fulton, R. Morrow, and D. O. Matson. 2002. Baculovirus expression and antigenic characterization of the capsid proteins of three Norwalk-like viruses. Arch. Virol. 147:119-130. [DOI] [PubMed] [Google Scholar]
- 8.Kitajima, M., E. Haramoto, C. Phanuwan, H. Katayama, and S. Ohgaki. 2009. Detection of genogroup IV norovirus in wastewater and river water in Japan. Lett. Appl. Microbiol. 49:655-658. [DOI] [PubMed] [Google Scholar]
- 9.La Rosa, G., M. Pourshaban, M. Iaconelli, and M. Muscillo. 2008. Detection of genogroup IV noroviruses in environmental and clinical samples and partial sequencing through rapid amplification of cDNA ends. Arch. Virol. 153:2077-2083. [DOI] [PubMed] [Google Scholar]
- 10.Martella, V., E. Lorusso, N. Decaro, G. Elia, A. Radogna, M. D'Abramo, C. Desario, A. Cavalli, M. Corrente, M. Camero, C. A. Germinario, K. Bànyai, B. Di Martino, F. Marsilio, L. E. Carmichael, and C. Buonavoglia. 2008. Detection and molecular characterization of a canine norovirus. Emerg. Infect. Dis. 14:1306-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martella, V., M. Campolo, E. Lorusso, P. Cavicchio, A. L. Bellacicco, N. Decaro, G. Elia, G. Greco, M. Corrente, C. Desario, S. Arista, K. Banyai, M. Koopmans, and C. Buonavoglia. 2007. Norovirus in captive lion cub (Panthera leo). Emerg. Infect. Dis. 13:1071-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peasey, A. E., M. Guillermo, G. M. Ruiz-Palacios, M. Qugley, W. Newsholme, J. Martinez, G. Rosales, X. Jiang, and U. J. Blumenthal. 2004. Seroepidemiology and risk factors for sporadic norovirus/Mexico strain. J. Infect. Dis. 189:2027-2036. [DOI] [PubMed] [Google Scholar]
- 13.Rockx, B., M. de Wit, H. Vennema, J. Vinje, E. de Bruin, Y. van Duynhoven, and M. Koopmans. 2002. Natural history of human calicivirus infection: a prospective cohort study. Clin. Infect. Dis. 35:246-253. [DOI] [PubMed] [Google Scholar]
- 14.van der Poel, W. H., R. van der Heide, F. Veschoor, H. Gelderblom, J. Vinje, and M. P. Koopmans. 2003. Epidemiology of Norwalk-like virus infections in cattle in The Netherlands. Vet. Microbiol. 92:297-309. [DOI] [PubMed] [Google Scholar]
- 15.Vinje, J., and M. P. Koopmans. 2000. Simultaneous detection and genotyping of “Norwalk-like viruses” by oligonucleotide array in a reverse line blot hybridization format. J. Clin. Microbiol. 38:2595-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, Q. H., M. Han, S. Cheethaam, M. Sousa, J. A. Funk, and L. J. Saif. 2005. Porcine noroviruses related to human noroviruses. Emerg. Infect. Dis. 11:1874-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wobus, C. E., S. M. Karst, L. B. Thackray, K. O. Chang, S. V. Sosnovtsev, G. Belliot, A. Krug, J. M. Mackenzie, K. Y. Green, and H. W. Virgin. 2004. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2:432-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng, D. P., T. Ando, R. L. Frankhouser, R. S. Beard, R. I. Glass, and S. S. Monroe. 2006. Norovirus classification and proposed strain nomenclature. Virology 346:312-323. [DOI] [PubMed] [Google Scholar]