Abstract
Recently, major species-specific antibody epitopes in three immunoreactive tandem repeat proteins (TRPs) of Ehrlichia chaffeensis, TRP32, TRP47, and TRP120, have been identified and molecularly characterized within tandem repeat (TR) regions. In this study, we mapped the major immunodeterminants of the E. chaffeensis 200-kDa ankyrin protein (Ank200) and the minor immunodeterminants in the N- and C-terminal regions of E. chaffeensis TRP47. Major antibody epitopes of Ank200 were localized to four polypeptide regions (18-mer, 20-mer, 20-mer, and 21-mer, respectively) in terminal acidic domains, which reacted with antibodies in sera from human monocytotropic ehrlichiosis (HME) patients and an E. chaffeensis-infected dog. Two minor epitope-containing regions were identified in the N terminus and the C terminus of TRP47. The sensitivities and specificities of synthetic peptides representing these and other well-defined major immunodeterminants of E. chaffeensis were determined by enzyme-linked immunosorbent assay (ELISA). Thirty-one HME patient serum samples that had detectable E. chaffeensis antibodies (titers from 64 to 8,192) by indirect fluorescent-antibody assay (IFA) were tested. All 31 serum samples reacted with at least one E. chaffeensis peptide, 30 (96.8%) with TRP120 peptides, 27 (87.1%) with TRP32 peptides, 24 (77.4%) with TRP47 peptides, 19 (61.3%) with Ank200 peptides, and 28 (90.3%) with recombinant TRP120-TR protein. A mixture of the two most sensitive peptides from TRP120 and TRP32 did not provide enhanced analytical sensitivity compared to that provided by TRP120 alone. Our results demonstrate that the TRP120 peptide can be utilized for development of standardized sensitive point-of-care and reference laboratory immunodiagnostics for HME. This is the first study to compare analysis of molecularly defined major antibody epitopes with IFA for diagnosis of HME.
Ehrlichia chaffeensis is a tick-transmitted obligately intracellular bacterium which causes the emerging zoonosis human monocytotropic ehrlichiosis (HME) (18). Clinical diagnosis of HME is usually confirmed retrospectively by detection of Ehrlichia-specific antibodies in patient sera using an indirect fluorescent-antibody assay (IFA) (7). However, the limitations of IFA include lack of standardization between laboratories, false-positive interpretations due to autoantibodies or antibodies directed at conserved bacterial proteins, and cross-reactive antibodies produced by related organisms (for example, Ehrlichia canis, Ehrlichia ewingii, and Anaplasma phagocytophilum) that can make identification of the specific etiologic agent difficult (2, 3, 5, 18, 24). Furthermore, IFA requires expensive microscopy equipment and highly skilled technicians to produce the antigen and interpret results. Molecular diagnostic methods such as PCR are useful for specific and sensitive detection of E. chaffeensis prior to development of reactive antibodies (4), but PCR is not useful after antibiotic therapy is initiated, and the clinical sensitivity of PCR in the primary care setting has not been unequivocally determined. Therefore, PCR is currently considered a valuable adjunct to IFA for diagnosis (26).
Advances in the immunomolecular characterization of E. chaffeensis have provided new opportunities to dramatically improve the sensitivity, specificity, and standardization of immunodiagnostics for the ehrlichioses. Major immunoreactive proteins of E. chaffeensis that have been molecularly characterized include P28 (OMP1), TRP32 (32-kDa tandem repeat-containing protein; formerly named VLPT [variable-length PCR target]), TRP47, TRP120, and Ank200 (200-kDa ankyrin protein), and all are strongly recognized by sera from HME patients and E. chaffeensis-infected dogs (6, 9-11, 17). Ehrlichia tandem repeat proteins (TRPs) are secreted, and two of these (TRP47 and TRP120) are differentially expressed by dense-cored ehrlichiae (6, 20). TRP47 is an effector protein that interacts with multiple host proteins associated with cell signaling, transcriptional regulation, and vesicle trafficking (25). Ank200 is the largest immunoreactive protein identified in E. chaffeensis and is translocated to the nuclei of infected monocytes, where it interacts with the mid-A-stretch of host promoter and intronic Alu elements (33).
Species-specific continuous epitopes have been identified in the tandem repeats (TRs) of E. chaffeensis TRP32, TRP47, and TRP120 (6, 9, 10). The E. chaffeensis TRP32 has two to six nonidentical 30-amino-acid TRs, and two major species-specific antibody epitopes (continuous and discontinuous) have been identified in the tandem repeats (10). Single major molecularly distinct continuous antibody epitopes (18 to 22 amino acids) have also been identified in TRP47 and TRP120 and corresponding orthologs of E. canis (6, 9). Although the molecular immunodeterminants of E. chaffeensis Ank200 have not been defined, the corresponding E. canis ortholog (Ank200) has multiple major species-specific immunodeterminants located in acidic N- and C-terminal domains (16).
In this study, we mapped and molecularly defined four major antibody epitopes in E. chaffeensis Ank200 and two minor antibody epitopes in the TRP47 N- and C-terminal regions and evaluated synthetic peptides representing the antibody epitopes from four E. chaffeensis immunodominant proteins, TRP32, TRP47, TRP120, and Ank200, for serologic diagnosis of HME by enzyme-linked immunosorbent assay (ELISA).
MATERIALS AND METHODS
Culture and purification of E. chaffeensis.
E. chaffeensis (Arkansas strain) was propagated in DH82 cells and purified by size exclusion chromatography as previously described (12, 21). The fractions containing bacteria were frozen and utilized for DNA and antigen preparation (14).
PCR amplification of the E. chaffeensis genes.
Oligonucleotide primers for the amplification of the E. chaffeensis Ank200 and TRP47 gene fragments were designed manually or by using PrimerSelect (Lasergene v5.08; DNAStar, Madison, WI) according to the sequences in GenBank (accession numbers YP_507490 and DQ085430, respectively) and synthesized (Sigma-Genosys, Woodlands, TX) (Table 1). Gene fragments corresponding to the different regions used for epitope mapping were amplified by PCR (Fig. 1 for Ank200; see Fig. 4A for TRP47).
TABLE 1.
Oligonucleotide primers for amplification of E. chaffeensis Ank200 and TRP47 gene fragments
| Fragment | Sequence (5′ to 3′) of indicated primer |
Size (bp) | |
|---|---|---|---|
| Forward primera | Reverse primer | ||
| Ank200 | |||
| N | CAACAAAATCCTAATTCGCAAG | CGATTTTATATCATTACCAGCA | 1,644 |
| N1 | CACCATGGCAGATCCAAAACAAG | TACCGCATACAATGGATCTTC | 384 |
| N2 | CACCCCTTTACCTAAAGGTCAAAG | ATCCCTAACACCTTCCC | 456 |
| N3 | CACCGCAGTTATTCATGATGAAGAG | CAATGGGGATTGATTTC | 468 |
| N4 | CACCCATGTTATGGTTCAGAACC | ATCATTACCAGCAACAGC | 354 |
| N5 | CACCATGGCAGATCCAAAACAAG | TTGCTGAGAAGGCAAATC | 195 |
| N6 | CACCGAAACAGGAGAAACTGTAGAA | TACCGCATACAATGGATCTTC | 189 |
| N7 | CACCGCAGTTATTCATGATGAAGAG | AGCTAAATGCAGTAATGTCATTAC | 246 |
| N8 | CACCGTAATGACATTACTGCATTTAGCT | CAATGGGGATTGATTTC | 246 |
| N9 | CACCGCAGTTATTCATGATGAAGAG | AATTTCTTCTAGATCTGGCTC | 123 |
| N10 | CACCGAGCCAGATCTAGAAGAAATT | AGCTAAATGCAGTAATGTCATTAC | 144 |
| I | TGTTCAGTTAAAGGACGTGTTC | AGCTAAATGCAGCGGTGTATC | 1,371 |
| C | TTTGCTGAAAAGGGTGTAAAAA | ATCTTCAGATGTAATAGGAGGTAGTCCC | 1,368 |
| C1 | TTTGCTGAAAAGGGTGTAAAAA | TCCATGTAGACCATGAACTGC | 822 |
| C2 | GCAGTTCATGGTCTACATGGA | TTTGCTCTGGCAAGAACTT | 639 |
| C3 | GCAGTTCATGGTCTACATGGA | CGCTGATGCACCTAGAGA | 318 |
| C4 | TCTCTAGGTGCATCAGCG | TTTGCTCTGGCAAGAACTT | 339 |
| C5 | TCTCTAGGTGCATCAGCG | ACCCTTATCAAATATTCCACT | 171 |
| C6 | AGTGGAATATTTGATAAGGGT | TTTGCTCTGGCAAGAACTT | 189 |
| TRP47 | |||
| N1 | ATGCTTCATTTAACAACAGAA | ATGATAACCACGATCAGGTTC | 135 |
| N2 | GAACCTGATCGTGGTTATCAT | AGGATCAACTAAGAAAGAAGC | 135 |
| N3 | GCTTCTTTCTTAGTTGATCCT | ATGATCATGTTCATTGTGATG | 132 |
| N4 | CATCACAATGAACATGATCATG | ATTTCCTTCAAGAACTGGAAC | 132 |
Linker sequences for cloning are underlined.
FIG. 1.
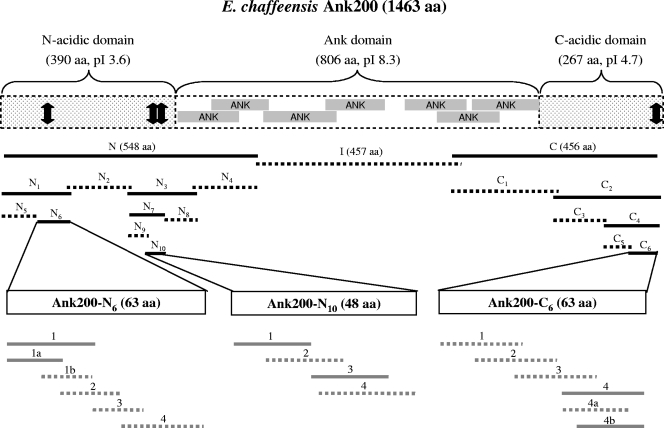
Schematic of E. chaffeensis Ank200 protein, showing domains, predicted isoelectric points (pIs), and the recombinant proteins and synthetic peptides used for epitope mapping. Predicted ankyrin domains are shown in shaded boxes. The recombinant proteins and synthetic peptides are shown in black lines and gray lines, respectively, and solid lines show regions containing an epitope(s), whereas dashed lines show regions which did not react or reacted weakly with anti-E. chaffeensis human and dog sera. The approximate locations of mapped epitopes are designated by arrows.
FIG. 4.
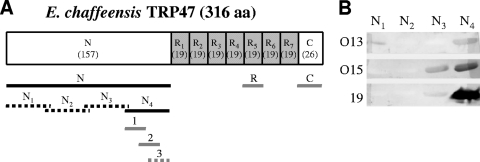
Schematic of E. chaffeensis TRP47 and immunoreactivities of recombinant TRP47-N proteins by Western immunoblotting. (A) Schematic of TRP47, showing domains, locations of TRs (number of amino acids in parentheses), and the recombinant proteins and synthetic peptides used for epitope mapping. The recombinant proteins and synthetic peptides are shown with black lines and gray lines, respectively, and the solid lines show regions containing an epitope(s). (B) Western immunoblot assay of four recombinant proteins representing TRP47-N probed with three HME patient serum samples (no. O13, O15, and 19).
PCR was performed with PCR HotMaster mix (Eppendorf, Westbury, NY) and E. chaffeensis genomic DNA as the template. The thermal cycling profile was as follows: 95°C for 3 min, 30 cycles of 94°C for 30 s, annealing temperature (1°C less than the lowest primer melting temperature [Tm]) for 30 s, and 72°C for the appropriate extension time (1 min/1,000 bp), followed by a 72°C extension for 10 min and a 4°C hold.
Expression and purification of the recombinant proteins.
The expression of the three largest E. chaffeensis Ank200 fragments (N, I, and C) was performed using the pUni/pRSET-E Echo vector system (Invitrogen, Carlsbad, CA). Expression of the recombinant proteins in Escherichia coli BL21(DE3)pLysS (Invitrogen) was induced by adding 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to cultures in log growth phase incubated for 4 h at 37°C. All other Ank200 fragments were expressed by pBAD/Thio-TOPO or pBAD102/D-TOPO vector (Invitrogen). Expression of the recombinant proteins in E. coli TOP10 (Invitrogen) was induced by adding 0.02% arabinose to 4 h cultures. All recombinant proteins were purified under native conditions using His-Select columns (Sigma, St. Louis, MO). The expression of the N-terminal region of E. chaffeensis TRP47 (TRP47-N) and the tandem repeat region of E. chaffeensis TRP120 (TRP120-TR; containing first two tandem repeats of TRP120 only) has been previously described (6, 28).
Synthetic peptides.
For E. chaffeensis Ank200, six, four, and six overlapping peptides corresponding to three regions (N6, N10, and C6) (see gray lines for locations in Fig. 1; see Fig. 3A to C, left, for sequences), respectively, were commercially synthesized (Bio-Synthesis, Lewisville, TX). For TRP47, the C-terminal peptide and three overlapping peptides corresponding to the N4 region (see Fig. 4A and 5A) were synthesized (Bio-Synthesis). All other synthetic peptides (TRP120-R-I1 [SKVEQEETNPEVLIKDLQDVAS], TRP47-R [ASVSEGDAVVNAVSQETPA], TRP32-R3 [SDLHGSFSVELFDPFKEAVQLGNDLQQSSD], TRP32-R4 [SDSHEPSHLELPSLSEEVIQLESDLQQSSN], and E. canis TRP36-2R [TEDSVSAPATEDSVSAPA], which contained two tandem repeat units of TRP36) used in this study have been described previously (6, 9, 10). All peptides were supplied as a lyophilized powder and resuspended in molecular biology grade water (1 mg/ml).
FIG. 3.
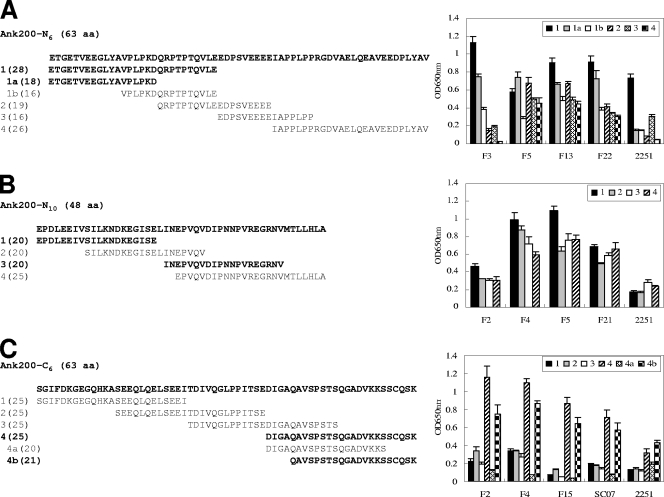
Immunoreactivities of overlapping synthetic peptides spanning the E. chaffeensis Ank200-N6, -N10, and -C6 fragments by ELISA. (A) Ank200-N6 peptides (left) reacted with four HME patient serum samples (no. F3, F5, F13, and F22) and an anti-E. chaffeensis dog serum sample derived from an experimentally infected dog (no. 2251). The OD readings of peptide N6-1 were significantly (P < 0.05) higher than those of N6-2, -3, and -4 for the dog serum sample and for most patient sera, and the OD readings of peptide N6-1a were significantly (P < 0.05) higher than those of N6-1b for all patient sera. (B) Ank200-N10 peptides (left) reacted with four HME patient serum samples (no. F2, F4, F5, and F21) and the dog serum sample. (C) Ank200-C6 peptides (left) reacted with four HME patient serum samples (no. F2, F4, F15, and SC07) and the dog serum sample. The OD readings of peptide C6-4 were significantly (P < 0.05) higher than those of C6-1, -2, and -3 for all sera, and OD readings of peptide C6-4b were significantly (P < 0.05) higher than those of C6-4a for all sera. The OD readings represent the mean values for three wells (± standard deviations), with the OD values of the buffer-only wells subtracted. Normal dog or human sera did not recognize these peptides (data not shown).
FIG. 5.
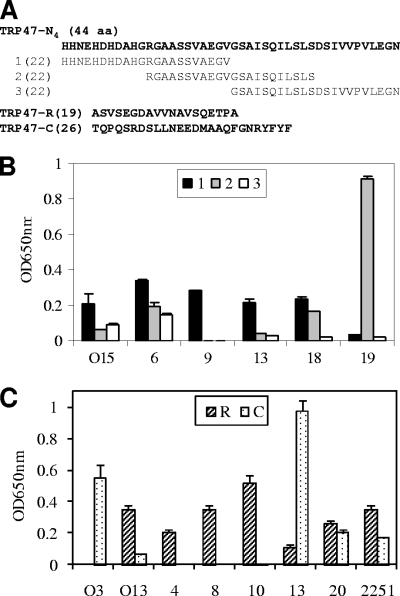
Immunoreactivities of overlapping synthetic peptides spanning E. chaffeensis TRP47-N4 and the synthetic TRP47-R and TRP47-C peptides, as determined by ELISA. (A) Sequences of three overlapping peptides spanning the TRP47-N4 fragment and the TRP47-R and TRP47-C peptides. (B) TRP47-N4 peptides reacted with five HME patient serum samples (no. O15, 6, 9, 13, 18, and 19) by ELISA. (C) The TRP47-R and TRP47-C peptides reacted with seven HME patient serum samples (no. O3, O13, 4, 8, 10, 13, and 20) and an anti-E. chaffeensis dog serum sample (no. 2251) by ELISA. The OD readings represent the mean values for three wells (± standard deviations), with the OD values of the buffer-only wells subtracted. The OD readings of peptide TRP47-R were significantly (P < 0.05) higher than those of TRP47-C for all patient sera except for sample no. O3 and 13, for which the OD readings of peptide TRP47-C were significantly (P < 0.05) higher than those of TRP47-R. The normal human or dog sera did not recognize TRP47 polypeptides (data not shown).
Antisera.
A convalescent-phase anti-E. chaffeensis dog serum sample was obtained from an experimentally infected dog (no. 2251). HME patient serum samples were kind gifts from Focus Technologies (Cypress, CA) and the Centers for Disease Control and Prevention (Atlanta, GA). Patient serum samples positive for Rickettsia spp. but negative for E. chaffeensis by IFA were kind gifts from Arkansas Public Health Laboratory (Little Rock, AR). Rabbit anti-Ank200-N6-1 antiserum was generated against the synthetic keyhole limpet hemocyanin-conjugated peptide Ank200-N6-1 by a commercial vendor (Bio-Synthesis).
Gel electrophoresis and Western immunoblotting.
Purified recombinant proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose, and Western immunoblotting was performed as previously described (13), except that primary dog sera were diluted 1:100, human sera were diluted 1:200, and rabbit antisera were diluted 1:1,000.
ELISA.
For epitope mapping, an ELISA was performed as previously described (9). For serologic diagnosis evaluation, an Immobilizer amino plate (Nunc, Roskilde, Denmark) was used to increase the signal-to-noise ratio. Immobilizer amino plates were coated with synthetic peptides or recombinant proteins (0.5 μg/well; 50 μl) suspended in 100 mM sodium carbonate buffer (pH 9.6) and incubated with gentle agitation at room temperature for 1 to 2 h or overnight at 4°C. The wells were washed four times with 300 μl phosphate-buffered saline containing 0.05% (vol/vol) Tween 20 (PBST; pH 7.2) by a plate washer (SkanWasher 400; Molecular Devices, Sunnyvale, CA). Dog or human sera diluted (1:100 or 1:200, respectively) in PBST were added to each well (50 μl) and incubated at room temperature for 1 h. The plates were washed again, and 50 μl alkaline phosphatase-labeled goat anti-dog or -human IgG(H+L) secondary antibody (Kirkegaard & Perry Laboratories, Gaithersburg, MD) diluted (1:5,000) in PBST was added and incubated at room temperature for 1 h. After the addition of substrate (BluePhos; Kirkegaard & Perry Laboratories), plates were incubated in the dark for 30 min, color development was determined on a microplate reader (VersaMax; Molecular Devices, Sunnyvale, CA) at A650, and data were analyzed by SoftMax Pro version 4.0 (Molecular Devices). Optical density (OD) readings represent the mean OD value for three wells (± standard deviations) after subtracting the OD value of the negative control wells. All sera negative for E. chaffeensis by IFA had readings of <0.05 OD unit; therefore, a positive sample threshold was set at >0.1 OD unit. In addition, a reading of 0.1 to 0.5 OD unit was considered a weak positive, and a reading of >0.5 OD unit was considered a strong positive.
IFA.
The anti-E. chaffeensis antibody status in HME patient sera was determined as described previously (13). Antigen slides were prepared from DH82 cells infected with E. chaffeensis (Arkansas strain) (12). Sera were diluted 2-fold in PBS, starting at 1:64.
Statistics.
The statistical differences between experimental groups were assessed with the two-tailed Student t test, and significance was indicated by a P value of <0.05.
Locus tag numbers of nucleotide sequences.
Ehrlichia gene locus tag numbers for the proteins in this study were previously available in the Integrated Microbial Genomes system (http://img.jgi.doe.gov/) (ECH_0170 for TRP32, ECH_0166 for TRP47, ECH_0039 for TRP120, ECH_0684 for Ank200, and Ecaj_0109 for TRP36).
RESULTS
E. chaffeensis Ank200 amino acid composition and domains.
The overall Ank200 composition (1,463 amino acids [aa]) was dominated by three hydrophobic amino acids (L, V, and A; 353 aa), three polar amino acids (S, G, and N; 362 aa), and two strongly acidic amino acids (E and D; 198 aa), resulting in a protein with an acidic nature (pI 4.6). Like for E. canis Ank200, three specific domains (N acidic, Ank, and C acidic) were identified, according to amino acid composition and conserved motifs (Fig. 1). The distal terminal polypeptides (N acidic, first 390 aa; C acidic, last 267 aa) exhibited a substantially larger proportion of strongly acidic amino acids (D and E; 22.6% in the N-acidic domain and 13.1% in the C-acidic domain) than the internal region (Ank domain, 806 aa [positions 391 to 1196]; 9.3% D and E) of the protein, where ankyrin repeats were located. In contrast, the Ank domain region contained more strongly basic amino acids (K and R; 10.2%) than strongly acidic amino acids. Consequently, the isoelectric points of two terminal domains were acidic (pI 3.6 and 4.7), whereas the internal Ank domain region was slightly basic (pI 8.3) (Fig. 1).
Immunoreactivities of the major E. chaffeensis Ank200 fragments.
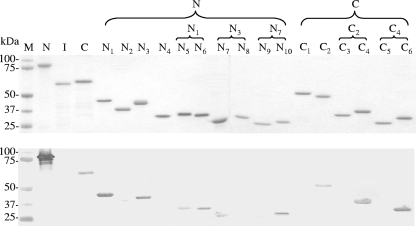
To determine the major epitope-containing regions of Ank200, the recombinant fragments corresponding to the N terminus (Ank200-N, aa 10 to 557), internal region (Ank200-I, aa 562 to 1018), and C terminus (Ank200-C, aa 984 to 1439), covering 98% of the open reading frame, were expressed (Fig. 1). By Western immunoblotting, the recombinant Ank200-N and Ank200-C (containing the N- and C-acidic domains, respectively) proteins reacted with an HME patient serum sample (no. SC07); however, recombinant protein of the Ank200-I (a majority of the Ank domain) did not react with the patient serum sample (Fig. 2). A similar result was obtained by Western blotting probed with the anti-E. chaffeensis dog serum sample derived from a dog (no. 2251) experimentally infected with E. chaffeensis (data not shown). Thus, the two immunoreactive fragments Ank200-N and Ank200-C were considered to contain antibody epitopes and were investigated further. The anti-E. chaffeensis patient or dog sera did not recognize thioredoxin protein, and the normal human or dog sera did not recognize these recombinant proteins by Western immunoblotting (data not shown).
FIG. 2.
Immunoreactivities of recombinant E. chaffeensis Ank200 proteins by Western immunoblotting. Top, SDS-PAGE and total protein staining of purified recombinant Ank200 proteins (N, N-terminal fragment; I, internal fragment; and C, C-terminal fragment). Bottom, corresponding Western immunoblot probed with a patient serum sample (no. SC07). The patient serum sample did not recognize thioredoxin protein, and the normal human serum sample did not recognize these recombinant proteins by Western immunoblotting (data not shown). M, Precision protein standard (Bio-Rad).
Major epitope-containing regions in Ank200-N.
The major epitope-containing region(s) in Ank200-N was identified by evaluating the immunoreactivities of four overlapping recombinant proteins (N1 to N4) and of some smaller overlapping recombinant proteins (N5 to N10) (Fig. 1). Western immunoblotting revealed that N1 and N3 fragments were reactive with the patient serum samples, whereas two other fragments (N2 and N4) of Ank200-N were not reactive or only weakly reactive (Fig. 2). Western blotting probed with anti-E. chaffeensis dog sera exhibited a similar result (data not shown). Therefore, smaller overlapping recombinant proteins (N5, N6, N7, and N8) representing N1 and N3 regions were expressed, and two fragments, N6 and N7, were immunoreactive with the patient sera or anti-E. chaffeensis dog sera by Western blotting, while the other two fragments (N5 and N8) were not immunoreactive or were weakly immunoreactive (Fig. 2). N7 was further divided into two overlapping polypeptides, N9 and N10, and polypeptide N10 was immunoreactive with the patient sera or anti-E. chaffeensis dog sera by Western blotting, while N9 was not immunoreactive (Fig. 2). Thus, the N6 (63-aa) and N10 (48-aa) sections were identified as the major epitope-containing regions of E. chaffeensis Ank200-N, which were located in a highly acidic domain and exhibited high glutamate content (22.2% and 14.6%, respectively) (Fig. 1 and 3A and B, left, for sequences).
Major epitope-containing region in Ank200-C.
The major epitope(s) in Ank200-C was identified by evaluating the immunoreactivities of six overlapping recombinant proteins. Ank200-C was divided into two overlapping fragments (C1 and C2), and Western immunoblotting revealed that the C2 fragment was immunoreactive with a patient serum sample, while C1 was not reactive (Fig. 2). Therefore, C2 fragment was further divided into two overlapping polypeptides (C3 and C4), and the C4 fragment was immunoreactive with a patient serum sample by Western blotting, while C3 was not reactive or was only weakly reactive (Fig. 2). Smaller overlapping polypeptides (C5 and C6) representing the C4 region were expressed, and the C6 fragment reacted with a patient serum sample by Western blotting, while C5 was not reactive or was weakly reactive (Fig. 2). A similar result was obtained by Western blotting probed with an anti-E. chaffeensis dog serum sample (data not shown). Thus, the C6 (63-aa) section of E. chaffeensis Ank200-C was identified as a major epitope-containing region, which was also located in a highly acidic domain and exhibited a high glutamate content (11.1%) (Fig. 1 and 3C, left, for the sequence).
Determination of the major immunodeterminants of E. chaffeensis Ank200 with synthetic peptides.
Synthetic peptides were used to localize the major epitope(s) in three immunoreactive regions (N6, N10, and C6) of Ank200. Four synthetic overlapping polypeptides (N6-1, -2, -3, and -4) (Fig. 3A, left) covering the sequence of Ank200-N6 (63 aa) were generated and reacted by ELISA with an anti-E. chaffeensis dog serum sample (no. 2251) and four HME patient serum samples (no. F3, F5, F13, and F22) that had detectable E. chaffeensis antibodies by IFA. Among five serum samples, peptide N6-2 did not react with two serum samples and reacted weakly with one and strongly with two, peptide N6-3 did not react with one serum sample and reacted weakly with four, and peptide N6-4 did not react with two serum samples and reacted weakly with three; however, peptide N6-1 was found to react strongly with all the anti-E. chaffeensis dog and patient sera, indicating that the N-terminal fragment (28 aa) of the Ank200-N6 region had a significantly (P < 0.05 for dog sera and most patient sera) stronger immunoreactivity than other fragments and contained a major antibody epitope (Fig. 3A, right). To further determine the amino acid sequence reactive with antibody, N6-1 was divided into two smaller overlapping peptides (N6-1a and N6-1b). By ELISA, peptide N6-1b did not react with the anti-E. chaffeensis dog sera and reacted weakly with four patient serum samples; however, although peptide N6-1a was also not reactive with antibodies in dog sera, it reacted strongly with all four patient serum samples, indicating that the N-terminal amino acids (ETGETVEEGLYA) contributed significantly (P < 0.05) to epitope reactivity with all patient sera (Fig. 3A, right). Therefore, N6-1a (18 aa; ETGETVEEGLYAVPLPKD) contained a major continuous antibody epitope of Ank200 for humans, but the longer sequence of peptide N6-1 (28 aa; ETGETVEEGLYAVPLPKDQRPTPTQVLE) exhibited the strongest immunoreactivity and was necessary for full reconstitution of the major antibody epitope of Ank200.
To identify the peptide sequence containing the immunodeterminant in the Ank200-N10 (48-aa) region, four overlapping peptides (N10-1, -2, -3, and -4) (Fig. 3B, left) covering the N10 region were reacted with an anti-E. chaffeensis dog serum sample (no. 2251) and four HME patient serum samples (no. F2, F4, F5, and F21). By ELISA, peptides N10-1 and -2 did not react, and peptides N10-3 and -4 reacted weakly with antibodies in the dog serum sample (Fig. 3B, right). Since the recombinant Ank200-N10 protein reacted strongly with the anti-E. chaffeensis dog serum sample by Western blotting (Fig. 2), the data suggested that the sequence longer than those of the above-mentioned peptides was required to reconstitute the major antibody epitope of Ank200 recognized by antibodies in the dog serum sample. By ELISA, peptide N10-2 reacted weakly with two patient serum samples and reacted strongly with two patient serum samples, and peptides N10-1, -3, and -4 reacted weakly with one patient serum sample but reacted strongly with three other patient serum samples, suggesting that N10 had two epitope-containing regions for humans, N10-1 (20 aa; EPDLEEIVSILKNDKEGISE) and N10-3 (20 aa; INEPVQVDIPNNPVREGRNV), and the C-terminal amino acids (MTLLHLA) of N10 had no substantial contribution to the epitope reactivity. Moreover, peptide N10-1 exhibited substantially stronger immunoreactivity than peptides N10-3 with three patient serum samples (Fig. 3B, right).
Four synthetic overlapping peptides (C6-1, -2, -3, and -4) (Fig. 3C, left) covering the sequence of Ank200-C6 (63 aa) were reacted by ELISA with an anti-E. chaffeensis dog serum sample (no. 2251) and four HME patient serum samples (no. F2, F4, F15, and SC07). Peptides C6-1, -2, and -3 were only weakly immunoreactive with one or two patient serum samples, but peptide C6-4 was found to react with the anti-E. chaffeensis dog serum sample and to react strongly with all patient sera, indicating that the C-terminal fragment (25 aa) of Ank200 contained a major antibody epitope, and the C-terminal sequence (QGADVKKSSCQSK, 13 aa) significantly (P < 0.05 for all sera) contributed to the epitope reactivity (Fig. 3C). To further determine the amino acid sequence reactive with antibody, two smaller overlapping peptides (C6-4a and -4b) representing fragment C6-4 reacted with anti-E. chaffeensis dog and patient sera by ELISA. The peptide C6-4a was not immunoreactive with all sera; however, peptide C6-4b was found to react with the anti-E. chaffeensis dog serum sample and to react strongly with all patient sera, indicating that the very distal C-terminal fragment C6-4b (21 aa; QAVSPSTSQGADVKKSSCQSK) contained a major continuous antibody epitope of Ank200. Moreover, the very distal C-terminal amino acids (SCQSK) contributed significantly (P < 0.05 for all sera) to the epitope immunoreactivity (Fig. 3C).
Identification of TRP47 antibody epitopes in the TR flanking terminal regions.
E. chaffeensis TRP47 has N (157-aa) and short C (26-aa) termini flanking the TR region (19 aa each) (Fig. 4A). In a previous study, we determined that the TR of TRP47 contained a major antibody epitope (6); however, the N- and C-terminal regions were not fully explored. The immunoreactivities of the TRP47-N and TRP47-C regions were further investigated using HME patient sera in this report. A large panel of 31 patient serum samples that had detectable E. chaffeensis antibodies by IFA was used to detect the recombinant TRP47-N protein by Western blotting; as a result, 13 of 31 serum samples reacted with TRP47-N, indicating that the N-terminal region of TRP47 contained a minor antibody epitope (data not shown).
To locate the epitope in TRP47-N, four recombinant overlapping proteins (TRP47-N1, -N2, -N3, and -N4) (Fig. 4A) covering the sequence of the whole TRP47-N region were expressed and reacted with three HME patient serum samples (no. O13, O15 and 19) that recognized TRP47-N by Western blotting. The recombinant N2 fragment did not react with the patient sera, N1 reacted weakly with one patient serum sample, and N3 reacted with two serum samples (one weakly), while the N4 fragment strongly reacted with all three serum samples, indicating that the TRP47-N4 fragment (44 aa) contained a minor antibody epitope (Fig. 4B). Three synthetic overlapping polypeptides (N4-1, -2, and -3) (Fig. 4A and 5A) covering the sequence of TRP47-N4 were generated and reacted with six HME patient serum samples (no. O15, 6, 9, 13, 18, and 19) that recognized TRP47-N by Western blotting. By ELISA, peptide N4-3 was not reactive with any tested sera, N4-1 was found to react with five serum samples (except for no. 19), N4-2 reacted with three serum samples (no. 6, 18, and 19), and the reaction with serum sample no. 19 was very strong (Fig. 5B). Therefore, the assembled sequences (33 aa) of N4-1 and -2 fragments contained the antibody epitope with the TRP47-N region.
Although the TR of TRP47 has previously been reported to react with anti-E. chaffeensis dog sera, its immunoreactivity with the HME patient sera has not been reported. Synthesized TR unit (TRP47-R; 19 aa) and the C terminus of TRP47 (TRP47-C; 26 aa) (Fig. 4A and 5A) were reacted by ELISA with sera from seven HME patients and one experimentally infected dog. Peptide TRP47-R was recognized by six patient serum samples and the dog serum sample; peptide TRP47-C was recognized by three patient serum samples but exhibited significantly (P < 0.05) stronger reactivity than TRP47-R with two serum samples (no. O3 and 13) (Fig. 5C). Hence, both TRP47-R and TRP47-C exhibited immunoreactivity with HME patient sera; however, TRP47-R had stronger overall immunoreactivity (P < 0.05 for most sera) than TRP47-C. Moreover, TRP47-R exhibited stronger immunoreactivity than TRP47-C with an anti-E. chaffeensis dog serum sample.
Evaluation of synthetic E. chaffeensis major immunodeterminants for serologic diagnosis of HME.
In order to examine and compare the immunoreactivities of E. chaffeensis major immunoreactive epitopes that have been characterized, a panel of 31 HME patient serum samples that had detectable E. chaffeensis antibodies by IFA (titers from 64 to 8,192) was used to examine and compare the sensitivities of synthetic epitopes from E. chaffeensis TRP32, TRP47, TRP120, and Ank200 with IFA. Epitopes for TRP32, TRP47, and TRP120 mapped in our previous studies were also included in this evaluation. An equal (wt/wt) mixture of TRP32-R3 (30-aa) and -R4 (30-aa) peptides was used for TRP32, an equal mixture of TRP47-N4-1 (22-aa), -R (19-aa), and -C (26-aa) peptides was used for TRP47, the TRP120-R-I1 (22-aa) peptide was used for TRP120, and an equal mixture of Ank200-N6-1a (18 aa), -N10-1 (20 aa), and -C6-4b (21 aa) was used for Ank200. In addition, a recombinant TRP120 TR protein (rTRP120-TR) and an equal mixture of TRP32-R3, TRP32-R4, and TRP120-R-I1 peptides were also tested. E. canis TRP36-2R (18 aa) was used as a negative control peptide. Patient sera (n = 10) negative for E. chaffeensis antibodies by IFA were also tested.
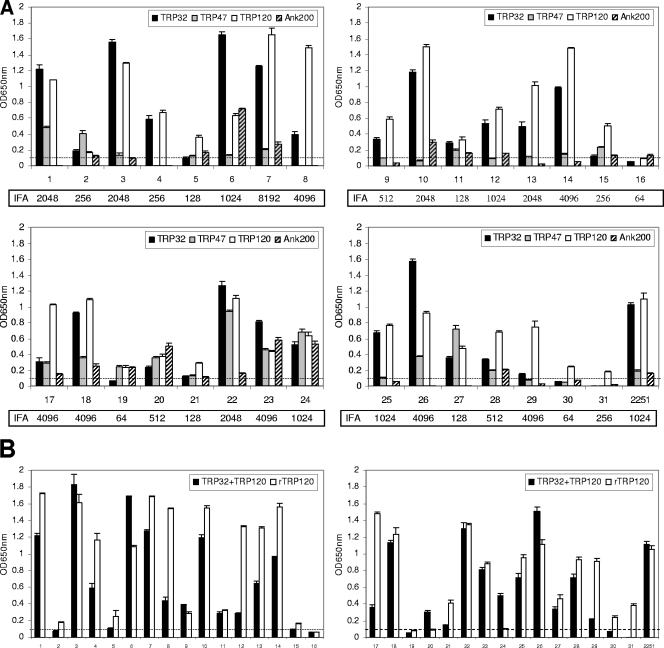
All 31 HME patient serum samples reacted with at least one E. chaffeensis peptide. A total of 30 serum samples (96.8%) reacted with the TRP120 peptide, 27 (87.1%) with TRP32 peptides, 24 (77.4%) with TRP47 peptides, and 19 (61.3%) with Ank200 peptides (Fig. 6A; Table 2). Only one serum sample (no. 16) with a low IFA titer (1:64) did not reach our established positive cutoff with the TRP120 peptide, and four serum samples (no. 16, 19, 30 and 31) with low IFA titers (three with 1:64 and one with 1:256) did not react with TRP32 peptides. The recombinant TRP120-TR protein was recognized by 28 (90.3%) serum samples, and a mixture of the TRP120 and TRP32 peptides was recognized by only 26 (83.9%) serum samples and did not provide enhanced sensitivity over TRP120 alone (Fig. 6B; Table 2). These results suggested that TRP120 is the best candidate for immunodiagnosis of HME, and a single synthetic peptide, TRP120-R-I1 from TRP120 repeats, exhibited higher sensitivity than the peptide mixture or recombinant TRP120-TR protein did with HME patient sera. Moreover, the peptides were not recognized by patient sera that were positive for Rickettsia spp. but not positive for E. chaffeensis by IFA, indicating that ELISA reactions between synthetic E. chaffeensis immunodeterminants and HME patient sera were specific (data not shown).
FIG. 6.
Immunoreactivities of major antibody epitopes from E. chaffeensis immunoreactive proteins with HME patient sera by ELISA. (A) Synthetic epitope peptides of TRP32 (R3 plus R4), TRP47 (N4-1 plus R plus C), TRP120 (R-I1), and Ank200 (N6-1a plus N10-1 plus C6-4b) reacted with 31 HME patient serum samples and an anti-E. chaffeensis dog serum sample (no. 2251). (B) An equal mixture of TRP32-R3, TRP32-R4, and TRP120-R-I1 peptides and the recombinant TRP120 TR protein (rTRP120-TR, containing the first two tandem repeats of TRP120 only) reacted with 31 HME patient serum samples and an anti-E. chaffeensis dog serum sample (no. 2251). The OD readings represent the mean values for three wells (± standard deviations), with the OD values of the negative control (E. canis TRP36-2R peptide) wells subtracted. The cutoff OD (0.1) established for the positive reading is shown by a dotted line. The normal human or dog sera did not recognize these peptides (data not shown).
TABLE 2.
Analytical sensitivities of synthetic antibody epitopes of E. chaffeensis immunoreactive proteins for immunodiagnosis of HME by ELISAa
| Antigen | No. of patients with detectable antibodies | % of patients with detectable antibodies |
|---|---|---|
| TRP32 | 27 | 87.1 |
| TRP47 | 24 | 77.4 |
| TRP120 | 30 | 96.8 |
| Ank200 | 19 | 61.3 |
| TRP32 plus TRP120 | 26 | 83.9 |
| Overall | 31 | 100 |
Synthetic epitope peptides of TRP32 (R3 plus R4), TRP47 (N4-1 plus R plus C), TRP120 (R-I1), and Ank200 (N6-1a plus N10-1 plus C6-4b) and an equal mixture of TRP32-R3, TRP32-R4, and TRP120-R-I1 peptides as well as rTRP120 (recombinant TRP120-TR protein, containing the first two tandem repeats of TRP120 only) reacted with 31 HME patient sera. For rTRP120, the number of patients with detectable antibodies was 28, and the percentage of patients with detectable antibodies was 90.3%. “Overall” refers to the overall number and percentage of patients with detectable antibodies against any tested synthetic peptide. A sample with a reading of 0.1 OD unit above the negative control absorbance was considered positive.
DISCUSSION
Many of the major immunoreactive proteins of E. chaffeensis and E. canis have been identified and molecularly characterized, and interestingly, most are members of a small group of tandem repeat- or ankyrin repeat-containing proteins, including the TRP32/TRP19, TRP47/TRP36, TRP120/TRP140, and Ank200 proteins (6, 11, 15, 23, 29, 31). Common features among these proteins include serine-rich TRs and acidic pIs (due to a predominance of glutamate/aspartate). Both recombinant and native proteins exhibit electrophoretic masses larger than those predicted by amino acid sequences, due to the acidic properties of the proteins and not to the addition of glycans posttranslationally (8-10). Notably, major continuous antibody epitopes of these proteins have been mapped to acidic domains, which are located in the TR regions in all TRPs or N- and C-terminal regions in E. canis Ank200, indicating that ehrlichial acidic domains, particularly those in TRs, are primary targets of the host humoral immune response (6, 9-11, 15, 16). The association of these acidic domains with the host immune response is interesting and unique and, to our knowledge, has not been described with respect to any other pathogen; however, the specific role of these domains in ehrlichial pathobiology or immunity is still unknown.
E. chaffeensis and E. canis Ank200 protein orthologs are the largest ehrlichial major immunoreactive proteins. They have identical chromosomal locations and exhibit ∼50% nucleic acid identity and ∼32% amino acid identity, and they lack the serine-rich TRs present in other ehrlichial major immunoreactive proteins (11). However, they have similar distal N- and C-terminal acidic domains flanking the centralized ankyrin domain containing numerous ankyrin repeats that may mediate protein-protein interactions (16). Like the ankyrin protein AnkA from Anaplasma phagocytophilum (19), E. chaffeensis Ank200 is also translocated to the nuclei of infected cells, where it interacts with the DNA motif Alu (33). In this study, major epitope-containing regions of E. chaffeensis Ank200 were mapped to the distal N- and C-terminal acidic (pI 3.6 and 4.7, respectively) domains, which is consistent with the location of the four epitopes mapped in the E. canis Ank200 N- and C-terminal acidic (pI 4 and 4.9, respectively) domains (16). The antibody epitopes in E. chaffeensis Ank200, which exhibited the strongest antibody reactivity with both dog and human sera, were localized to four polypeptides, N6-1a, N10-1, N10-3, and C6-4b (18-mer, 20-mer, 20-mer, and 21-mer, respectively), with three in the N-terminal domain and only one in the C-terminal domain, demonstrating that the N-terminal domain has multiple epitopes and, thus, is the immunodominant region. The lengths of the Ank200 epitopes were similar and consistent in size (around 20-mer) with those described of other molecularly characterized continuous ehrlichial epitopes (6, 9, 10, 15, 16). However, a smaller six-amino-acid epitope has been reported for the Anaplasma marginale MSP1a protein (1). One conformational epitope has been mapped in TRP32-R4 (10), and there may be other conformational epitopes associated with these major immunoreactive proteins that were not determined, although the host response to the continuous major epitopes in ehrlichial immunodominant proteins is strong and suggests the absence of dominant conformational epitopes.
We previously reported a major epitope in the TR regions of TRP47 and the corresponding ortholog (TRP36) in E. canis (6). However, a comprehensive analysis of the regions flanking the TR was not performed. Hence, in this study, HME patient sera were used to fully explore these regions, and all three regions exhibited immunoreactivity with patient sera. Two additional epitope-containing regions were identified in the N and C termini of TRP47, respectively, but TRP47-TR exhibited stronger overall immunoreactivity than TRP47-N and -C and was more consistently recognized by antibodies in HME patient sera. Therefore, TRP47 TR appears to be the major antibody epitope, and minor epitopes are located in the N and C termini. Similarly, minor cross-reactive antibody epitopes have been identified in N- and C-terminal regions of TRP120 and TRP140 (9). Some HME patients developed only antibodies to one or more of the TRP47 minor epitopes and not to the TR epitope. This could be related to diversity in the TR of TRP47, which has been described in the Arkansas and Sapulpa strains (6, 32). This is in contrast to other TRPs, such as TRP120 and TRP32, in which the TR epitopes appear to be more conserved (27, 32). Therefore, the increased sensitivity attained with a peptide mixture containing all the TRP47 epitopes compared to that attained with the TR epitope alone is likely related to the antigenic diversity of this protein. Additional characterization of TRP47 variants could provide an explanation for the decreased sensitivity of this protein compared to that of TRP120 or TRP32 as well as information regarding the kinetics of the antibody response in HME patients.
All of the ehrlichial major immunoreactive protein orthologs (TRP32/TRP19, TRP47/TRP36, and TRP120/TRP140) identified and characterized recently are antigenically distinct and elicit species-specific antibodies (6, 9, 10, 15). The five major antibody epitopes characterized in E. canis Ank200 are also molecularly distinct (16). Consistent with these findings, the amino acid alignments of the mapped epitopes in Ank200 identified no significant homology with E. canis Ank200 or other proteins from organisms in closely related genera; moreover, antisera against recombinant E. chaffeensis or E. canis Ank200-N did not cross-react (data not shown), indicating that these epitopes appear to be species specific primarily and could be utilized for species-specific diagnostic development. We have previously reported that minor antibody epitope-containing regions in the N and C termini of E. chaffeensis TRP120 and E. canis TRP140 are cross-reactive, further suggesting that cross-reactive antibodies generated between closely related Ehrlichia spp. were directed at some minor epitopes rather than major epitopes (9).
Previous studies have concluded that TRP120 is a sensitive immunodiagnostic antigen for HME (30). We also concluded in this study that TRP120 is the most sensitive immunodiagnostic antigen for HME. It is becoming increasingly evident that all of the major immunoreactive proteins of Ehrlichia spp. have molecularly distinct epitopes which can be used to serologically identify etiologic agents, a task that has been routinely difficult or impossible to accomplish (6, 9, 10, 15, 16). The TRP epitopes are molecularly distinct, and therefore, serologic responses specific to E. chaffeensis can be distinguished from those against closely related agents or conserved bacterial proteins using these immunodeterminants. We previously determined serologically that TRP120-R-I1 is a species-specific epitope, and lack of serologic cross-reactivity with E. canis was related to divergence at the amino acid level (9). In addition, TRP120 has very limited amino acid homology with two A. phagocytophilum repeat-containing proteins, GE100 and GE130; however, the TRP120-R-I1 peptide does not have any amino acid homology with these two proteins (22). Compared with those of TRP32 and TRP47, TRP120 has less molecular variation among examined E. chaffeensis strains, and this trait is shared with an ortholog, E. canis TRP140 (32). The synthetic TRP120-R-I1 peptide exhibited even more sensitive reactivity than recombinant TRP120-TR with patient sera, demonstrating that high purity of the immunodeterminant could contribute to enhanced sensitivity of ELISA and effectively replace recombinant proteins. However, as we observed with other immunoreactive peptides from Ehrlichia spp. (our unpublished data), in some cases, but not all, a mixture of the TRP120 and TRP32 peptides does not provide enhanced sensitivity compared to that of TRP120 alone, indicating that mixed peptides could compete with each other, resulting in decreased sensitivity.
This is the first study to compare multiple molecularly defined major antibody epitopes of E. chaffeensis for serodiagnosis of HME in a solid-phase assay. The advantages provided by synthetic peptides rather than recombinant proteins is that they can be produced consistently in highly pure forms without contaminating E. coli proteins that can result in false-positive reactions when utilizing recombinant proteins. In addition, peptides can be produced quickly and efficiently without costly and laborious purification procedures and the need for defined expression vectors and hosts. The development of standardized and commercially available assays will be advanced by molecularly defined polypeptide epitopes that provide comparable or better sensitivity than IFA and analytical and clinical specificity that is much higher than that of IFA. We demonstrated that a single synthetic peptide from TRP120 can provide highly sensitive and specific diagnosis of HME infection comparable to the “gold standard” IFA and can be utilized to develop standardized, consistent, reliable, sensitive, and specific point-of-care and reference laboratory immunodiagnostics for HME.
Acknowledgments
This work was supported by National Institutes of Health grant AI 071145, AI 069270, and the Clayton Foundation for Research.
Footnotes
Published ahead of print on 2 December 2009.
REFERENCES
- 1.Allred, D. R., T. C. McGuire, G. H. Palmer, S. R. Leib, T. M. Harkins, T. F. McElwain, and A. F. Barbet. 1990. Molecular basis for surface antigen size polymorphisms and conservation of a neutralization-sensitive epitope in Anaplasma marginale. Proc. Natl. Acad. Sci. U. S. A. 87:3220-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter, C. F., T. K. Gandhi, L. K. Kong, G. R. Corey, S. M. Chen, D. H. Walker, J. S. Dumler, E. Breitschwerdt, B. Hegarty, and D. J. Sexton. 1999. The incidence of ehrlichial and rickettsial infection in patients with unexplained fever and recent history of tick bite in central North Carolina. J. Infect. Dis. 180:900-903. [DOI] [PubMed] [Google Scholar]
- 3.Chen, S. M., J. S. Dumler, H. M. Feng, and D. H. Walker. 1994. Identification of the antigenic constituents of Ehrlichia chaffeensis. Am. J. Trop. Med. Hyg. 50:52-58. [PubMed] [Google Scholar]
- 4.Childs, J. E., J. W. Sumner, W. L. Nicholson, R. F. Massung, S. M. Standaert, and C. D. Paddock. 1999. Outcome of diagnostic tests using samples from patients with culture-proven human monocytic ehrlichiosis: implications for surveillance. J. Clin. Microbiol. 37:2997-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comer, J. A., W. L. Nicholson, J. G. Olson, and J. E. Childs. 1999. Serologic testing for human granulocytic ehrlichiosis at a national referral center. J. Clin. Microbiol. 37:558-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doyle, C. K., K. A. Nethery, V. L. Popov, and J. W. McBride. 2006. Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infect. Immun. 74:711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumler, J. S., J. E. Madigan, N. Pusterla, and J. S. Bakken. 2007. Ehrlichioses in humans: epidemiology, clinical presentation, diagnosis, and treatment. Clin. Infect. Dis. 45(Suppl. 1):S45-S51. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Ortega, L., V. De los Rios, A. Martinez-Ruiz, M. Onaderra, J. Lacadena, A. Martinez del Pozo, and J. G. Gavilanes. 2005. Anomalous electrophoretic behavior of a very acidic protein: ribonuclease U2. Electrophoresis 26:3407-3413. [DOI] [PubMed] [Google Scholar]
- 9.Luo, T., X. Zhang, and J. W. McBride. 2009. Major species-specific antibody epitopes of the Ehrlichia chaffeensis p120 and E. canis p140 orthologs in surface-exposed tandem repeat regions. Clin. Vaccine Immunol. 16:982-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo, T., X. Zhang, A. Wakeel, V. L. Popov, and J. W. McBride. 2008. A variable-length PCR target protein of Ehrlichia chaffeensis contains major species-specific antibody epitopes in acidic serine-rich tandem repeats. Infect. Immun. 76:1572-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McBride, J. W., J. E. Comer, and D. H. Walker. 2003. Novel immunoreactive glycoprotein orthologs of Ehrlichia spp. Ann. N. Y. Acad. Sci. 990:678-684. [DOI] [PubMed] [Google Scholar]
- 12.McBride, J. W., R. E. Corstvet, E. B. Breitschwerdt, and D. H. Walker. 2001. Immunodiagnosis of Ehrlichia canis infection with recombinant proteins. J. Clin. Microbiol. 39:315-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McBride, J. W., R. E. Corstvet, S. D. Gaunt, C. Boudreaux, T. Guedry, and D. H. Walker. 2003. Kinetics of antibody response to Ehrlichia canis immunoreactive proteins. Infect. Immun. 71:2516-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McBride, J. W., R. E. Corstvet, S. D. Gaunt, J. Chinsangaram, G. Y. Akita, and B. I. Osburn. 1996. PCR detection of acute Ehrlichia canis infection in dogs. J. Vet. Diagn. Investig. 8:441-447. [DOI] [PubMed] [Google Scholar]
- 15.McBride, J. W., C. K. Doyle, X. Zhang, A. M. Cardenas, V. L. Popov, K. A. Nethery, and M. E. Woods. 2007. Identification of a glycosylated Ehrlichia canis 19-kilodalton major immunoreactive protein with a species-specific serine-rich glycopeptide epitope. Infect. Immun. 75:74-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nethery, K. A., C. K. Doyle, X. Zhang, and J. W. McBride. 2007. Ehrlichia canis gp200 contains dominant species-specific antibody epitopes in terminal acidic domains. Infect. Immun. 75:4900-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohashi, N., N. Zhi, Y. Zhang, and Y. Rikihisa. 1998. Immunodominant major outer membrane proteins of Ehrlichia chaffeensis are encoded by a polymorphic multigene family. Infect. Immun. 66:132-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paddock, C. D., and J. E. Childs. 2003. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin. Microbiol. Rev. 16:37-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park, J., K. J. Kim, K. S. Choi, D. J. Grab, and J. S. Dumler. 2004. Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell. Microbiol. 6:743-751. [DOI] [PubMed] [Google Scholar]
- 20.Popov, V. L., X. Yu, and D. H. Walker. 2000. The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb. Pathog. 28:71-80. [DOI] [PubMed] [Google Scholar]
- 21.Rikihisa, Y., S. A. Ewing, J. C. Fox, A. G. Siregar, F. H. Pasaribu, and M. B. Malole. 1992. Analyses of Ehrlichia canis and a canine granulocytic Ehrlichia infection. J. Clin. Microbiol. 30:143-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Storey, J. R., L. A. Doros-Richert, C. Gingrich-Baker, K. Munroe, T. N. Mather, R. T. Coughlin, G. A. Beltz, and C. I. Murphy. 1998. Molecular cloning and sequencing of three granulocytic Ehrlichia genes encoding high-molecular-weight immunoreactive proteins. Infect. Immun. 66:1356-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sumner, J. W., J. E. Childs, and C. D. Paddock. 1999. Molecular cloning and characterization of the Ehrlichia chaffeensis variable-length PCR target: an antigen-expressing gene that exhibits interstrain variation. J. Clin. Microbiol. 37:1447-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unver, A., S. Felek, C. D. Paddock, N. Zhi, H. W. Horowitz, G. P. Wormser, L. C. Cullman, and Y. Rikihisa. 2001. Western blot analysis of sera reactive to human monocytic ehrlichiosis and human granulocytic ehrlichiosis agents. J. Clin. Microbiol. 39:3982-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wakeel, A., J. A. Kuriakose, and J. W. McBride. 2009. An Ehrlichia chaffeensis tandem repeat protein interacts with multiple host targets involved in cell signaling, transcriptional regulation, and vesicle trafficking. Infect. Immun. 77:1734-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker, D. H., and the Task Force on Consensus Approach for Ehrlichiosis. 2000. Diagnosing human ehrlichioses: current status and recommendations. ASM News 66:287-290. [Google Scholar]
- 27.Yabsley, M. J., S. E. Little, E. J. Sims, V. G. Dugan, D. E. Stallknecht, and W. R. Davidson. 2003. Molecular variation in the variable-length PCR target and 120-kilodalton antigen genes of Ehrlichia chaffeensis from white-tailed deer (Odocoileus virginianus). J. Clin. Microbiol. 41:5202-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu, X. J., P. Crocquet-Valdes, L. C. Cullman, and D. H. Walker. 1996. The recombinant 120-kilodalton protein of Ehrlichia chaffeensis, a potential diagnostic tool. J. Clin. Microbiol. 34:2853-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu, X. J., P. Crocquet-Valdes, and D. H. Walker. 1997. Cloning and sequencing of the gene for a 120-kDa immunodominant protein of Ehrlichia chaffeensis. Gene 184:149-154. [DOI] [PubMed] [Google Scholar]
- 30.Yu, X. J., P. A. Crocquet-Valdes, L. C. Cullman, V. L. Popov, and D. H. Walker. 1999. Comparison of Ehrlichia chaffeensis recombinant proteins for serologic diagnosis of human monocytotropic ehrlichiosis. J. Clin. Microbiol. 37:2568-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu, X. J., J. W. McBride, C. M. Diaz, and D. H. Walker. 2000. Molecular cloning and characterization of the 120-kilodalton protein gene of Ehrlichia canis and application of the recombinant 120-kilodalton protein for serodiagnosis of canine ehrlichiosis. J. Clin. Microbiol. 38:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu, X. J., J. W. McBride, and D. H. Walker. 2007. Restriction and expansion of Ehrlichia strain diversity. Vet. Parasitol. 143:337-346. [DOI] [PubMed] [Google Scholar]
- 33.Zhu, B., K. A. Nethery, J. A. Kuriakose, A. Wakeel, X. Zhang, and J. W. McBride. 2009. Nuclear translocated Ehrlichia chaffeensis ankyrin protein interacts with a specific adenine-rich motif of host promoter and intronic Alu elements. Infect. Immun. 77:4243-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]